Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Detection of haplotype mutations of the MD-2 gene promoter associated with Der p2-induced allergy using a nanostructured biosensor
Authors Chin Y, Liao E, Wu C, Wang G , Tsai J
Received 13 December 2013
Accepted for publication 22 January 2014
Published 12 March 2014 Volume 2014:9(1) Pages 1403—1412
DOI https://doi.org/10.2147/IJN.S59151
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Yu-Ting Chin,1 En-Chih Liao,2 Chia-Che Wu,1 Gou-Jen Wang,1,3,4 Jaw-Ji Tsai2
1Department of Mechanical Engineering, National Chung-Hsing University, 2Department of Medical Education and Research, Taichung Veterans General Hospital, 3Graduate Institute of Biomedical Engineering, 4PhD Program in Tissue Engineering and Regenerative Medicine, National Chung-Hsing University, Taichung, Taiwan
Abstract: Group 2 allergens (Der p2) have been reported to be a major cause of the human immune response to dust mite allergens. In this study, we have demonstrated for the first time the effective differentiation between haplotype mutation and normal genes in the MD-2 gene promoter using a nanostructured biosensor. A 70-mer gene fragment containing the haplotype of two single nucleotide polymorphisms in the MD-2 gene promoter region was used as a probe to detect haplotype mutations associated with Der p2-induced allergy. Discrimination was achieved using electrochemical impedance spectroscopy. The discrimination experiments employed 30 haplotype mutation samples and 30 normal target samples. The haplotype mutation samples and normal target samples could be clearly discriminated, even using samples produced by a five-cycle polymerase chain reaction process. The time and cost of sample preparation for the polymerase chain reaction process in the clinical setting can thus be reduced.
Keywords: haplotype mutation detection, nanostructured biosensor, MD-2 gene promoter, electrochemical impedance spectroscopy
Introduction
Allergens produced by the house dust mite are a common cause of allergic symptoms, such as asthma and allergic rhinitis. The pathogenic principle is the inflammatory response due to adhesion of allergens from the air to the epithelium of the respiratory tract. It is estimated that as many as 1.2 billion people are chronically sensitive to the house dust mite.1 There are reportedly 21 mite allergens that are separated into groups. The group 2 allergens have been reported to be the major source of the human immune response to these mite allergens.2 Approximately 87.8% of all asthma patients and 80% of pediatric asthma patients have been reported to be allergic to Der p2.3,4 The group 2 allergens are proteins that tend to bind with the lymphocyte antigen 96 protein (also known as MD-2) associated with Toll-like receptor 4 on respiratory epithelial cells, leading to respiratory disease.2 The MD-2 protein in humans is encoded by the LY96 gene.5–7 It has been reported that mutations of the LY96 gene promoter on chromosome 8 reduce the human innate response.8 There are six commonly seen single nucleotide polymorphisms (SNPs) in the MD-2 gene promoter, including rs11465996, rs1809442, rs1809441, rs1809440, rs16938754, and rs7842342. Recently, our group has discovered that the −441 G/G and −442 C/T SNPs, separated by 29 nucleotides, represent a haplotype of the MD-2 gene promoter. A haplotype is a set of SNPs on a single chromosome of a chromosome pair that mutates concurrently.
Several analytical methods, including DNA sequencing,9 mass spectrometry,10 capillary electrophoresis,11 electrochemical analysis,12 electrochemical impedance spectroscopy (EIS),13 denaturing high-pressure liquid chromatography,14 gel electrophoresis,15 and hybridization analysis,16 have been reported to be effective for detection of known SNPs. However, aside from the widely used but time-consuming and labor-intensive polymerase chain reaction (PCR) combined with electrophoresis, the clinical detection of haplotype mutations has been discussed in very few reports.
A DNA-based piezoelectric biosensor has been developed for haplotype detection of Southeast Asian α-thalassemia using a three-primer system for PCR coupling.17 The haplotype was differentiated by measuring the natural frequency variations of the sample when different SNPs were immobilized. A gel electrophoresis-based haplotype identification method for concurrent sensing of two adjacent polymorphic sites, an SNP, and a long polymorphism within 1–2 kb has been reported by Ronai et al.18 Successful genotyping of the −521 C/T SNP and the 120 base pair duplication in the 5′-upstream region of the dopamine D4 receptor (DRD4) gene was demonstrated. Another haplotyping approach of integrating double-tube allele-specific PCR, restriction fragment length polymorphism, and capillary gel electrophoresis analysis has been described by the same group.19 In addition to haplotype investigation of the −616 C/G and −521 C/T SNPs of the DRD4 gene, coincidental genotyping of the variable number of 48 base pair tandem repeats of the third exon of the same gene was also demonstrated. Song et al used temperature analysis to identify the haplotype of four SNPs in the tumor necrosis factor alpha promoter region.20 A rapid and cost-effective screen using the LightCycler™ assay (Roche Diagnostics, Basel, Switzerland) that permits a one-step analysis of ten functional haplotype combinations of the basic RET promoter has been developed by Görgens et al.21
Label-free detection using EIS to enable rapid detection of rare molecules in an analyte has been applied in many biomedical sensing areas.22–25 Owing to their distinctive characteristics, including high surface activity, high catalytic efficiency, and strong adsorbability, nanomaterials have been widely used for biomedical detection. Among these, gold nanoparticles (GNP), which are generally biocompatible and can be stably immobilized with biomolecules to preserve their bioactivity, have been a very useful material for biosensor electrodes.26–29
In this study, we developed an EIS-based nanostructured biosensor for efficient detection of the haplotype of two SNPs in the MD-2 gene promoter. A barrier layer with a nanohemisphere array of anodic aluminum oxide (AAO) was used as the substrate for the biosensor. A gold thin film was sputtered onto the AAO barrier layer surface as the electrode for GNP deposition. A 70-mer gene fragment of the genome that contains the haplotype of two SNPs in the MD-2 gene promoter region was used as a probe for detection of haplotype mutations in allergy patients. Complementary targets, comprising 156-mer nucleotides, were prepared from the fresh blood of patients with allergic disease. EIS was implemented to discriminate matching and mismatching hybridizations of the capture probes and complementary targets.
Materials and methods
Sensor fabrication
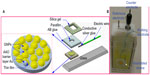
Figure 1 shows the structure of the EIS-based nanostructured biosensor used for detection of the haplotype of two SNPs in the MD-2 gene promoter. Figure 1A is a schematic illustration and Figure 1B shows the set-up of the EIS connection of the sensing device.
The AAO films were prepared using a well-known anodizing process. A 20×20 mm2 square of aluminum foil (99.9995%, 175 μm thick) was prepared and placed in a custom-designed Teflon fixture, as shown in Figure 2, to assure the consistency of the anodization process. AAO films, with a nanopore diameter of approximately 60 nm and a thickness of 50 μm, were obtained by anodizing the polished aluminum foil in a 0.3 M oxalic acid solution with a voltage of 90 V at 0°C±1°C applied for 2 hours. The remaining aluminum under the barrier layer was then removed using an aqueous CuCl2-HCl solution (CuCl2 powder 13.45 g, 35 wt% hydrochloric acid 100 mL). A honeycomb-like surface barrier layer with convex honeycomb structures (φ=80 nm) was obtained. The surface of the barrier layer was immersed in 30 wt% phosphoric acid for 40 minutes to further modify the shape of the honeycomb structures. A 10 nm thick gold electrode was sputtered onto the modified barrier layer surface using a radiofrequency magnetron sputter. An annealing process was then applied (target temperature 120°C, heating rate 5°C per minute, holding time 60 minutes, cooling to room temperature). To ensure that the sensing area of each biosensor was the same, a φ=6 mm hole was punched into the center of a 2.5×2.5 cm2 piece of Parafilm, the bottom surface of the square was coated with a thin layer of AB glue, and the square was bonded to the nanostructured electrode on the AAO barrier layer surface. Finally, a GNP layer was electrochemically deposited onto the nanostructured gold thin film electrode (applied voltage −0.7 V, temperature 25°C, processing time 180 seconds). Electrochemical deposition of the GNPs was performed using an SP-150 electrochemical analyzer (Bio-Logic, Knoxville, TN, USA). The electrolyte was produced by dissolving 1 mL of a 0.02 M HAuCl4 (Sigma-Aldrich, St Louis, MO, USA) solution in 39 mL of deionized water.
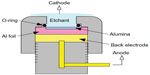  | Figure 2 Anodic aluminum oxide membrane fabrication setup. |
Preparation of capture probe and complementary target DNA
A 70-mer gene fragment containing the haplotype of two SNPs in the MD-2 gene promoter region was used to probe for haplotype mutations in allergy patients. The sequence of the capture probe, as shown in the first row of Table 1, comprises 70 nucleotides. The 20th (G) and 50th (C) nucleotides, indicated in blue, are the haplotype mutation locations. Synthetic probes for the single-stranded DNA (ssDNA) in the MD-2 gene promoter were purchased from Tri-I Biotech Inc. (Taipei, Taiwan). The complementary target DNAs, listed in the second row of Table 1, comprising 156 nucleotides, were prepared according to the genomic DNA mini kit protocol for fresh blood (Geneaid Biotech, Taipei, Taiwan). The 49th (G) and 79th (T) nucleotides, indicated in red, are the mutation locations, and should be (C) and (G), respectively, for a normal person. Hybridization between the capture probe and the complementary target is illustrated in the third row of Table 1. PCR reactions for various numbers of cycles (5, 10, and 20) were tested to determine the number of cycles required to produce large enough amounts of the complementary target to permit efficient detection of the haplotype mutations using the proposed nanostructured biosensor.
  | Table 1 Base sequences of the probe and target oligonucleotide sequences |
Immobilization of ssDNA probe and hybridization of complementary target
The sequential process for ssDNA probe immobilization included probe immobilization and blocking with 6-mercapto-1-hexanol. A probe ssDNA solution (10 μM) was diluted 100-fold with phosphate-buffered saline, followed by uniform shaking. A 30 μL aliquot of the diluted probe ssDNA solution was then deposited onto the nanostructured electrode surface of the biosensor and incubated for one hour. 6-mercapto-1-hexanol solution (2 mM) was added to the probe ssDNA-immobilized electrode to block the nonimmobilized area. The sample was then incubated for 30 minutes and rinsed three times with phosphate-buffered saline.
The hybridization process for the haplotype complementary target involved denaturation of the target DNAs and hybridization of the denatured target ssDNAs with the capture probes. The target double-stranded DNA (dsDNA) solution was denatured at 95°C for 10 minutes to yield ssDNA. The solution of denatured target DNA was cooled to 50°C, followed by uniform shaking. A 30 μL aliquot of the diluted target DNA solution was then deposited onto the biosensor and incubated at 50°C for 30 minutes, followed by rinsing with phosphate-buffered saline.
In the absence of the −441 G/G and −442 C/T haplotype mutations in the target DNA, all of the nucleotides in the specific DNA fragment match their corresponding nucleotides in the synthetic probe DNA fragment. However, if the haplotype mutations are present, two of the nucleotides in the selected DNA fragment will fail to bind to the corresponding nucleotides in the selected probe DNA fragment. The measured electrical properties of matching and mismatching samples will be different.
Electrochemical analysis
An SP-150 potentiostat (Bio-Logic) instrument was used to perform EIS to discriminate between allergy patients and normal subjects by haplotype mutation detection. The working electrode, counter electrode, and reference electrode were the electrodes of the nanostructured biosensor, Pt film, and Ag/AgCl, respectively. The buffer solution was a mixture of 5 mM Fe(CN)64−, 5 mM Fe(CN)63−, and 0.1 M KCl in 100 mM 2-(N-morpholino) ethanesulfonic acid (pH 6.0). The applied DC power and AC power were 0 mV and 10 mV, respectively. The scanning AC frequency was between 0.01 Hz and 100 kHz.
Results and discussion
Sensor fabrication results
A scanning electron microscopic image of the hemisphere array surface electrochemically deposited with uniformly distributed GNPs is shown in Figure 3. The diameter of the gold thin film-coated hemispheres of the AAO barrier layer was estimated to be about 100 nm, while the diameter of the deposited GNPs was less than 10 nm. To fulfill an effective PCR free detection of hepatitis B virus (HBV) DNA, the biosensor requires a relatively high sensitivity.
  | Figure 3 Scanning electron microscopic image of the GNP-deposited nanostructured electrode. |
GNPs were estimated to be 10–15 nm. Since there is only a two base pair difference between matching and mismatching hybrids, the difference in measured electric signal between them is slight and its detection requires a highly sensitive biosensor. The nanostructured electrode as shown in Figure 3 provides a sensor with an enlarged electrode area due to the uniformly distributed GNPs, thereby enhancing the sensitivity of the biosensor.
A cyclic voltammogram trace can be used to estimate the effective area of a sensing device for adhesion of the analytes. Figure 4A shows the cyclic voltammograms for the gold thin film-coated nanostructured electrode (AAO/Au thin film) and the GNP-deposited (AAO/Au thin film/GNP) electrode. Figure 4B shows the corresponding current versus time curves for Figure 4A. By integrating the area under the reducing peak in Figure 4B, the total electric charge of the AAO/Au thin film electrode and the AAO/Au thin film/GNP electrode are 154.94 μC and 514.91 μC, respectively. Since a charge of 386 μC per centimeter of gold electrode is required to form AuO, the effective area of the AAO/Au thin film electrode and the AAO/Au thin film/GNP electrode were calculated to be 0.401 cm2 and 1.334 cm2, respectively. Because the flat area of the sensing device is 0.283 cm2 (φ=0.6 cm), the uniformly distributed GNP on the nanohemisphere array enhance the sensing area by about 4.71-fold. Although there are a number of relatively simple ways to obtain a surface enlargement of 4.7-fold, the distinguishing feature of our sensor scheme is that both the hemispheres and the electrochemically deposited GNP are uniformly distributed, allowing the ssDNA probes to be neatly arranged onto the sensor surface. Hence the sensing sensitivity can be significantly enhanced.
Capture probe immobilization
For efficient detection, it is desirable that the electrode area be completely covered by the probe ssDNA. EIS was used to determine a suitable amount of probe ssDNA for complete coverage of the electrode. The original 10 μM probe ssDNA solution was diluted 10-fold, 100-fold, and 1,000-fold, and the optimal concentration of probe ssDNA solution was determined in terms of the charge transfer resistance measured by EIS.
In EIS, an equivalent circuit model that faithfully models the electric properties of the sensing device is required. Figure 5A depicts the fitting result of the capture probe binding using the Randles circuit (shown in the inset), that models total impedance by an active electrolyte resistance Rs in series with the parallel combination of an active charge transfer resistance Rct and a double-layer capacitance Cdl. It can be observed that the Randles circuit is not precise enough to model the nanostructured biosensor used in our study. Figure 5B depicts the fitting result using a more suitable equivalent circuit (shown in the inset). In this circuit model, Rs is the solution resistance, Cdl is the double-layer capacitance, Rct is the charge transfer resistance, Zw models the constant phase element, W is the Warburg element, and Rn represents the nanostructure resistance. A good fit is revealed by the small value of the chi-squared test.
  | Figure 5 Equivalent circuit model for the nanostructured biosensor. (A) Randles circuit and (B) the proposed circuit model. |
To minimize the influence of structural variations between the different sensor electrodes, the Rctp-ratio as shown in Equation (1) was used as an indicator for evaluation of the optimal concentration for probe ssDNA on the nanostructured electrode, where Rctp denotes the charge transfer of the probe ssDNA-immobilized electrode and Rctb is the charge transfer resistance of the original bare nanostructured electrode.
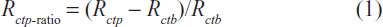
Figure 6 shows various Rctp-ratio values with respect to their corresponding probe ssDNA concentrations. It is observed that the Rctp-ratio value increases with the concentration of the probe ssDNA solution. The Rctp-ratio for the 10-fold and 100-fold diluted probe ssDNA solutions is similar, indicating that the probe ssDNA solution diluted100-fold was sufficient to cover the surface of the nanostructured electrode.
  | Figure 6 Optimization of capture probe immobilization. |
Complementary target hybridization
PCR is a necessary and effective technique for magnification of complementary target DNA extracted from the blood of a patient. In general, a PCR process with at least 30 cycles is required to produce a sufficient amount of complementary target DNA for valid haplotype detection.17–21 With the aim of investigating the detection limitation of our nanostructured biosensor so that the time-consuming PCR process could be reduced, various complementary target solutions prepared with different numbers of PCR cycles were used for hybridization experiments. When the hybridization reached a stable condition, hybridized DNA pairs were detached by sonication. A 30-cycle PCR process was applied to a solution containing the primers for the complementary target and the sonicated DNA to further magnify the complementary targets. Finally, the complementary targets were identified by gel electrophoresis. Figure 7 shows the results of gel electrophoresis for three samples of the complementary target. The gel electrophoresis results indicate that hybridization of complementary targets produced by a five-cycle PCR process is feasible.
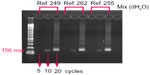  | Figure 7 Gel electrophoresis confirmation of complementary target hybridization. |
Elemental analysis of ssDNA probe immobilization and complementary target hybridization
X-ray photoelectron spectra for the GNP-deposited, ssDNA probe-immobilized, and complementary target-hybridized electrode surfaces are shown in Figure 8. X-ray photoelectron spectral peaks for C, O, P, and N with the atomic orbitals of C1s, O1s, P2p, and N1s can be observed. These results indicate covalent immobilization of phosphorus. The double peaks for C1s show that the binding structures of carbon are C=C and C−H. Successful hybridization of the complementary target was confirmed by the intensity and shift in binding energy.30
Discrimination between haplotype mutation and normal target samples
With approval from the institutional review board of Taichung Veterans General Hospital, Taichung, Taiwan, 30 haplotype mutation target samples and 30 normal target samples obtained clinically, with their haplotype mutation locations having already been indicated by reverse transcriptase PCR, were used for our differentiation experiments. Each sample was subjected to five-cycle, ten-cycle, and 20-cycle PCR processes. The haplotype mutation locations for the gene-mutated samples were verified by gel electrophoresis. Each experiment was repeated six times. The scanning time for each EIS was measured to be 77.4±2.4 seconds.
Table 2 tabulates the average value of the fitted parameters for the equivalent circuit model shown in Figure 5B corresponding to the discrimination experimentations, in which ΔRct denotes the charge transfer difference between the complementary target-immobilized electrode (Rctt) and the charge transfer resistance of the capture probe-immobilized electrode (Rctp). It can be observed that ΔRct is more substantial when compared with the change in the other parameters. Hence the ΔRct can be a suitable indicator for normal and haplotype mutation discrimination. To eliminate the effect of structural variations between different sensor electrodes, the Rctt-ratio shown in Equation (2) was used as the indicator for discrimination between haplotype mutation samples and normal target samples.
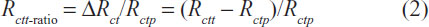
  | Table 2 Fitting parameters for discrimination experiments with (A) matched samples and (B) nonmatched samples |
Figure 9 shows the results of the discrimination experiments for the complementary targets resulting from five-cycle, ten-cycle, and 20-cycle PCR processes, in which the vertical axis and the horizontal axis represent the Rctt-ratio and the PCR cycles for the target DNA samples, respectively. As mentioned above, a PCR process with at least 30 cycles is required to magnify a sufficient amount of target DNA for valid haplotype detection. Our experimental results indicate that haplotype mutation samples and normal target samples can be discriminated by the nanostructured biosensor, even for samples produced by a five-cycle PCR process. The Rctt-ratio increases with increasing number of PCR cycles performed for production of the complementary targets. The sensitivities of the haplotype mutation samples and the normal target samples were calculated to be 0.16 and 0.045 per PCR cycle, respectively. We infer that the uniformly distributed GNP on the hemispheric array increase the surface area and binding sites of the electrode so that an increased number of ssDNA probes can be immobilized onto the electrode, accordingly enhancing the effective hybridization between the capture probes and the complementary targets. Even though the difference in charge transfer between a haplotype mutation and normal DNA is only two base pairs, the haplotype mutation and the normal target DNA can be clearly discriminated using this nanostructured biosensor.
  | Figure 9 Discrimination between haplotype-mutated and normal target samples. |
To further illustrate the specificity of the assay, we used a 96-base pair fragment of HBV DNA (3,020–3,320 base pairs) as the complementary target. The complementary target ssDNAs were prepared by Chun-Ying Wu of the Taichung Veterans General Hospital, Taiwan, according to the genome sequences of HBV from the National Center for Biotechnology Information. The Rctt-ratio of HBV DNA (10,000 copies/mL) immobilization was measured to be 0.044±0.006. Compared with the Rctt-ratio value for normal samples of the MD-2 promoter under five PCR cycles (Rctt-ratio =0.217), it is much lower, implying that the single-stranded HBV DNA segment did not immobilize to the 70-mer probe. This result confirms the specificity of the assay.
Because the nanostructured biosensor can discriminate mutated from normal genes using target samples produced by a five-cycle PCR process, the time and cost of the sample preparation PCR process can be reduced. Furthermore, high linearity of the calibration curves for both haplotype mutation samples and normal samples can be observed. We propose that the sensing scheme presented in this study is feasible for clinical detection of haplotype mutations, such as that in the MD-2 gene promoter.
Conclusion
This study demonstrates effective discrimination between the haplotype mutation gene and the normal gene in the MD-2 gene promoter using a nanostructured biosensor. A synthetic 70-mer gene fragment containing the haplotype of two SNPs in the MD-2 gene promoter region was used as a probe for detection of the haplotype mutations associated with Der p2-induced allergy. Complementary 156-mer targets were prepared using fresh blood from patients with allergic disease. Discrimination between haplotype mutation samples and normal target samples was achieved by EIS. A total of 60 combined target samples from patients, including 30 haplotype mutation samples and 30 normal samples, were used for the discrimination experiments. Our results indicate that haplotype mutation samples and normal target samples can be discriminated by the nanostructured biosensor, even for samples produced by a five-cycle PCR process. The sensing scheme presented in this study may be feasible for clinical detection of haplotype mutations, such as that in the MD-2 gene promoter.
Acknowledgment
The authors are grateful to the Department of Health in Taiwan for its financial support of this research under grants DOH101-TD-N-111-007 and NSC-101-2212-E-005-002-MY3.
Disclosure
The authors report no conflicts of interest in this work.
References
Basagana X, Sunyer J, Kogevinas M, et al. Socioeconomic status and asthma prevalence in young adults – The European Community Respiratory Health Survey. Am J Epidemiol. 2004;160:178–188. | |
Osterlund C, Gronlund H, Polovic N, Sundstrom S, Gafvelin G, Bucht A. The non-proteolytic house dust mite allergen Der p 2 induce NF-kappa B and MAPK dependent activation of bronchial epithelial cells. Clin Exp Allergy. 2009;39:1199–1208. | |
Thomas WR, Smith WA, Hales BJ, Mills KL, O’Brien RM. Characterization and immunobiology of house dust mite allergens. Int Arch Allergy Immunol. 2002;129:1–18. | |
Tsai JJ, Shen HD, Chua KY. Purification of group 2 dermatophagoides pteronyssinus allergen and prevalence of its specific IgE in asthmatics. Int Arch Allergy Immunol. 2000;121:205–210. | |
Shimazu R, Akashi S, Ogata H, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. | |
Abreu MT, Vora P, Faure E, Thomas LS, Arnold ET, Arditi M. Decreased expression of toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J Immunol. 2001;167:1609–1616. | |
Re F, Strominger JL. Monomeric recombinant MD-2 binds Toll-like receptor 4 tightly and confers lipopolysaccharide responsiveness. J Biol Chem. 2002;277:23427–23432. | |
Miyake K. Innate recognition of lipopolysaccharide by toll-like receptor 4-MD-2. Trends Microbiol. 2004;12:186–192. | |
Altshuler D, Pollara VJ, Cowles CR, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. | |
Griffin TJ, Smith LM. Genetic identification by mass spectrometric analysis of single-nucleotide polymorphisms: ternary encoding of genotypes. Anal Chem. 2000;72:3298–3302. | |
Drabovich AP, Krylov SN. Identification of base pairs in single-nucleotide polymorphisms by MutS protein-mediated capillary electrophoresis. Anal Chem. 2006;78:2035–2038. | |
Huang Y, Zhu J, Li G, et al. Electrochemical detection of point mutation based on surface hybridization assay conjugated allele-specific polymerase chainreaction. Biosens Bioelectron. 2013;42:526–531. | |
Palchetti I, Berti F, Laschi S, Marrazza G, Mascini M. In: Malcovati P, Baschirotto A, d’Amico A, Natale C, editors. Sensors and Microsystems. Dordrecht, The Netherlands: Springer; 2010;54:181–184. | |
Meierhofer D, Mayr JA, Fink K, Schmeller N, Kofler B, Sperl W. Mitochondrial DNA mutations in renal cell carcinomas revealed no general impact on energy metabolism. BrJ Cancer. 2006;94:268–274. | |
Schmalzing D, Belenky A, Novotny MA, et al. Microchip electrophoresis: a method for high-speed SNP detection. Nucleic Acids Res. 2000;28:E43. | |
Hacia JG, Fan JB, Ryder O, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet. 1999;22:164–167. | |
Vattanaviboon P, Sangseekhiow K, Winichagoon P, Promptmas C. Detection and haplotype differentiation of Southeast Asian alpha-thalassemia using polymerase chain reaction and a piezoelectric biosensor immobilized with a single oligonucleotide probe. Transl Res. 2008;151:246–254. | |
Ronai Z, Guttman A, Nemoda Z, Gervai J, Sasvari-Szekely M. Direct haplotype detection of adjacent polymorphic sites in the regulatory region of the dopamine D4 receptor (DRD4) gene. Electrophoresis. 2002;23:1512–1516. | |
Szantai E, Szilagyi A, Guttman Ad, Sasvari-Szekely Ma, Ronai Z. Genotyping and haplotyping of the dopamine D4 receptor gene by capillary electrophoresis. J Chromatogr A. 2004;1053:241–245. | |
Song Y, Araki J, Zhang L, et al. Haplotyping of TNFα gene promoter using melting temperature analysis: detection of a novel −856(G/A) mutation. Tissue Antigens. 2005;66:284–290. | |
Görgens H, Fitze G, Roesner D, Schackert HK. One-step analysis of ten functional haplotype combinations of the basic RET promoter with a LightCycler assay. Clin Chem. 2004;50:1693–1695. | |
Wang Y, Li C, Li X, Li Y, Kraatz HB. Unlabeled hairpin-DNA probe for the detection of single-nucleotide mismatches by electrochemical impedance spectroscopy. Anal Chem. 2008;80:2255–2260. | |
Kafka J, Panke O, Abendroth B, Lisdat F. A label-free DNA sensor based on impedance spectroscopy. Electrochim Acta. 2008;53:7467–7474. | |
Huang J, Yang G, Meng W, Wu L, Zhu A, Jiao X. An electrochemical impedimetric immunosensor for label-free detection of Campylobacter jejuni in diarrhea patients’ stool based on O-carboxymethylchitosan surface modified Fe3O4 nanoparticles. Biosens Bioelectron. 2010;25:1204–1211. | |
Chen YS, Wu CC, Tsai JJ, Wang GJ. Electrochemical impedimetric biosensor based on a nanostructured polycarbonate substrate. Int J Nanomedicine. 2012;7:133–140. | |
Myroshnychenko V, Rodríguez-Fernández J, Pastoriza-Santos I, et al. Modelling the optical response of gold nanoparticles. Chem Soc Rev. 2008;37:1792–1805. | |
Ensafi AA, Taei M, Rahmani HR, Khayamian T. Sensitive DNA impedance biosensor for detection of cancer, chronic lymphocytic leukemia, based on gold nanoparticles/gold modified electrode. Electrochim Acta. 2011;56:8176–8183. | |
Holland JT, Lau C, Brozik S, Atanassov P, Banta S. Engineering of glucose oxidase for direct electron transfer via site-specific gold nanoparticle conjugation. J Am Chem Soc. 2011;133:19262–19265. | |
Zhao J, Bo B, Yin YM, Li GX. Gold nanoparticles-based biosensors for biomedical application. Nano Life. 2012;02:1230008. | |
Ye YK, Zhao JH, Yan F, Zhu YL, Ju HX. Electrochemical behavior and detection of hepatitis B virus DNA PCR production at gold electrode. Biosens Bioelectron. 2003;18:1501–1508. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.