Back to Journals » Infection and Drug Resistance » Volume 16
Diagnosis of Legionnaires’ Disease Assisted by Next-Generation Sequencing in a Patient with COVID-19
Authors Huang PH, Huang YT, Lee PH, Tseng CH , Liu PY, Liu CW
Received 5 November 2022
Accepted for publication 3 January 2023
Published 20 January 2023 Volume 2023:16 Pages 355—362
DOI https://doi.org/10.2147/IDR.S396254
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Po-Hsiu Huang,1,* Yao-Ting Huang,2,* Po-Hsin Lee,3– 6 Chien-Hao Tseng,7 Po-Yu Liu,5– 7 Chia-Wei Liu7
1Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan; 2Department of Computer Science and Information Engineering, National Chung Cheng University, Chia-Yi, Taiwan; 3Division of Chest Medicine, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan; 4College of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan; 5Ph.D. Program in Translational Medicine, National Chung Hsing University, Taichung, Taiwan; 6Rong Hsing Research Center for Translational Medicine, National Chung Hsing University, Taichung, Taiwan; 7Division of Infectious Diseases, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan
*These authors contributed equally to this work
Correspondence: Po-Yu Liu; Chia-Wei Liu, Tel +886 4 2359 2525, Fax +886 4 2359 2525 2111, Email [email protected]; [email protected]
Abstract: Coinfection in COVID-19 patients is associated with worsening outcome. Among patients with COVID-19, Legionella pneumophila, a common cause of pneumonia, has been reported as a co-occurring respiratory infection. A nonspecific clinical presentation, however, makes an early diagnosis difficult. Bronchoalveolar lavage fluid was collected from a patient suffering from COVID-19 and presenting with pneumonia and sent for metagenomic analysis. Differential abundance analysis was carried out by comparing each taxon reads per million between the bronchoalveolar lavage fluid sample and the negative control. Two replicates of metagenomic sequencing were conducted on bronchoalveolar lavage fluid samples. Within each replicated sequencing, one negative control was sequenced for comparison of taxon abundance in the BALF sample. In both replicates, Legionella pneumophila was the only taxon with significantly higher abundance when compared with the negative control. PCR of the bronchoalveolar further confirmed the presence of L. pneumophila. Several studies have estimated that the incidence of Legionnaires’ disease co-infection in patients with COVID-19 infection is approximately 0% to 1.5%. There are some common characteristics of COVID-19 and co-infection with Legionnaires’ disease, making it difficult to diagnose bacterial infection early. The diagnosis of these cases is important due to the different treatments used. Current diagnostic tests for Legionnaires’ disease include conventional culture, urinary antigen for L. pneumophila serogroup 1, polymerase chain reaction, direct fluorescent antibody stain, and paired serology. The current study demonstrated that metagenomics is a promising approach that facilitated the diagnosis of Legionnaires’ disease.
Keywords: Legionnaires’ disease, COVID-19, co-infection, next-generation sequencing
Introduction
Coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, led to a global pandemic. To date, it has infected six hundred million people and has led to more than six million deaths.1 Older adults and people who have certain underlying medical conditions2 are at increased risk for severe illness from COVID-19.
Since the initial outbreak in 2019, various co-infections have been reported in patients with COVID-19. These patients were found to have higher rates of receiving invasive mechanical ventilation, prolonged hospital admissions, and higher mortality.3 Bacterial co-pathogens reported by studies include Mycoplasma pneumoniae, Legionella pneumophila, Streptococcus pneumoniae, and Chlamydia pneumoniae. Many other viral and fungal co-pathogens were also identified.4 However, it is hard to differentiate between viral infection and bacterial infection, and it is also hard to distinguish co-infection from secondary infection due to the similarity of their symptoms.
Legionnaires’ disease, one of the co-infections with COVID-19, is caused by the gram-negative bacteria Legionella pneumophila. It is a common form of severe pneumonia and shares many similarities with COVID-19. Its features include fever, non-productive cough, headache, myalgias, rigors, dyspnea, diarrhea, and delirium.5 People who are at increased risk of developing dangerous symptoms of COVID-19 are also more susceptible to Legionnaires’ disease.6
Current diagnostic tests for Legionnaires’ disease include culture of the respiratory tract, urinary antigen for L. pneumophila serogroup 1, polymerase chain reaction (PCR), direct fluorescent antibody stain, and paired serology. More than 50 species of Legionella have been identified;7 among them, Legionella pneumophila is the most common, causing community-acquired and nosocomial infections. Although research has shown that L. pneumophila serogroup 1, which can be detected by urine antigen tests, is responsible for most cases of Legionnaires’ disease, 20–50% of cases of Legionnaires’ disease would remain undiagnosed if no additional testing methods are used.7 Although culture can detect all species and serogroups, it is technically difficult and requires a long turn-around time. PCR assays vary by laboratory and may not be available in some. Therefore, information on respiratory diseases related to other species and serogroups of Legionella is limited. In addition to this limitation, similarities in the clinical presentation of COVID-19 and Legionnaires’ disease make an accurate and timely diagnosis of Legionnaires’ disease challenging. In these years, next-generation sequencing technique is crucial for us to detect such pathogen like Legionella. It can be used to detect pathogen quickly and accurately than conventional detection tools, and it can bring advantage for diagnosis and therapy in clinical setting. Higher detection rate, quicker clinical symptom resolution, and accurately change in antibiotic use are noted in using next-generation sequencing technique.
Materials and Methods
Case Presentation and Study Sample
A 58-year-old man with a history of hypertension developed fever, cough, and muscle aches 5 days after being diagnosed with mild COVID-19. CXR consisted of right lower lobe pneumonia. Empiric piperacillin/tazobactam was started for four days without improvement. To investigate the etiology of his pneumonia, bronchoalveolar lavage was performed. In addition to the routine work-up, the specimen was submitted for metagenomic analysis and L. pneumophila was identified. Levofloxacin was started which rapidly resolved symptoms/signs. A real-time RT-PCR assay of the bronchoalveolar lavage fluid (BALF) also confirmed the presence of L. pneumophila.
Sample Preparation and Sequencing
The tissue sample was first ground and mixed with 1 g of 0.5-mm diameter glass beads and then placed on a vortex mixer for 30 min at 3000 rpm. DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) was used for DNA extraction in 300 μL of the sample following the manufacturer’s instructions. We used an enzymatic method to fragment the DNA into 150–200 bp in length. The DNA library was built through an end-repaired adapter and polymerase chain reaction amplification. We applied the DNA Qubit Assay (Thermo Fisher) to determine the DNA concentrations and used an Agilent 2100 system (Agilent Technologies, Santa Clara, CA) to evaluate DNA quality electrophoretically. The DNA library was built through an end-repaired adapter and polymerase chain reaction amplification using MGIEasy FS DNA Library Prep Kit (MGI). We then transformed the single-strand circularized DNA library into DNA nanoballs (DNBs) and sequenced them by DNBSeq-G50 with an average read length equal to 50bp.
Bioinformatics Analysis
The sequencing reads were preprocessed by removing low-quality (ie, reads <80% Phred score Q30), duplicated, and short-length (<35bp) reads. The entire workflow of bioinformatics analysis is shown in Supplementary Figure S1. Briefly, the high-quality reads were BWA-aligned against the human genome (hg38) to remove human-derived sequences.8 The non-human reads were BWA-aligned to the NCBI microbial reference genomes (RefSeq) for taxonomic identification. The abundance of each taxon was computed and normalized by Reads per Million (RPM). Differential abundance analysis was carried out by comparing each taxon RPMs between the BALF sample and the negative control. The taxa with statistically higher abundance than the controls were identified by the Chi-square test. We only output the differential-abundant taxa concordantly identified in all replicates.
Results
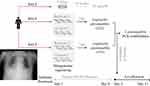
We sequenced the BALF sample with two replicated metagenomic sequencing (Figure 1). Within each replicated sequencing, one negative control was sequenced for comparison of taxon abundance in the BALF sample. We identified the pathogens of significantly higher abundance in the BALF sample than in the negative control. Only the significant-abundant taxon concordantly identified in the two replicates was retained. In both replicates, Legionella pneumophila was the only taxon with significantly higher abundance when compared with the negative control. For instance, the numbers of reads of L. pneumophila 252 and 353 in the two replicates, which corresponds to Reads per Million (RPM) of 21.23 and 14.44 after normalization of sequencing yields, respectively. When compared with the two negative controls (Figure 2), no reads of L. pneumophila were detected. Moreover, the remaining detected species (eg, Pseudomonas monteilii) all exhibited similar abundance between the BALF samples and controls (eg, 4.06 vs 1.36 RPM in Replicate 1, 2.82 vs 2.83 RPM in Replicate 2). Upon receiving the metagenomic diagnosis (Day 5), the antibiotic was switched to levofloxacin. Clinical symptom gradually subsided after changing antibiotic. The patient recovered and was discharged.
 |
Figure 2 Comparison of taxa abundance between the BALF and control samples of L. pneumophila and P. monteilii. |
Discussion
It is estimated that the prevalence of co-infection of influenza and bacteria is around 11% to 35%. The most common bacterial pathogen was Streptococcus pneumoniae, which represented 35% of cases, and the second most common bacteria was Staphylococcus aureus, which accounted for 28% of infections.9 Bacterial co-infection with the SARS-CoV-2 virus was less frequent, with an estimated prevalence of only 1.2–3.5%.10,11 Bacterial secondary infection had a slightly higher prevalence than that of co-infection and accounted for about 14.3%. Although the co-infection rate was relatively low, about two-thirds of the patients received antibiotic treatment.10
In addition to bacterial co-infection with SARS-CoV-2, several pathogen co-infections have been reported. Patients diagnosed with tuberculosis are at increased risk of developing COVID-19 and tend to have a worse prognosis.12,13 Some invasive fungal infections were also documented, although the true prevalence is believed to have been underestimated because few bronchoscopies were performed.14,15 The risk factors for fungal co-infection among COVID-19 patients were older age, diabetes, immunosuppression, racial or ethnic minority status, and smoking.15 For the early detection of a fatal invasive fungal infection, other fungal markers may be helpful.14
We report a case of a 58-year-old man co-infected with Legionnaires’ disease and COVID-19. Herein, we also review a number of case series and case reports, which includes discussion of nine articles covering sixteen cases. The basic characteristics, diagnostic parameters, and clinical characteristics are listed in Tables 1–3. According to the basic characteristics listed in Table 1, most cases were reported in Europe. Most cases had existing comorbidities, with diabetes mellitus and hypertension being the most commonly reported. Regarding the diagnostic parameters listed in Table 2, the urine antigen is the most commonly used. With respect to the clinical characteristics listed in Table 3, the mortality rate was approximately 33%, which is higher than in previous studies. This may be due to the limited available data.
 |
Table 1 Basic Character of Cases |
 |
Table 2 Diagnosis Parameters of Cases |
 |
Table 3 Clinical Characteristics of Cases |
There are some common characteristics of COVID-19 and co-infection with Legionnaires’ disease. Almost every patient had respiratory tract symptoms or signs, such as productive cough, and dyspnea. In terms of extrapulmonary symptoms, fever, and general malaise were also noted in almost every case. Diarrhea was also mentioned in some cases.16–18 In terms of laboratory data, hyponatremia was observed in some cases,16,17,19 which is consistent with common findings among patients with Legionnaires’ disease. Inflammation and infection parameters, including white blood cells and C-reactive protein, may or may not be in the normal range. Regarding chest radiographs, findings ranged from non-significant to consolidation or massive infiltration.16 Chest CT was surveyed in some cases,16,18,20 and there were also some cases presenting with ground-glass opacity, which is not common in Legionnaires’ disease, but has been noted in previous studies with COVID-19 infection.21 Antibiotics are essential for these patients, and there are different categories of antibiotics used in these cases, which are summarized in Table 3. Fluoroquinolone, macrolide, and cephalosporin are the most commonly used. The outcomes of these patients varied from mortality to moderate illness.16,20,22
Diagnostic tools are crucial in COVID-19 patients with a co-infection. In these cases, the diagnostic tools for COVID-19 involve PCR-based techniques, one of which is termed loop-mediated isothermal amplification (LAMP).19 Regarding the diagnosis of Legionnaires’ disease, one of the cases was diagnosed by serum antigen (IgM),23 and the rest of the cases were diagnosis on the basis of urine antigen, which is widely used as a diagnostic tool for Legionnaires’ disease. In the aforementioned studies, it is emphasized that the diagnosis of these cases is important but difficult. In most of the cases, COVID-19 infection was found first, and then Legionnaires’ disease was detected later. This could be due to a lack of screening for Legionnaires’ disease in patients with COVID-19. Upon diagnosis with co-infection of Legionnaires’ disease, broad-spectrum antibiotics and a prolonged admission period can be avoided.24 Except for cases with a specific related history, the diagnosis of Legionnaires’ disease is not considered in the differential diagnosis of patients with COVID-19 infection. However, many studies conclude that Legionnaires’ disease should be considered in patients with COVID-19 infection, especially with moderate to critical diseases. There may be useful information that can be gleaned from co-infected patients. As mentioned in one study,17 unilobar consolidation, which does not appear to be common in COVID-19 infection, may be a clue suggesting the presence of co-infection. Furthermore, in one study, some cardiovascular complications were noted, including COVID-19-related stress cardiomyopathy.20
The incidence of co-infection with Legionnaires’ disease in patients with COVID-19 infection ranges from 0% to 1.5%, based on estimates reported in different studies.6,25,26 Due to the limited sensitivity of the current diagnostic tools, especially urine antigen, further diagnostic tool applications should be considered.
In addition to Legionnaires’ disease, there are also different kinds of pathogens in patients with community acquired pneumonia. And metagenomics assessments had been used in these years. In one study,27 patients with CAP were divided into two groups which next-generation sequencing (NGS)-based metagenomics was added in one group. Compared to control group, rates of pathogen detection were higher in NGS-based group (79.9% vs 37%, P < 0.001). In addition, higher rate of complete symptom resolution (55.9% vs 7.0%, P < 0.001), and shorter duration of mechanical ventilation (7.4 days vs 17.3 days, P < 0.05) were also noted. In another study28 which metagenomic next-generation sequencing (mNGS) applied in bronchoalveolar lavage fluid collected from children with community-acquired pneumonia. Positive coincidence rate of the mGNS group was higher than conventional test group (86.78% vs 66.94%, P < 0.001). It also showed that conventional methods combined with mNGS could confirm the diagnosis in 99.17% of the patients. About metagenomic next-generation sequencing tool, there is some different types of platforms. In one study,29 bronchoalveolar lavage fluid from suspected community acquired pneumonia was used. And included two types of platforms (including Illumina and Nanopore), both of these two types had higher coincidence rate than conventional culture group (Illumina 57.81%, Nanopore 59.38%, Culture 25%, respectively). About Legionnaires’ disease, co-detected of Legionella pneumophila by mNGS in both BALF and blood samples was noted in one patient in previous study.27 Antibiotic was changed to quinolones. The clinical outcome was resolved in this case.
In the present case, we used metagenomic sequencing and a number of studies have applied metagenomic tools in the diagnosis of Legionnaires’ disease. This technique has been utilized in the recent COVID-19 pandemic period,30–32 and it can be quickly modified for other emerging infectious diseases in the future.
Conclusion
The incidence of co-infection with Legionnaires’ disease in patients with COVID-19 infection is estimated to be between 0% and 1.5% in different studies. Due to the limited sensitivity of the current diagnostic tools, alternative diagnostic measures are warranted, such as metagenomic sequencing. The current study demonstrated that metagenomic is a promising approach that facilitated the diagnosis of Legionnaires’ disease. In this case, the use of metagenomic analysis as a diagnostic tool for Legionnaires’ disease resulted in a change of antibiotics and improvement of patient’s clinical symptoms.
Abbreviations
COVID-19, Coronavirus disease, SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; PCR, polymerase chain reaction; BALF, bronchoalveolar lavage fluid; MGI, MGIEasy FS DNA Library Prep Kit; DNBs, DNA nanoballs; RefSeq, reference genomes; RPM, Reads per Million; LMAP, loop-mediated isothermal amplification.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Document.
Ethics Approval and Informed Consent
This study was approved by the Institutional Review Board of Taichung Veterans General Hospital (CE20261A).
Consent for Publication
Written informed consent was obtained from the patient for publication of this article and any accompanying images.
Author Contributions
CHT, PYL, and YTH conceived and designed the study. PHH, CHT, PYL, YTH, and CWL contributed to comprehensive research and sample collection. PHH, PYL, YTH, and CWL wrote the paper. PHH, PYL, YTH, and CWL participated in manuscript editing. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
YTH was supported in part by Taiwan’s Ministry of Science and Technology (109-2221-E-194 −038 -MY3). PYL was supported in part by Taiwan’s Ministry of Science and Technology (110-2314-B-075A-011), Taichung Veterans General Hospital (TCVGH-1113901D, TCVGH-1113901C).
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. Weekly epidemiological update on COVID-19-7 December 2022; 2022.
2. Wingert A, Pillay J, Gates M, et al. Risk factors for severity of COVID-19: a rapid review to inform vaccine prioritisation in Canada. BMJ open. 2021;11(5):e044684. doi:10.1136/bmjopen-2020-044684
3. Swets MC, Russell CD, Harrison EM, et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet. 2022;399(10334):1463–1464. doi:10.1016/S0140-6736(22)00383-X
4. Lai CC, Wang CY, Hsueh PR. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect. 2020;53(4):505–512. doi:10.1016/j.jmii.2020.05.013
5. Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15(3):506–526. doi:10.1128/CMR.15.3.506-526.2002
6. Verhasselt HL, Buer J, Dedy J, et al. COVID-19 co-infection with Legionella pneumophila in 2 tertiary-care hospitals, Germany. Emerg Infect Dis. 2021;27(5):1535–1537. doi:10.3201/eid2705.203388
7. Pierre DM, Baron J, Yu VL, Stout JE. Diagnostic testing for Legionnaires’ disease. Ann Clin Microbiol Antimicrob. 2017;16(1):59. doi:10.1186/s12941-017-0229-6
8. Li H: Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997; 2013.
9. Klein EY, Monteforte B, Gupta A, et al. The frequency of influenza and bacterial coinfection: a systematic review and meta‐analysis. Influenza Other Respi Viruses. 2016;10(5):394–403. doi:10.1111/irv.12398
10. Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26(12):1622–1629. doi:10.1016/j.cmi.2020.07.016
11. Karaba SM, Jones G, Helsel T, et al. Prevalence of co-infection at the time of hospital admission in COVID-19 patients, A multicenter study. Open Forum Infect Dis. 2020;8(1):1.
12. Mousquer GT, Peres A, Fiegenbaum M. Pathology of TB/COVID-19 co-infection: the phantom menace. Tuberculosis. 2021;126:102020. doi:10.1016/j.tube.2020.102020
13. Tadolini M, Codecasa LR, García-García J-M, et al. Active tuberculosis, sequelae and COVID-19 co-infection: first cohort of 49 cases. Eur Respir J. 2020;56(1):1.
14. Pemán J, Ruiz-Gaitán A, García-Vidal C, et al. Fungal co-infection in COVID-19 patients: should we be concerned? Rev Iberoam Micol. 2020;37(2):41–46. doi:10.1016/j.riam.2020.07.001
15. Heaney AK, Head JR, Broen K, et al. Coccidioidomycosis and COVID-19 co-infection, United States, 2020. Emerg Infect Dis. 2021;27(5):1266. doi:10.3201/eid2705.204661
16. Arashiro T, Nakamura S, Asami T, et al. Furukawa K: SARS-CoV-2 and Legionella co-infection in a person returning from a Nile cruise. J Travel Med. 2020;27(3):taaa053. doi:10.1093/jtm/taaa053
17. Argemí G, Somoza M, Andrés M, Llunell A. SARS-CoV-2 and Legionella pneumophila coinfection. Enferm Infecc Microbiol Clin. 2021;2021:1.
18. Subedi Y, Haas CJ. Legionella coinfection in a patient with COVID-19 pneumonia. Cureus. 2021;13(8):6.
19. Shimizu M, Chihara Y, Satake S, et al. Co-infection with Legionella and SARS-CoV-2: a case report. JA Clin Rep. 2021;7(1):62. doi:10.1186/s40981-021-00467-3
20. Elikowski W, Fertala N, Zawodna-Marszalek M, et al. Concomitance of COVID-19 and legionnaires’ disease - A case series. Pol Merkur Lekarski. 2022;50(295):30–36.
21. Cozzi D, Cavigli E, Moroni C, et al. Ground-glass opacity (GGO): a review of the differential diagnosis in the era of COVID-19. Jpn J Radiol. 2021;39(8):721–732. doi:10.1007/s11604-021-01120-w
22. Chalker VJ, Adler H, Ball R, et al. Fatal co-infections with SARS-CoV-2 and Legionella pneumophila, England. Emerg Infect Dis. 2021;27(11):2950. doi:10.3201/eid2711.204121
23. Alhuofie ST. An elderly COVID-19 patient with community-acquired Legionella and mycoplasma coinfections: a rare case report. In: Healthcare. MDPI; 2021:1598.
24. Hughes S, Troise O, Donaldson H, Mughal N, Moore L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26(10):1395–1399. doi:10.1016/j.cmi.2020.06.025
25. Rothe K, Feihl S, Schneider J, et al. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis. 2021;40(4):859–869. doi:10.1007/s10096-020-04063-8
26. Rothe K, Spinner CD, Panning M, et al. Evaluation of a multiplex PCR screening approach to identify community-acquired bacterial co-infections in COVID-19: a multicenter prospective cohort study of the German competence network of community-acquired pneumonia (CAPNETZ). Infection. 2021;49(6):1299–1306. doi:10.1007/s15010-021-01720-8
27. Xie F, Duan Z, Zeng W, et al. Clinical metagenomics assessments improve diagnosis and outcomes in community-acquired pneumonia. BMC Infect Dis. 2021;21(1):1–9. doi:10.1186/s12879-021-06039-1
28. Guo W, Cui X, Wang Q, et al. Clinical evaluation of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid collected from children with community-acquired pneumonia. Front Med. 2022;9:1. doi:10.3389/fmed.2022.952636
29. Zhang J, Gao L, Zhu C, et al. Clinical value of metagenomic next-generation sequencing by Illumina and Nanopore for the detection of pathogens in bronchoalveolar lavage fluid in suspected community-acquired pneumonia patients. Front Cell Infect Microbiol. 2022;2022:1449.
30. Borthong J, Omori R, Sugimoto C, Suthienkul O, Nakao R, Ito K. Comparison of database search methods for the detection of Legionella pneumophila in water samples using metagenomic analysis. Front Microbiol. 2018;9:1272. doi:10.3389/fmicb.2018.01272
31. Wang Y, Dai Y, Lu H, et al. Case report: metagenomic next-generation sequencing in diagnosis of Legionella pneumophila pneumonia in a patient after umbilical cord blood stem cell transplantation. Front Med. 2021;8:643473. doi:10.3389/fmed.2021.643473
32. Yue R, Wu X, Li T, Chang L, Huang X, Pan L. Early detection of Legionella pneumophila and Aspergillus by mNGS in a critically ill patient with Legionella Pneumonia after extracorporeal membrane oxygenation treatment: case report and literature review. Front Med. 2021;8:686512. doi:10.3389/fmed.2021.686512
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.