Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
Detection of Novel Pathogenic Variants in Two Families with Recurrent Fetal Congenital Heart Defects
Authors Cai R , Tan Y, Wang M, Yu H, Wang J, Ren Z, Dong Z, He Y, Li Z, Lin L, Gu Y
Received 27 October 2022
Accepted for publication 17 February 2023
Published 8 March 2023 Volume 2023:16 Pages 173—181
DOI https://doi.org/10.2147/PGPM.S394120
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Rongqin Cai,1,* Ya Tan,1,* Mingming Wang,2 Huijun Yu,1 Jing Wang,1 Zhuo Ren,1 Zhe Dong,1 Yiwen He,1 Zhi Li,1 Li Lin,1 Ying Gu1,3
1Department of Obstetrics and Gynecology, Peking University International Hospital, Beijing, 102206, People’s Republic of China; 2Be Creative Lab (Beijing) Co. Ltd, Beijing, 101111, People’s Republic of China; 3Lianyungang Maternal and Child Health Hospital, Lianyungang, Jiangsu, 222000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Li Lin; Ying Gu, Department of Obstetrics and Gynecology, Peking University International Hospital, Beijing, People’s Republic of China, Tel +86-13683047582 ; +86-18705130302, Email [email protected]; [email protected]
Background: Congenital heart disease (CHD) is the most common birth defect with strong genetic heterogeneity. To date, about 400 genes have been linked to CHD, including cell signaling molecules, transcription factors, and structural proteins that are important for heart development. Genetic analysis of CHD cases is crucial for clinical management and etiological analysis.
Methods: Whole-exome sequencing (WES) was performed to identify the genetic variants in two independent CHD cases with DNA samples from fetuses and their parents, followed by the exclusion of aneuploidy and large copy number variations (CNVs). The WES results were verified by Sanger sequencing.
Results: In family A, a compound heterozygous variation in PLD1 gene consisting of c.1132dupA (p.I378fs) and c.1171C>T (p.R391C) was identified in the fetus. The two variants were inherited from the father (c.1132dupA) and the mother (c.1171C>T), respectively. In family B, a hemizygous variant ZIC3: c.861delG (p.G289Afs*119) was identified in the fetus, which was inherited from the heterozygous mother. We further confirmed that these variants PLD1: c.1132dupA and ZIC3: c.861delG were novel.
Conclusion: The findings in our study identified novel variants to the mutation spectrum of CHD and provided reliable evidence for the recurrent risk and reproductive care options to the affected families. Our study also demonstrates that WES has considerable prospects of clinical application in prenatal diagnosis.
Keywords: congenital heart disease, whole-exome sequencing, PLD1, ZIC3, prenatal diagnosis
Introduction
Congenital heart defects (CHD) are abnormal anatomical structures caused by abnormal formation of the heart and/or great vessels during embryonic development or failure to close the channels that should be closed postnatally.1 CHD is the most common birth defect, affecting approximately 1% of all live births2 and 10% stillbirths.3 Although both genetic and environmental contributions have been recognized, the underlying pathogenesis of CHD remains poorly understood.4
Risk factors that have been implicated in CHD include maternal conditions (maternal age older than 35 years, diabetes mellitus, collagen vascular disease, exposure to a teratogen, pregnancy conceived with in vitro fertilization and so on), genetic factors and other environmental factors. Prenatal diagnosis of CHD can refer the pregnant woman to a tertiary care facility capable of providing neonatal diagnosis and management. In addition, appropriate genetic testing and counseling provide essential information for parents including prognosis.5,6
The prenatal diagnosis rate for CHD is low, which ranges between 30% and 60%.7–9 Thanks to the advances in genomics and sequencing technology, some genetic etiological aspects of CHD have been discovered. The earliest identified genetic causes of CHD were aneuploidies, affecting 9–18% CHD cases.10 Copy number variation (CNV) is also the genetic basis for some clinical syndromes presenting with CHD4 and contributes to 10% −15% of CHD.11 So far, about 400 genes were identified as being associated with CHD pathogenesis, but only a minority of them were confirmed with the Mendelian inheritance pattern.4,12
Phospholipase D1, is a signal transduction enzyme that hydrolyses the membrane lipid phosphatidic acid,13 which is encoded by the PLD1 gene (MIM *602382). In 2017, Ta-shma et al were the first to find that homozygous or compound heterozygous variations in the PLD1 gene were associated with the cardiac valvular defect, a developmental (CVDD, MIM #212093) disorder.14 According to Human Gene Mutation Database (HGMD), 35 PLD1 gene variations have been described so far, most of which are missense variants. The zinc-finger in cerebellum 3 (ZIC3, MIM *300265), is a member of the GL1 transcription factor superfamily. It contains five highly conserved C2H2-type zinc finger domains required for DNA-binding and the subsequent transcriptional activation.15–18 Mutation in ZIC3 gene results in the X-linked recessive genetic disease heterotaxy-1 (HTX1, MIM #306955) or the isolated congenital heart defects-1 (CHTD1, MIM #306955) or VACTERL syndromes (MIM #314390) conditions.
In this study, two families with recurrent fetal congenital heart defects were enrolled and submitted to a clinical and genetic analysis. A sequential detection with conventional karyotyping, chromosomal microarray assay (CMA) and whole-exome sequencing (WES) was performed to detect the causative variations. Sanger sequencing was used as a validation method for suspected variants. A compound heterozygous variation in PLD1 gene and a hemizygous variant in ZIC3 gene were identified in two families, respectively. The conservatism of amino acid (AA) residues affected by missense variants was also analyzed to support the corresponding pathogenicity.
Materials and Methods
This study was approved by the ethics committee of Lianyungang Maternal and Child Health Hospital, and written informed consent was obtained from the parents. All procedures performed in the present study were in accordance with the Declaration of Helsinki 1964 and its later amendments or comparable ethical standards.
Subjects
Two families with recurrent prenatal CHD cases were recruited at our outpatient department. In family A, the proband was the third fetus of a healthy non-consanguineous couple and similar congenital heart defects occurred in all 3 pregnancies. Ultrasonographic examination for the first fetus suggested hypoplastic right heart syndrome. The second child was a male who was born at term. Fifteen days after birth, he was diagnosed with pulmonary valve atresia, dysplastic tricuspid valve, patent ductus arteriosus, patent foramen ovale, and minor atrial septal defect (ASD). He eventually died at 100 days of age. At the 21st gestational week of the third pregnancy, the fetus was presented with pulmonary valve atresia, dysplastic tricuspid valve, atrial septal defect, and reversed flow of ductus arteriosus.
In family B, the proband was the second fetus of a healthy non-consanguineous couple. The first fetus was male and died in pregnancy due to multiple cardiac malformations including hypoplastic left heart, atrial septal defect and pulmonary vein ectopic. The second fetus in this study was also male, and ultrasound testing at 23 weeks suggested he had severe defects, including double outlet right ventricle, pulmonic stenosis, single atrium, and pericardial effusion.
Amniocentesis was conducted during the process of induced termination of these two pregnancies to obtain fetal specimens. Peripheral blood samples from both couples were also collected for genetic testing.
Chromosome Karyotyping
The amniotic fluid cells of the proband were cultured and harvested. G banding was performed on the chromosome after slide-making.19 We then refer to International System for Human Cytogenetic Nomenclature (ISCN) 2016 for karyotype analysis.20
DNA Extraction
According to the genomic DNA kit’s manufacturer’s protocol, parents’ DNA was extracted from the peripheral blood and fetal DNA was extracted from the 109 cells out of the amniotic fluid samples.
Chromosome Microarray Analysis
The chromosome microarray analysis was performed using Affymetrix CytoScan 750K platform (Affymetrix, Thermo Fisher, USA) according to the manufacturer’s instructions. The procedure included genomic DNA extraction, digestion, amplification, PCR product purification, quantification and fragmentation, labeling, hybridization, rinsing, staining and scanning. The Chromosome Analysis Suite software (Affymetrix) was used to analyze the raw data. The copy number variant (CNV) reporting threshold was set at 100kb with marker count ≥50.
Whole Exome Sequencing
The genomic DNA was mechanically sheared into fragments, and then the phosphate group was added to the 5’ end and Poly A tail was added to the 3’ end. A complete DNA fragment library was constructed after a round of PCR amplification. These DNA fragments were hybridized with the capture chip to obtain the exonic fragments, which were further amplified by PCR and subjected to next-generation sequencing (NGS). Sequenced reads were aligned to UCSC hg19/GRCh37 human reference genome. Base quality score recalibration, single-nucleotide variants’ (SNVs) detection, insertions/deletions (indels) were performed the using Genome Analysis Toolkit (GATK) Haplotype Caller. Variant calling was performed with the Verita Trekker® Variants Detection system (v2.0; Berry Genomics, Beijing, China) and Genome Analysis Tool Kit (https://software.broadinstitute.org/gatk/). Then, variants were annotated and interpreted using ANNOVAR (v2.0) and Enliven® Variants Annotation Interpretation systems (Berry Genomics), based on the common guidelines by ACMG (American College of Medical Genetics and Genomics). To assist in the interpretation of variant pathogenicity, we referred to 3 frequency databases (ExAC_EAS, gnomAD_exome_EAS, 1000G_2015aug_eas) and HGMD (Human Gene Mutation Database) pro V2021.10; Revel score (a combined method of pathogenicity prediction) and pLI score (representing the tolerance for truncating variants) were also employed. Potential pathogenic variants were screened out according to the quality score, frequencies, variants’ position, etc. We focused on genes linked to heart disease, using the databases (OMIM, Clinvar, PubMed) to find the corresponding diseases, clinical symptoms and functions of the candidate variants. Also, we determined where the variants come from and ascertained its pattern of inheritance. Finally, sanger sequencing was performed on specific variants as a validation method with the 3730 DX Genetic Analyzer.
Results
Clinical Data Analysis
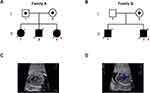
In family A, the non-consanguineous couple had three pregnancies and the third fetus was the proband (Figure 1A). Common clinical symptoms in all three fetuses were dysplastic tricuspid valve, pulmonary valve atresia and atrial septal defect. In family B, congenital heart defects occurred in two pregnancies of the non-consanguineous couple (Figure 1B). The major clinical manifestations and information of these pregnancies are included in Figure 1C and D and Table 1.
 |
Table 1 Summary of Clinical Manifestations and Information of Two Recruited Families |
Genetic Findings
The karyotypes (Figure 2A and B) and CMA results (Figure 2C and D) of the two probands were normal. To further investigate the cause of CHD, whole-exome sequencing (WES) was performed to the probands and their parents. In family A, a compound heterozygous variation in PLD1 gene comprising the c.1132dupA (p.I378fs) and c.1171C>T (p.R391C) variants were identified, which were inherited from the father (c.1132dupA) and mother (c.1171C>T), respectively. Additionally, the second child of this couple carried the same variation according to the Sanger sequencing result (Figure E). In family B, an X-linked hemizygous variant, ZIC3: c.861delG (p.G289Afs*119), was identified in the fetus, which was inherited from the mother (Figure 2F). All the variants were confirmed by Sanger sequencing.
Bioinformatic Analysis
In family A, the novel variant PLD1: c.1132dupA (p.I378fs) is a frameshift variant, and the population frequency is none in the 1000 Genomes Project, Genome Aggregation Database (gnomAD) and Exome Aggregation Consortium (ExAC) databases. The result of NCBI blasting showed that the PLD1: I378 amino acid residue was conserved among multiple species (Figure 3A). This novel variant was recognized as “Pathogenic” (PVS1+PM2+PP3+PP4) according to the American College of Medical Genetics and Genomics (ACMG) criteria.21 Another variant PLD1: c.1171C>T (p.R391C) has been reported, and the population frequency is 3.3×10−5 (http://exac.broadinstitute.org/). Also, the NCBI blasting showed that the PLD1: R391 residue was conserved among species (Figure 3A). So, we interpreted c.1171C>T as “Likely pathogenic” (PS4+PM2+PP3+PP4). In family B, the novel variant ZIC3: c.861delG (p.G289Afs*119) is a frameshift variant which results in the protein translation terminated prematurely. The population frequency in both gnomAD and internal databases was none. The ZIC3: G289 residue was evolutionarily conserved among species (Figure 3B). We then interpreted c.861delG as “Pathogenic” (PVS1+PM2+PP3+PP4) according to the ACMG criteria.
Discussion
The clinical detection of CHD has increased over the past fifty years, partly owing to the development of fetal sonography examination and improvements in ultrasound technology.22 The underlying causes of CHD are complex, with ~56% of all cases not clear,4 10% cases related to environmental factors,23 and the rest may be associated with genetic factors including aneuploidy,10 CNV,11 or single nucleotide variants (SNV).24–26 The incidence of aneuploidy is 33–42% in CHD fetuses.27 Specifically, CHD is observed in 35–50% of liveborns with trisomy 21, 60–80% of liveborns with trisomy 13 and trisomy 18, and 33% with monosomy X.4 Several well-characterized CNVs associated with CHD include del22q11, del8p23,28 del7q1129,30 and del11q24-25.31,32 Recent analyses of larger cohorts of patients with CHD found several recurrent CNVs associated with CHD, including 1q21.1, 3p25.1, 16p13.11, 15q11.2 and 2p13.3.33 The explosion of technological approaches as next-generation sequencing has opened a door for understanding the genetic etiology of complex disease such as CHD. To date, mutations in over 400 genes have been reported to be associated with CHD.12
The mammalian PLD1 gene was cloned from Hela cell library in 1995.34 PLD1 encodes a 1074 amino acid protein that hydrolyses phosphatidylcholine to generate free choline and phosphatidic acid. In 2017, Ta-Shma et al14 for the first time identified biallelic deleterious mutations of PLD1 gene in patients with malformations of the pulmonic, tricuspid and mitral valves for the first time. The functional study showed that PLD1 protein was specifically expressed in chick embryo cardiac. In PLD1 knockout mice, the thickened pulmonic valve leaflets and impaired function of the tricuspid and pulmonic valves were observed.14 In 2021, a study reported 21 unrelated families from various ancestries with biallelic PLD1 variants who presented predominantly with congenital cardiac valve defects.35 The majority of the variants in the 21 cases were located at the HKD1 and HKD2 domain. In our study, the compound heterozygous variants in Family A, I378fs and R391C, were also located at the HKD1 domain. As expected, mutations within both HKD motifs abolished PLD activity.36 Additionally, the evolutionary conservatism of R391 residue supports the pathogenicity of this missense R391C variant. In terms of the previous two pregnancies of Family A, the fetuses showed dysplastic tricuspid valve, pulmonary valve atresia and atrial septal defect, which might also be caused by the PLD1 variation. Theoretically, the couple still has a 25% risk of future pregnancy involvement, so care should be taken to refer to counseling as well as interventional reproductive options, such as prenatal diagnosis and preimplantation diagnosis.
Mutations in ZIC3 can cause X-linked heterotaxy-1, the isolated congenital heart defects-1 and VACTERL syndromes. Heterotaxy is a developmental condition characterized by randomization of the placement of visceral organs, including the heart, lungs, liver, spleen, and stomach. The organs are oriented randomly with respect to the left-right axis and with respect to one another.37 VACTERL is an acronym for vertebral anomalies, anal atresia cardiac malformations, tracheoesophageal fistula, renal anomalies and limb anomalies.38 Additional ZIC3 variants were identified in patients with isolated congenital heart defects including transposition of the great arteries, atrial septal defect, ventricular septal defect and pulmonary valve stenosis.39–41 In family B, we identified a novel variant ZIC3: c.861delG (p.G289Afs*119) which resulted in the isolated congenital heart defects in the male fetus. In the first pregnancy, the male fetus showed the similar clinical manifestations, which may also be related to this pathogenic variant. So, the novel variant ZIC3: c.861delG (p.G289Afs*119) may result in isolated congenital heart defects-1 (CHTD-1). Since the mother in the family is a heterozygous carrier, future male fetuses still have a 50% chance of getting affected. Therefore, interventional reproductive methods such as those described above should also be recommended.
The main limitation of this study is that there was no experimental analysis to determine the function of specific variants, which limited our understanding of how these variants caused the abnormal protein function. However, the published work related to these two genes strongly supports the pathogenic role of these variants in CHD.
Conclusion
In summary, our study identified the causative variations in two families with recurrent fetal CHD. It is the first time to report cardiac valvular defects, developmental (CVDD) caused by PLD1 variation in Chinese population. These findings provided reliable evidence for the recurrent risk and reproductive options to these families and strengthened the adaptability of WES in prenatal diagnosis.
Data Sharing Statement
The data that support the findings of this study are available in the Figshare repository at https://doi.org/10.6084/m9.figshare.22046615.
Ethics Statement
The probands’ parents were consented for sample collection and subsequent analysis, and written informed consent was obtained from the probands’ parents for publication and accompanying images.
Acknowledgments
The authors are grateful to the patients and the family members who participated in this study.
Funding
This work was supported by the Project of Peking University International Hospital [grant number YN2020QN02].
Disclosure
The authors have declared no conflicts of interest in this work.
References
1. Triedman JK, Newburger JW. Trends in congenital heart disease: the next decade. Circulation. 2016;133(25):2716–2733. doi:10.1161/CIRCULATIONAHA.116.023544
2. Van der linde D, Konings EE, Slager MA, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–2247. doi:10.1016/j.jacc.2011.08.025
3. Song MS, Hu A, Dyamenahalli U, et al. Extracardiac lesions and chromosomal abnormalities associated with major fetal heart defects: comparison of intrauterine, postnatal and postmortem diagnoses. Ultrasound Obstet Gynecol. 2009;33(5):552–559. doi:10.1002/uog.6309
4. Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res. 2017;120(6):923–940. doi:10.1161/CIRCRESAHA.116.309140
5. Hopkins MK, Dugoff L, Kuller JA. Congenital heart disease: prenatal diagnosis and genetic associations. Obstet Gynecol Surv. 2019;74(8):497–503. doi:10.1097/OGX.0000000000000702
6. Meller CH, Grinenco S, Aiello H, et al. Congenital heart disease, prenatal diagnosis and management. Arch Argent Pediatr. 2020;118(2):e149–e161. doi:10.5546/aap.2020.eng.e149
7. Van Velzen CL, Clur SA, Rijlaarsdam ME, et al. Prenatal detection of congenital heart disease--results of a national screening programme. BJOG. 2016;123(3):400–407. doi:10.1111/1471-0528.13274
8. Ramaekers P, Mannaerts D, Jacquemyn Y. Re: prenatal detection of congenital heart disease--results of a national screening programme. BJOG. 2015;122(10):1420–1421. doi:10.1111/1471-0528.13416
9. Van Velzen CL, Ket JCF, Van de Ven PM, et al. Systematic review and meta-analysis of the performance of second-trimester screening for prenatal detection of congenital heart defects. Int J Gynaecol Obstet. 2018;140(2):137–145. doi:10.1002/ijgo.12373
10. Hartman RJ, Rasmussen SA, Botto LD, et al. The contribution of chromosomal abnormalities to congenital heart defects: a population-based study. Pediatr Cardiol. 2011;32(8):1147–1157. doi:10.1007/s00246-011-0034-5
11. Kim DS, Kim JH, Burt AA, et al. Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg. 2016;151(4):1147–1151 e4. doi:10.1016/j.jtcvs.2015.09.136
12. Williams K, Carson J, Lo C. Genetics of congenital heart disease. Biomolecules. 2019;9(12):897–901. doi:10.3390/biom9120879
13. Nelson RK, Frohman MA. Physiological and pathophysiological roles for phospholipase D. J Lipid Res. 2015;56(12):2229–2237. doi:10.1194/jlr.r059220
14. Ta-Shma A, Zhang K, Salimova E, et al. Congenital valvular defects associated with deleterious mutations in the PLD1 gene. J Med Genet. 2017;54(4):278–286. doi:10.1136/jmedgenet-2016-104259
15. Zhu L, Zhou G, Poole S, Belmont JW. Characterization of the interactions of human ZIC3 mutants with GLI3. Hum Mutat. 2008;29(1):99–105. doi:10.1002/humu.20606
16. Aruga J, Yokota N, Hashimoto M, et al. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J Neurochem. 1994;63:1880–1890. doi:10.1046/j.1471-4159.1994.63051880.x
17. Pavletich N, Pabo C. Crystal structure of a five-finger GLI-DNA complex new perspectives on zinc fingers. Science. 1993;261(24):1701–1707. doi:10.1126/science.8378770
18. Mizugishi K, Aruga J, Nakata K, et al. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem. 2001;276(3):2180–2188. doi:10.1074/jbc.m004430200
19. Arsham MS, Barch MJ, Lawce HJ. The AGT Cytogenetics Laboratory Manual.
20. McGowan-Jordan J, Simons A, Schmid M. An International System for Human Cytogenomic Nomenclature (2016). Basel(CH): Karger; 2016.
21. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi:10.1038/gim.2015.30
22. Friedberg MK, Silverman HN, Moon-Grady AJ, et al. Prenatal detection of congenital heart disease. J Pediatr. 2009;155(1):26–31, e1. doi:10.1016/s0084-3954(09)79601-0
23. Jenkins KJ, Correa A, Feinstein JA, et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):2995–3014. doi:10.1161/CIRCULATIONAHA.106.183216
24. Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi:10.1038/nature12141
25. Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi:10.1126/science.aac9396
26. Sifrim A, Hitz MP, Wilsdon A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48(9):1060–1065. doi:10.1038/ng.3627
27. Wimalasundera RC, Gardiner HM. Congenital heart disease and aneuploidy. Prenat Diagn. 2004;24(13):1116–1122. doi:10.1002/pd.1068
28. Pehlivan T, Pober BR, Brueckner M, et al. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83:201–206. doi:10.1002/(sici)1096-8628(19990319)83:3<201::aid-ajmg11>3.0.co;2-v
29. Nickerson E, Greenberg F, Keating MT, et al. Deletions of the elastin gene at 7q11.23 occur in approximately 90% of patients with Williams syndrome. Am J Med Genet. 1995;56:1156–1161.
30. Li DY, Toland AE, Boak BB, et al. Elastin point mutations cause an obstructive vascular disease, supravalvular aortic stenosis. Hum Mol Genet. 1997;6(7):1021–1028. doi:10.1093/hmg/6.7.1021
31. Ye M, Coldren C, Liang X, et al. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum Mol Genet. 2010;19(4):648–656. doi:10.1093/hmg/ddp532
32. Grossfeld PD, Mattina T, Lai Z, et al. The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet A. 2004;129A(1):51–61. doi:10.1002/ajmg.a.30090
33. Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41(8):931–935. doi:10.1038/ng.415
34. Hammond SM, Altshuller YM, Sung TC, et al. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270(50):29640–29643. doi:10.1074/jbc.270.50.29640
35. Lahrouchi N, Postma AV, Salazar CM, et al. Biallelic loss-of-function variants in PLD1 cause congenital right-sided cardiac valve defects and neonatal cardiomyopathy. J Clin Invest. 2021;131(5). doi:10.1172/JCI142148
36. Sung TC, Roper RL, Zhang Y, et al. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 1997;16(15):4519–4530. doi:10.1093/emboj/16.15.4519
37. Srivastava D. Left, right…which way to turn? Nat Genet. 1997;17:252–254. doi:10.1038/ng1197-252
38. Khoury MJ, Cordero JF, Greenberg F, et al. A population study of the VACTERL association: evidence for its etiologic heterogeneity. Pediatrics. 1983;71(5):815–820. doi:10.1016/s0022-5347(17)50302-x
39. Gebbia M, Ferrero GB, Pilia G, et al. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17:305–308. doi:10.1038/ng1197-305
40. Megarbane A, Salem N, Stephan E, et al. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur J Hum Genet. 2000;8704–8708. doi:10.1038/sj.ejhg.5200526
41. Ware SM, Peng J, Zhu L, et al. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am J Hum Genet. 2004;74(1):93–105. doi:10.1086/380998
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.