Back to Journals » OncoTargets and Therapy » Volume 12
Diagnostic and prognostic roles of circ-SHPRH for solid cancers: a meta-analysis
Authors Li F, Huang Q, Gong Z , Wang H, Chen J
Received 8 January 2019
Accepted for publication 4 April 2019
Published 31 May 2019 Volume 2019:12 Pages 4351—4357
DOI https://doi.org/10.2147/OTT.S200755
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Feng Li,* Qilin Huang,* Zhenyu Gong, Hongxiang Wang, Juxiang Chen
Department of Neurosurgery, Changzheng Hospital, Second Military Medical University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Background: Circ-SHPRH is a novel circular RNA which was down-regulated in various cancer tissues. Several recent studies hinted that circ-SHPRH might function as a diagnostic and prognostic biomarker for solid cancers. This meta-analysis was performed to determine the diagnostic and prognostic roles of circ-SHPRH for these solid cancers.
Methods: Publications were searched in PubMed, MEDLINE, Web of Science, Embase, China National Knowledge Infrastructure (CNKI), and Wanfang database. Pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the summary receiver operating characteristic curve (SROC) (AUC) with 95% confidence interval (95% CI) were calculated to evaluate the diagnostic value of circ-SHPRH. Pooled hazard ratio (HR) with 95% CI was applied to assess the prognostic value of circ-SHPRH.
Results: Nine eligible studies, including 4 for diagnosis and 5 for prognosis, were included in this meta-analysis. Pooled sensitivity, specificity, PLR, NLR, DOR, and AUC were 0.79 (95% CI: 0.73–0.84), 0.75 (95% CI: 0.69–0.81), 3.20 (95% CI: 2.53–4.06), 0.28 (95% CI: 0.22–0.36), 11.27 (95% CI: 7.57–16.79), and 0.84 (95% CI: 0.80–0.87), respectively. Pooled HR indicated that low circ-SHPRH expression was significantly associated with poor overall survival (OS) (low vs high, HR=2.22, 95% CI: 1.60–3.09).
Conclusion: The results of this meta-analysis indicate that circ-SHPRH might be an important diagnostic and prognostic biomarker for various solid cancers.
Keywords: Circ-SHPRH, diagnosis, prognosis, cancer, meta-analysis
Introduction
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs which are characterized by highly conserved and stable covalently closed continuous loop without free 5ʹ cap and 3ʹ tail.1,2 CircRNAs were regarded as by-products of splicing mistakes without biological functions in the past few decades.3 As the deep high-throughput sequencing technology and bioinformatic approaches are being widely applied, plenty of circRNAs have been detected in many different tissues.4 Recent studies have indicated that circRNAs can function as microRNA sponges and participate in various physiological or pathological processes, including carcinogenesis.5–8 Circ-SHPRH (circBase ID: hsa_circ_0001649, circ2Traits ID: hsa_circ_001599), a transcription product of SNF2 Histone Linker PHD RING Helicase (SHPRH) gene, is located at chr6: 146209155–146216113. Mature circ-SHPRH is derived from SHPRH gene exons 26–29 and only contains 440 nucleotides by intron shearing.9 Recently, circ-SHPRH was reported as a novel cancer-related circRNA and served as a potential diagnostic and prognostic biomarker in several types of cancers.10–18 However, the limitations of these publications to determine the roles of circ-SHPRH in cancer diagnosis and prognosis were that they all had a small sample size. Therefore, we performed a meta-analysis to evaluate the diagnostic and prognostic value of circ-SHPRH in human solid cancers.
Materials and methods
Search strategy
The current meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.19 The publication searches were conducted in PubMed, MEDLINE, Web of Science, Embase, China National Knowledge Infrastructure (CNKI), and Wanfang database. The key terms applied for electronic databases were (“circ-SHPRH” or “hsa_circ_0001649” or “hsa_circ_001599”. The publication searches were updated to December 10, 2018.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) expression level of circ-SHPRH was detected in any type of human cancers; (2) the association between circ-SHPRH expression level and cancer diagnosis or prognosis was investigated.
The exclusion criteria were as follows: (1) duplicate publications; (2) reviews, case reports, or letters; (3) articles without available data.
Data extraction
Two authors (Feng Li and Qilin Huang) independently extracted the data from all eligible studies. The following information was extracted from studies for diagnostic meta-analysis: first author, publication year, cancer types, sample types, detection methods, reference standard, the number of patients, the number of controls, sensitivity, specificity, and area under the curve (AUC). The following information was extracted from studies for prognostic meta-analysis: first author, publication year, cancer types, sample types, detection methods, circ-SHPRH expression level, cut-off values, number of patients in low expression groups, number of patients in high expression groups, outcome, follow-up, multivariate analysis, and hazard ratios (HRs) as well as corresponding 95% confidence interval (CI) for overall survival (OS).
Quality assessment
Two authors (Feng Li and Qilin Huang) independently assessed the quality of included studies for diagnostic and prognostic meta-analysis by using Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2)20 and Newcastle–Ottawa Scale (NOS),21 respectively. QUADAS-2 comprises 4 domains: patient selection, index test, reference standard, and flow and timing. Each item was assessed as “yes”, “no”, or “unclear”. The answer of “yes” indicated low risk of bias, whereas “no” or “unclear” suggested high risk of bias. Total scores of NOS were ranged from 0 to 9, and studies getting higher than 6 were considered high-quality studies.
Statistical analysis
Statistical analyses were conducted using STATA 12.0 software and Review Manager 5.3 software. Heterogeneity test was performed by I-squared statistics. If I-squared value>50% and P-value<0.05, we judged that there was significant heterogeneity among the included studies and we performed a random effects model to assess the pooled results. Otherwise, there was no significant heterogeneity and a fixed effects model was performed. Statistical significance was defined as P-value<0.05.
For diagnostic meta-analysis, pooled results of sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the summary receiver operating characteristic curve (SROC) were calculated to evaluate the diagnostic accuracy of circ-SHPRH expression. The potential publication bias was assessed by using Deeks’ funnel plot asymmetry test. We judged that there was no publication bias if P-value>0.1 for Deeks’ test.
For prognostic meta-analysis, pooled HR as well as 95% CI was used to describe the prognostic value of circ-SHPRH expression. The potential publication bias was estimated by using Begg’s funnel plot. Sensitivity analysis was performed to assess the stability of pooled HR. We judged that there was no publication bias if P-value>0.1 for Begg’s test.
Results
Search strategy results
A total of 39 articles were identified from PubMed, MEDLINE, Web of Science, Embase, CNKI, and Wanfang database. After removing 26 duplicate publications, 13 studies were included for further screening. After screening the titles, abstracts, and full texts, 9 eligible studies, including 4 for diagnosis and 5 for prognosis, were finally included in this meta-analysis based on the inclusion and exclusion criteria (Figure 1).
 | Figure 1 Flow chart of the articles selection process. |
Main characteristics and quality assessment of eligible studies
Publication years of all included studies ranged from 2016 to 2018. The expression of circ-SHPRH was detected by quantitative real-time polymerase chain reaction (qRT-PCR). All included studies showed circ-SHPRH was down-regulated in cancer tissues compared to paired paracancerous normal tissues. Three types of cancers were included in the diagnostic meta-analysis: hepatocellular carcinoma (HCC),17 gastric cancer (GC),16 and colorectal cancer (CRC).15,18 Five types of cancers were included in the prognostic meta-analysis: pancreatic ductal adenocarcinoma (PDAC),14 non-small cell lung cancer (NSCLC),13 glioma,12 retinoblastoma (RB),11 and HCC10 Follow-up period in the studies for prognostic meta-analysis ranged from 45 to 60 months. The main characteristics and quality assessment of eligible studies for diagnostic meta-analysis are shown in Table 1 and Figure 2, respectively. The main characteristics and quality assessment of eligible studies for prognostic meta-analysis are summarized in Table 2.
 | Table 1 Main characteristics of eligible studies for diagnosis |
 | Table 2 Main characteristics and quality assessment of eligible studies for prognosis |
 | Figure 2 Quality assessment of eligible studies for diagnostic meta-analysis. |
Diagnostic role of circ-SHPRH expression
Overall, 279 patients with their cancer tissues and paired paracancerous normal tissues from 4 eligible studies were included for diagnostic meta-analysis. As shown in Figure 3, both I-squared value <50% and P-value >0.05, we judged that there was no significant heterogeneity among the included studies and we performed a fixed effects model to assess the pooled results; the pooled sensitivity and specificity were 0.79 (95% CI: 0.73–0.84) and 0.75 (95% CI: 0.69–0.81), respectively. As shown in Figure 4, all I-squared value <50% & P-value >0.05, we judged that there was no significant heterogeneity among the included studies and we performed a fixed effects model to assess the pooled results; the pooled PLR, NLR, and DOR were 3.20 (95%CI: 2.53–4.06), 0.28 (95%CI: 0.22–0.36), and 11.27 (95%CI: 7.57–16.79), respectively. As shown in Figure 5, the area under the SROC was 0.84 (95% CI: 0.80–0.87). All the above results showed that circ-SHPRH had a moderate-high diagnostic accuracy for solid cancers.
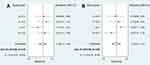 | Figure 3 (A) Forest plots for pooled sensitivity. (B) Forest plots for pooled specificity. |
 | Figure 4 (A) Forest plots for pooled positive likelihood ratio (PLR). (B) Forest plots for pooled negative likelihood ratio (NLR). (C) Forest plots for pooled diagnostic odds ratio (DOR). |
 | Figure 5 Summary receiver operating characteristic (SROC) curve based on circ-SHPRH. |
Prognostic role of circ-SHPRH expression
A total of 312 patients from 5 eligible studies were included for prognostic meta-analysis. As shown in Figure 6, the fixed effects model was conducted due to there was no significant heterogeneity among the 5 studies (I2=0%, P=0.748). The results showed a significant association between circ-SHPRH expression and OS; low expression of circ-SHPRH predicted poor OS (low vs high, pooled HR=2.22, 95% CI: 1.60–3.09). In other words, low circ-SHPRH expression is a risk factor for solid cancers prognosis.
 | Figure 6 Forest plot for estimating hazard ratios (HRs) of low circ-SHPRH expression in solid cancers. |
Publication bias and sensitivity analysis
The potential publication bias of diagnostic meta-analysis was assessed by Deeks’ funnel plot asymmetry test. As shown in Figure 7A, the Deeks’ funnel plot showed no asymmetry, and the P-value of Deeks’ test was 0.486, suggesting that there was no significant publication bias. The potential publication bias of prognostic meta-analysis was assessed by Begg’s test. However, as shown in Figure 7B, the Begg’s funnel plot showed asymmetric, and the P-value of Begg’s test was 0.001, suggesting that publication bias did exist in the prognostic meta-analysis. Meanwhile, sensitivity analysis was performed to evaluate the influence of the omitted study on the pooled HR; as shown in Figure 7C, omitting any study had no significant effect on the pooled results. The sensitivity analysis indicated that pooled HR of OS was reliable.
 | Figure 7 (A) Deeks’ funnel plot. (B) Begg’s funnel plot. (C) Sensitivity analysis for evaluating the influence of the omitted study on the pooled hazard ratio (HR). |
Discussion
Research on circRNAs is still in infancy, and we just start to explain its functional roles in carcinogenesis. Evidence so far indicates that circRNAs can not only function as valuable biomarkers for various cancers but also serve as potential targets in cancer therapy.8 There are obvious advantages in applying circRNAs as diagnostic and prognostic biomarkers even therapeutic targets: 1) circRNAs are superior to linear RNAs in terms of stability; 2) circRNAs can cover the shortage of low organ specificity of traditional biomarkers; 3) circRNAs are non-invasive biomarkers, which can be easily detected in blood and body fluid.22
SHPRH, a cancer suppressor gene, functions in DNA repair and regulates ribosomal RNA transcription.23–25 Circ-SHPRH is a circular RNA derived from SHPRH exons via back-splicing. A recent study reported that a novel protein termed SHPRH-146aa encoded by circ-SHPRH was a tumor suppressor in human glioblastoma. In other words, circ-SHPRH is a protein-coding circRNA and plays a vital role in glioma tumorigenesis.9
To our knowledge, this meta-analysis is the first try to estimate correlation of circ-SHPRH expression with the diagnosis and prognosis of patients with solid cancers. A total of 9 eligible studies were included following the publication selection process. Results of pooled sensitivity, specificity, PLR, NLR, DOR, and AUC indicated that circ-SHPRH was a remarkable diagnostic biomarker for solid cancers. We were convinced that low circ-SHPRH expression was associated with poor prognosis after performing a fixed effects model to estimate the pooled HR and the 95% CI (low vs high, pooled HR=2.22, 95% CI: 1.60–3.09).
However, there were several limitations in our meta-analysis that should be noted. First, we only enrolled 4 studies in diagnostic meta-analysis and 5 studies in prognostic meta-analysis. A small number of studies limited the widespread clinical applicability of the meta-analysis. Moreover, because of the small number of included studies, it was hard to accurately assess the publication bias by funnel plot. And we were unable to perform a subgroup analysis to explore sources of heterogeneity due to the limited number of included studies. Therefore, more comprehensive studies are of great need to confirm the value of circ-SHPRH in diagnosis and prognosis for solid cancers. Second, heterogeneities might exist among the included studies due to the cancer types, cut-off values, and the definitions of poor OS were different in each study. Third, there were only a few published studies about circ-SHPRH; coincidentally, all studies included in this meta-analysis tested circ-SHPRH expression among Chinese population, it is unclear whether our results might be applicable to other populations.
Conclusion
The results of diagnostic meta-analysis provided evidence that circ-SHPRH has moderate-high diagnostic accuracy for various solid cancers. The results of prognostic meta-analysis suggested that low expression of circ-SHPRH is significantly associated with poor OS. Therefore, circ-SHPRH could be a novel diagnostic and prognostic biomarker for various solid cancers. Due to the limitations, further better-designed and independent studies are needed to verify the roles of circ-SHPRH in human cancers.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No.81872072 and 81572501).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
2. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
3. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559
4. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi:10.1186/s13059-014-0409-z
5. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384. doi:10.1038/nature11993
6. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480.
7. Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
8. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2017;37:555. doi:10.1038/onc.2017.361
9. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi:10.1038/s41388-017-0019-9
10. Zhang XW, Qiu SL, Luo P, et al. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018;22(1):135–142. doi:10.3233/CBM-171109
11. Xing LC, Zhang LM, Feng YL, Cui Z, Ding L. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;105:326–333. doi:10.1016/j.biopha.2018.05.141
12. Wang Y, Sui X, Zhao H, et al. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117–122. doi:10.1016/j.gene.2018.07.037
13. Liu TM, Song Z, Gai YL. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503(3):1503–1509. doi:10.1016/j.bbrc.2018.07.070
14. Jiang YH, Wang T, Yan L, Qu LJ. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88–93. doi:10.1016/j.gene.2018.06.099
15. Ji WX, Qiu CL, Wang M, Mao N, Wu SF, Dai YH. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497(1):122–126. doi:10.1016/j.bbrc.2018.02.036
16. Li WH, Song YC, Zhang H, et al. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. doi:10.1155/2017/4587698
17. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi:10.3233/CBM-150552
18. Ji WX, Qiu CL, Bao JM, Dai YH, Song YC. [The expression level of hsa_circ_0001649 in colorectal cancer and its clinical value]. J Xian Jiaotong Univ. 2019;40(1):92–95.
19. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi:10.1371/journal.pmed.1000097
20. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi:10.7326/0003-4819-155-8-201110180-00009
21. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi:10.1007/s10654-010-9491-z
22. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98. doi:10.1186/s13045-018-0643-z
23. Tomi NS, Davari K, Grotzky D, et al. Analysis of SHPRH functions in DNA repair and immunoglobulin diversification. DNA Repair (Amst). 2014;24:63–72. doi:10.1016/j.dnarep.2014.09.010
24. Lee D, An J, Park YU, et al. SHPRH regulates rRNA transcription by recognizing the histone code in an mTOR-dependent manner. Proc Natl Acad Sci U S A. 2017;114(17):E3424–e3433. doi:10.1073/pnas.1701978114
25. Lee D, Park JH, Kim S, Lee SG, Myung K. SHPRH as a new player in ribosomal RNA transcription and its potential role in homeostasis of ribosomal DNA repeats. Transcription. 2018;9(3):190–195. doi:10.1080/21541264.2017.1381795
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
