Back to Journals » Cancer Management and Research » Volume 10
Difference of molecular alterations in HER2-positive and HER2-negative gastric cancers by whole-genome sequencing analysis
Authors Zhou C, Feng X, Yuan F, Ji J, Shi M, Yu Y, Zhu Z, Zhang J
Received 30 April 2018
Accepted for publication 24 July 2018
Published 26 September 2018 Volume 2018:10 Pages 3945—3954
DOI https://doi.org/10.2147/CMAR.S172710
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Chenfei Zhou,1,* Xiaojing Feng,2,* Fei Yuan,3 Jun Ji,4 Min Shi,1 Yingyan Yu,4 Zhenggang Zhu,1,4 Jun Zhang1
1Department of Oncology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 2Department of Clinical Laboratory, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 3Department of Pathology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 4Department of Surgery, Shanghai Institute of Digestive Surgery, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Objective: The aim of this study was to compare the molecular profiling, including somatic mutation and somatic copy number variation (SCNV), between human epidermal growth factor receptor 2 (HER2)-positive (HER2+) and HER2-negative (HER2–) gastric cancer patients.
Patients and methods: Tumor samples were collected from 15 gastric cancer patients, including 10 HER2+ samples and five HER2– samples, which were diagnosed by immunohistochemistry. Whole-genome sequencing was performed by Illumina HiSeq PE150 instrument, along with somatic single nucleotide variant (SNV), somatic structural variation (SV) and SCNV analyses.
Results: The average number of somatic SNVs and mutation spectrum were similar between HER2+ and HER2– samples. Transition of C>T was the main type of mutation. For somatic SV, number of intrachromosomal translocation (2,850.3±1,260.4 vs 1,157±586.6, P=0.015) and insertion of large fragment (1,125.6±457.4 vs 500±138.9, P=0.002) in HER2+ samples were higher than those in HER2– samples. For all samples, lysine methyltransferase 2C (KMT2C), ZNF91, TAF1 and MAP4 genes were identified as new significant mutated driver genes. KMT2C gene mutations were mainly detected in HER2+ samples (7/10), which were correlated with the lysine degradation pathway. SERF2 gene mutations were more common in HER2– samples (3/5) than in HER2+ samples (1/10). Copy number gain was the major type of SCNV in both groups, and the average number of SCNVs was similar. In the HER2+ samples, by using the GISTIC algorithm, amplification of known driver genes cyclin-dependent kinase 12 (CDK12, 6/10) and RARA (5/10) was mainly observed, and other amplifications including JUP, GJD3, KRT39, CDC6, RAPGEFL1, WIPF2, FAM65C, KLF5, DACH1 and PIBF1 genes were also observed. Amplifications of solute carrier family 12 member 7 (SLC12A7, 5/5), TTC40 (4/5) and GALNT9 (4/5) genes were mainly detected in HER2– samples.
Conclusion: Differences in genomic landscape between HER2+ and HER2– gastric cancer samples were revealed in this study. KMT2C mutation and CDK12 amplification were mainly detected in HER2+ gastric cancer, whereas SERF2 mutation and SLC12A7 amplification were detected in HER2– gastric cancer.
Keywords: gastric cancer, HER2, whole-genome sequencing, gene mutation, gene amplification
Introduction
Gastric cancer is one of the most malignant diseases worldwide. Both its incidence and mortality rate are on the second place among all malignant neoplasms in China1. For gastric cancer patients with local advanced or distant metastatic diseases (advanced gastric cancer [AGC]), chemotherapy is the main strategy to prolong patients’ survival and improve quality of life.2 However, the efficacy of chemotherapy is still unsatisfied.
The results of ToGA clinical trial demonstrated that trastuzumab, a monoclonal antibody targeting human epidermal growth factor receptor 2 (HER2), in combination with chemotherapy has become the standard strategy for HER2-positive (HER2+) AGC patients.3 Positive rate of HER2 in gastric cancer is ~10%–20%, and it is mainly detected in cardiac and gastro-esophageal junction carcinoma.4 However, for Chinese gastric cancer patients, carcinoma located in antrum and pylori accounts for the major proportion. Furthermore, in ToGA trial, there were about 50% of the HER2+ patients in whom the tumor did not shrink after treatment of trastuzumab plus chemotherapy, which suggests that a large number of patients still might not benefit from HER2-targeted therapy in China.
Currently, whole-genome sequencing (WGS) by next-generation sequencing technology becomes a useful tool to explore and identify molecular characteristics of malignant diseases.5 The molecular classification of gastric cancer had been revealed by The Cancer Genome Atlas program (TCGA) and other researchers.6 The application of molecular classification in clinical practices of gastric cancer is now widely investigated. However, the difference in molecular events between HER2+ and HER2-negative (HER2–) gastric cancer samples had not been fully elucidated. Therefore, in this study, we aimed to explore and identify the molecular events of gastric cancer with different HER2 status, which can provide useful information for investigations of molecular biology and targeted therapy of gastric cancer.
Patients and methods
Cases and sample preparation
Paraffin-embedded gastrectomy specimens and paired nonneoplastic gastric samples derived from 15 gastric cancer patients were collected for WGS. All patients received no pervious treatment and underwent radical resection (D2 dissection) in the Department of Gastrointestinal Surgery, Ruijin Hospital. Pathological diagnosis confirmed that the tumor samples were all adenocarcinoma, and pathological TNM staging was all stage III (IIIA: n=10, IIIB: n=5, according to Cancer Staging Manual of American Joint Committee on Cancer, eighth edition).
Immunohistochemistry (IHC) staining
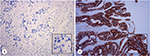
IHC staining was performed on 4-μm-thick slices following EnVision two-step procedure of the Dako REAL™ Envision™ Detection System. Slides were incubated by primary antibody of HER2, followed by incubation with horseradish peroxidase-labeled secondary antibody and were visualized by diaminobenzidine. IHC scoring of ToGA trial was used to identify HER2 status in this study.3 Ten cases were HER2+ (IHC 3+), and five cases were HER2– (IHC 0). Typical staining of HER2 status is shown in Figure 1.
Whole-genome library construction and sequencing
DNA of each case was extracted, and those samples with an OD value of 1.8–2.0 and content of 1.5 µg were used. DNA samples were randomly broken into ~350 bp insert length to construct library following the protocol of TruSeq Library Construction Kit. Samples were sequenced by Illumina HiSeq PE150 instrument. Average depth of tumor tissues and paired nonneoplastic gastric tissues was 64.5× and 33.4×, respectively. For tumor samples, average 99.6% of reference human genome was covered by ≥10×, and 98.2% was covered by ≥10× in nonneoplastic tissues.
Primary sequence analysis
Raw FASTQ files were processed and filtered to acquire clean reads for downstream analysis. Filtered clean reads were aligned to the reference human genome (B37) using BWA and Samblaster software (v0.1.22). For tumor and nonneoplastic gastric samples, single-nucleotide polymorphism (SNP) and insertion and deletion (INDEL) were analyzed by SAM tools (v1.0). On average, 3,550,069 and 3,635,355 SNPs were called in nonneoplastic and tumor samples, respectively. The average heterozygous to homozygous SNP ratio was 1.33 in nonneoplastic and 1.35 in tumor samples, and average transition/transversion ratios were 2.076 and 2.065, respectively.
Somatic mutation analysis
Somatic mutations of tumor samples were analyzed by comparing with their paired nonneoplastic gastric samples as reference. For tumor samples, single nucleotide variants (SNVs) were analyzed by muTect (v1.1.4), and somatic INDEL were analyzed by Strelka (v1.0.13) software. The average number of SNVs in the region of coding DNA sequence (CDS) was 746 and 671 in HER2+ and HER2– samples, respectively. Somatic structural variations (SVs) were detected by using crest software (v0.0.1). SV information of tumor samples was compared with that of nonneoplastic gastric samples. The average number of SVs was 24,316 and 14,069 in HER2+ and HER2– samples, respectively.
Driver gene prediction and significantly mutated gene analysis
To identify the driver-mutated genes in tumor samples, the results of SNV were compared with known driver genes from the database of Cancer Gene Census and reports of Vogelstein et al,7 Kandoth et al8 and Tamborero et al.9 Among these matched somatic mutations, those identified by at least three data sets were included. OncodriveCLUST (0.4.1) software was used to predict the potential driver gene.
Significantly mutated gene was identified by MuSic software by combining results of SNV and INDEL. Convolution test, Fisher’s combined P-value test and likelihood ratio test were used to detect mutated genes whose frequency was significantly higher than the background mutation rate.
Copy number data analysis
Somatic copy number variations (SCNV) in tumor samples were analyzed by Control-FreeC software (v6.7), and B-allele frequency (BAF) was also detected. The recurrence of copy number variations (CNVs) was assessed by GISTIC software (2.0). The average number of gains was 434 and 224 in HER2+ and HER2– samples, respectively, and average number of losses was 23 and 12, respectively. Significantly recurring CNVs were determined by GISTIC.
Statistical analysis
Differences in clinicopathological characteristics of HER2+ and HER2– samples were analyzed by the chi-squared test. Differences in genomic variables between HER2+ and HER2– samples were analyzed by Student’s t-test when variables were continuous. A P-value of <0.05 was considered to be statistically significant. Statistical analysis was performed by SPSS software (v19.0; IBM Corporation, Armonk, NY, USA).
Ethics approval
All participants provided their written informed consent as per the ethics protocol approved by the institutional review board of Ruijin Hospital, Shanghai Jiaotong University School of Medicine (Shanghai, People’s Republic of China).
Results
Clinicopathological characteristics of gastric cancer patients
The clinicopathological characteristics of gastric cancer patients enrolled in this study are shown in Table 1, and there was no significant difference between the two groups. In both HER2+ and HER2– groups, most patients were male (8/10 and 4/5). Median age of the patients in these two groups was similar. All patients in the HER2– group have lymph node metastasis, whereas two patients with HER2+ were lymph node negative. Most patients in the HER2+ group were stage IIIA (8/10). In the HER2– group, two patients were IIIA and three patients were IIIB. For tumor site, lesions of six patients with HER2+ were located in antrum (6/10), whereas there was one patient in the HER2– group (1/5).
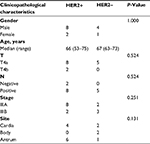  | Table 1 Clinicopathological characteristics of gastric cancer patients Abbreviation: HER2, human epidermal growth factor receptor 2. |
Somatic SNVs in HER2+ and HER2– patients
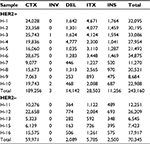
A total of 2.8 million somatic SNVs were identified in 15 samples (average: 187,338 per sample). The number of somatic SNVs in HER2+ samples was higher than that in HER2– samples (211,029±71,244.2 vs 139,954.6±45,834.1), although a statistical significance was not achieved (P=0.065). Most variants were located in the intronic region (Figure 2A). The total number of somatic mutations located in coding regions or essential splice sites in all patients (Table 2) included 6,324 missense mutations, 427 stop codon gains, five stop codon losses, 383 splice sites, 25 INDEL and 3,687 synonymous mutations. There was no significant difference between two groups for the number of mutations located in coding sequence (CDS; 746.4±259.4 vs 671.8±496.5, P=0.703). The ratio of nonsynonymous to synonymous mutation was 2.06±0.49 and 1.69±0.61 in HER2+ and HER2– samples, respectively (P=0.229). For mutation spectrum analysis, C>T transition was identified as the major type. The pattern of mutation spectrum was not much different between HER2+ and HER2– samples (Figure 2B).
  | Table 2 Somatic SNVs in coding DNA sequence of 15 gastric cancer samples Abbreviations: HER2, human epidermal growth factor receptor 2; INDEL, insertion and deletion; SNV, single nucleotide variant. |
Somatic SVs in HER2+ and HER2– groups
The average number of somatic SVs in HER2+ and HER2– groups was 24,316±9,030.5 and 14,069±8,158.9 (P=0.053, Table 3), respectively. Interchromosomal translocation (CTX) was the most common type of somatic SV, and deletion of large fragment (DEL), intrachromosomal translocation (ITX) and insertion of large fragment (INS) were also detected (Figure 2C). Interchromosomal inversion was rarely detected in both HER2+ and HER2– patients. Number of ITX (2,850.3±1,260.4 vs 1,157±586.6, P=0.015) and INS (1,125.6±457.4 vs 500±138.9, P=0.002) in the HER2+ group were significantly higher than those in the HER2– group.
Significantly mutated gene analysis
In all 15 samples, 26 mutated genes were found in four or more samples, with P<0.05 and false discovery rate ≤0.2 (Table 4 and Figure 2D). By searching TCGA database, TTN, TP53 and MUC16 were also identified as the most common mutated genes in gastric cancer. Among those 26 mutated genes, TP53, lysine methyltransferase 2C (KMT2C), ZNF91, TAF1 and MAP4 genes were identified as driver genes by comparing with reported driver mutation data set.7–9
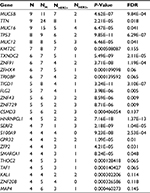  | Table 4 Significant mutated gene analysis in HER2+ and HER2– samples Abbreviations: FDR, false discovery rate; HER2, human epidermal growth factor receptor 2; KMT2C, lysine methyltransferase 2C. |
Upon comparing significant gene mutations in HER2+ and HER2– samples, KMT2C gene mutations were only identified in HER2+ samples (7/10), whereas they were not detected in HER2– samples. Among those seven samples, six samples have single mutation and one sample had two mutations, and all were missense mutation. The mutation sites of KMT2C gene in those samples were highly heterogeneous. Only c.1042G>A (D348N) mutation in exon8 was found in both H-7 and H-9 samples (Table 5). This mutation was located in one of the zinc fingers (344–389 aa) of KMT2C protein. By driver mutation analysis, c.12383G>T (S4128I) mutation was identified as a driver mutation, whereas it could not be located in any known domains of KMT2C protein. KMT2C was correlated with the lysine degradation pathway by pathway enrichment analysis.
  | Table 5 KMT2C and SERF2 mutations in HER2+ and HER2– samples Abbreviations: HER2, human epidermal growth factor receptor 2; KMT2C, lysine methyltransferase 2C. |
For HER2– samples, SERF2 gene mutations were found in three of five patients. Only one HER2+ sample carried SERF2 mutation. Sample H-16 had three mutations of SERF2 gene and H-13 had two mutations. All mutations were missense mutation. Mutation of c.388C>T (P130S) in exon3 was found in all four samples, and other three mutations were also located in exon3 (Table 5). By OncodriveCLUST analysis, SERF2 gene was predicted as a putative driver-mutated gene. No pathway was correlated with SERF2 gene in pathway enrichment analysis.
SCNV analysis
SCNV gain was the dominant type in both HER2+ and HER2– patients. The number of CNV gains in both groups is much higher than CNV losses. There was no significant difference in average number of CNV gains between HER2+ and HER2– samples, although absolute count was higher in HER2+ patients (434.2±307.9 vs 224.4±130.0, P=0.174; Table 6).
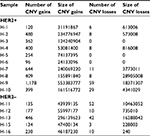  | Table 6 Somatic CNV analysis in HER2+ and HER2– samples Abbreviations: CNV, copy number variation; HER2, human epidermal growth factor receptor 2. |
In HER2+ samples, frequent gains were observed on chromosomes 7, 8q, 9 p, 10 p, 11q, 13q, 17q and 20. In HER2– samples, frequent gains were observed on chromosomes 1 p, 3q, 5 p, 7 p, 10q, 11 p, 12, 16 p, 18q, 19q, 20q and 22q. The CNV gains of 17q12, 17q21.2, 20q13.13 and 13q22.1 fragments were only observed in HER2+ samples. In HER2– samples, CNV gains of 5p15.33 (5/5), 10q26.3 (4/5) and 12q24.33 (4/5) were more common than those in HER2+ samples.
By using the GISTIC algorithm, recurrence of CNV and genes located in those cytobands were analyzed. A total of nine known driver genes including ERBB2, cyclin-dependent kinase 12 (CDK12), RARA, INTS1, CCNE1, MDM2, PTPRB, HMGA2 and FRG1B were identified. The CNV gain of ERBB2 (10/10), CDK12 (6/10) and RARA (5/10) was only observed in HER2+ samples. Other genes detected in HER2+ samples include JUP, GJD3, KRT39, CDC6, RAPGEFL1, WIPF2, FAM65C, KLF5, DACH1 and PIBF1. In HER2– samples, no known driver genes were identified. In highly frequent CNV gain, solute carrier family 12 member 7 (SLC12A7), TTC40 and GALNT9 genes were more commonly detected in HER2– samples (Table 7 and Figure 3). The function of those genes has not been reported in gastric cancer.
Discussion
In this study, we performed WGS to analyze and compare the genomic difference between HER2+ and HER2– gastric cancer samples. Patients with similar clinicopathological characteristics but different HER2 status were collected. The results showed that for somatic mutation, the number of SNVs and mutation spectrum were similar between HER2+ and HER2– groups, whereas the specific gene mutations were different. For SCNV analysis, the distribution of CNV gain was different between those two groups, as well as gene amplifications.
WGS and analysis of gastric cancer had been reported by several research groups. By TCGA project, four subtypes based on molecular characteristics of gastric cancer had been established.6 HER2 amplification was mainly detected in chromosomal instability subtype and also found in genomically stable and Epstein–Barr virus-positive subtypes. The distribution of HER2 amplification in those subtypes suggested that a distinct molecular difference may not exist between HER2+ and HER2– samples. In this study, significant difference in SNV, SV and mutation spectrum analysis between HER2+ and HER2– samples was not detected.
Although the molecular landscape between HER2+ and HER2– samples was similar, different significantly mutated genes were detected in both groups, respectively. Commonly mutated genes reported by other studies including TTN, TP53 and MUC16 were also detected in both HER2+ and HER2– samples in the study.10 On the other hand, KMT2C gene mutations were mainly detected in HER2+ samples, whereas SERF2 gene mutations were detected in HER2– samples.
Missense mutations of KMT2C gene in gastric cancer had not been fully investigated by previous studies. In this study, c.12383G>T (S4128I) mutation of KMT2C was identified as a driver mutation.7–9 KMT2C is a member of the myeloid/lymphoid or mixed-lineage leukemia family and encodes a nuclear protein. It is a catalytic subunit of the MLL2/3 complex, which participates in methylation of histone H3 and is involved in transcriptional coactivation.11 KMT2C mutations have been detected in several malignant diseases. In gastric cancer, SNPs, rs6943984 and rs4725443, of KMT2C were associated with increased gastric cancer risk.12 KMT2C frameshift mutations were also detected in gastric cancer samples with high microsatellite instability and resulted in low protein expression.13 It was reported that inactivation of KMT2C resulted in tumor formation of ureter epithelium, which suggested its role as a tumor suppressor gene. In this study, new mutations of KMT2C were detected. Missense mutations of exon8 D348N and exon8 K339N were located in one of the zinc finger motifs of KMT2C protein (344–389 aa). Although the biological function of these mutations found in this study is currently unclear, those results suggested a potential role of KMT2C in gastric cancer development and progression.
It has been reported that KMT2C mutation might be a prognostic biomarker for cancer patients, but the results are controversial in different kinds of tumors. In cutaneous squamous cell carcinoma, KMT2C mutation was correlated with bone invasion and shorter recurrence-free survival.14 In pancreatic cancer, its mutation resulted in low expression and was associated with good outcome of patients.15 Je et al13 reported that KMT2C mutation trended toward reduced overall survival (OS) in gastric cancer. Due to the limited sample size, its correlation with patient’s outcome can hardly be analyzed in this study. However, prognostic analysis by searching TCGA database showed that OS of gastric cancer patient with KMT2C mutation was not significantly different from those with wild-type KMT2C. In this study, KMT2C mutation was only found in HER2+ patients. Although the correlation between HER2 expression and outcome of gastric cancer patients has been widely investigated, the combination of HER2 status and KMT2C mutation has not been assessed as an independent parameter. Therefore, the prognostic role of KMT2C mutation in HER2+ patients should be further investigated.
In five HER2– samples, SERF2 mutation was detected in three of them and was identified as a significant mutated gene. Small EDRK-rich factor 2 (SERF2) protein is ubiquitously expressed in organs, including prostate, adrenal, stomach, colon and brain. However, its mutation and biological function have not been reported. In this study, all SERF2 mutations were located in exon 3 of isoform 2. Although SERF2 was predicted as a putative driver gene by OncodriveCLUST analysis, mutations that can interfere the biological function of SERF2 protein are currently unknown. Therefore, function of SERF2 mutation should be further analyzed to understand its role in development or progression of HER2– gastric cancer.
For SCNV analysis, although the number of CNV gains was similar between HER2+ and HER2– samples, the distribution of frequent SCNV was different. In HER2+ samples, frequent gain of 17q12 cytoband that includes HER2 amplicon was demonstrated, as well as that of 17q21.2, 20q13.13 and 13q22.1 cytobands. In HER2– patients, frequent gain of 5p15.33, 10q26.3 and 12q24.33 was detected. According to GISTIC analysis, genes located in those cytobands were identified.
CDK12 was located in cytoband 17q12. The CNV gain of CDK12 was detected in 60% of the HER2+ samples (6/10). In breast cancer, ~70% ERBB2 amplicon also contained CDK12 gene.16 CDK12 belongs to transcription-associated subfamily of CDKs that can phosphorylate the C-terminal domain of the large subunits of RNA polymerase II, thereby acting as a key regulator of transcription elongation. The downstream genes regulated by CDK12 include BRCA1, ATR, FANC1 and FANCD2, which participate in homologous recombination repair and maintenance of genomic stability.17,18 In tumor, the activity of those genes has been correlated with resistance to platinum-containing chemotherapy.19 Cisplatin plus capecitabine (XP) were standard regime to combine with trastuzumab for HER2+ gastric cancer patients.3 Therefore, it is possible that coamplification of CDK12 with ERBB2 in HER2+ gastric cancer might reduce the efficacy of XP regime. High expression of BRCA1, one of the CDK12 downstream genes, was correlated with sensitivity to microtubule targeting agents in breast cancer, including paclitaxel and vinorelbine.20 For those patients with ERBB2 and CDK12 amplification, taxane-containing regimes could be a better partner other than XP regime to combine with trastuzumab. Several studies had evaluated the efficacy of combination of trastuzumab and taxane in gastric cancer. Kagawa et al21 reported the results of the combination of trastuzumab with docetaxel and S-1 as first-line therapy in 23 HER2+ metastatic gastric cancer patients. Median progression-free survival (PFS) and OS were 6.7 months and 17.5 months, respectively. In another Phase II study, 47 metastatic gastric cancer patients received paclitaxel plus trastuzumab as second-line therapy, and median PFS and median OS were 5.1 months and 17.1 months, respectively.22 These results suggested the potential efficacy of trastuzumab plus taxane-containing regime for HER2+ metastatic gastric cancer. In future perspective study, the predictive value of CDK12 amplification to choose chemotherapy regime to combine with trastuzumab should be further investigated.
Amplification of SLC12A7, also known as KCC4, was mainly found in HER2– samples. SLC12A7 acts as an electroneutral potassium chloride cotransporter activated by cell swelling, which participates in regulating cell osmotic homeostasis and cell volume.23 SLC12A7 amplification was also detected in adrenocortical carcinoma.24 Its overexpression was found in cervical cancer, ovarian cancer and breast cancer. In tumor cells, SLC12A7 participates in regulating cell proliferation, invasion and metastasis.25 Shen et al26 reported that SLC12A7 could be recruited to cell membrane by Insulin-like growth factor 1 and EGF stimulation, which functioned as a membrane scaffold facilitating the cytoskeletal reorganization required for cellular invasiveness. Furthermore, its role in regulating cell volume control and Matrix metalloproteinase activity also involved in tumor invasion. Amplification of SLC12A7 in HER2– gastric cancer suggested that it might participate in regulating malignant behavior of HER2– gastric cancer. Currently, expression of SLC12A7 in gastric cancer and its correlation with HER2 expression had not been reported, as well as its biological function. Further study should be performed to elucidate its role in HER2– gastric cancer.
Conclusion
In this study, differences in genomic landscape between HER2+ and HER2– gastric cancers were revealed. For somatic mutation, new significant mutated gene KMT2C was detected in HER2+ samples, and SERF2 mutations were detected in HER2– samples. Amplifications of CDK12 and SLC12A7 were identified in HER2+ and HER2– gastric cancers, respectively.
Acknowledgments
This study was supported by National Science Foundation of China (81672327, 81802319 and 81602411), Program of Shanghai Academic/Technology Research Leader (17XD1402600), Program for Outstanding Medical Academic Leader and Shanghai Municipal Education Commission – Gaofeng Clinical Medicine Grant Support (20161410) and Development Grant for Clinical Trial (SHDC12017X06).
Disclosure
The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Waddell T, Verheij M, Allum W, et al. Gastric cancer: ESMO-ESSO-ESTRO clinical practice guidelines for diagnosis, treatment and follow-up. Eur J Surg Oncol. 2014;40(5):584–591. | ||
Bang YJ, van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–697. | ||
Kelly CM, Janjigian YY. The genomics and therapeutics of HER2-positive gastric cancer-from trastuzumab and beyond. J Gastrointest Oncol. 2016;7(5):750–762. | ||
Nakagawa H, Fujita M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018;109(3):513–522. | ||
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. | ||
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. | ||
Kandoth C, Mclellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. | ||
Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. | ||
Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. | ||
Cho YW, Hong T, Hong S, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. | ||
Li B, Liu HY, Guo SH, Sun P, Gong FM, Jia BQ. Mll3 genetic variants affect risk of gastric cancer in the chinese han population. Asian Pac J Cancer Prev. 2013;14(7):4239–4242. | ||
Je EM, Lee SH, Yoo NJ, Lee SH. Mutational and expressional analysis of MLL genes in gastric and colorectal cancers with microsatellite instability. Neoplasma. 2013;60(2):188–195. | ||
Pickering CR, Zhou JH, Lee JJ, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20(24):6582–6592. | ||
Dawkins JB, Wang J, Maniati E, et al. Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016;76(16):4861–4871. | ||
Paculová H, Kohoutek J. The emerging roles of CDK12 in tumorigenesis. Cell Div. 2017;12:7. | ||
Blazek D, Kohoutek J, Bartholomeeusen K, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25(20):2158–2172. | ||
Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem. 2014;289(13):9247–9253. | ||
O’Grady S, Finn SP, Cuffe S, Richard DJ, O’Byrne KJ, Barr MP. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev. 2014;40(10):1161–1170. | ||
Bajrami I, Frankum JR, Konde A, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74(1):287–297. | ||
Kagawa S, Muraoka A, Kambara T, et al. A multi-institution phase II study of docetaxel and S-1 in combination with trastuzumab for HER2-positive advanced gastric cancer (DASH study). Cancer Chemother Pharmacol. 2018;81(2):387–392. | ||
Nishikawa K, Takahashi T, Takaishi H, et al. Phase II study of the effectiveness and safety of trastuzumab and paclitaxel for taxane- and trastuzumab-naïve patients with HER2-positive, previously treated, advanced, or recurrent gastric cancer (JFMC45-1102). Int J Cancer. 2017;140(1):188–196. | ||
Weng TY, Chiu WT, Liu HS, et al. Glycosylation regulates the function and membrane localization of KCC4. Biochim Biophys Acta. 2013;1833(5):1133–1146. | ||
Brown TC, Juhlin CC, Healy JM, et al. DNA copy amplification and overexpression of SLC12A7 in adrenocortical carcinoma. Surgery. 2016;159(1):250–258. | ||
Chen YF, Chou CY, Wilkins RJ, Ellory JC, Mount DB, Shen MR. Motor protein-dependent membrane trafficking of KCl cotransporter-4 is important for cancer cell invasion. Cancer Res. 2009;69(22):8585–8593. | ||
Shen MR, Lin AC, Hsu YM, et al. Insulin-like growth factor 1 stimulates KCl cotransport, which is necessary for invasion and proliferation of cervical cancer and ovarian cancer cells. J Biol Chem. 2004;279(38):40017–40025. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.