Back to Journals » International Journal of Nanomedicine » Volume 18
Dental Pulp Stem Cell-Derived Exosomes Regulate Anti-Inflammatory and Osteogenesis in Periodontal Ligament Stem Cells and Promote the Repair of Experimental Periodontitis in Rats
Authors Qiao X, Tang J, Dou L, Yang S, Sun Y, Mao H, Yang D
Received 11 May 2023
Accepted for publication 9 August 2023
Published 17 August 2023 Volume 2023:18 Pages 4683—4703
DOI https://doi.org/10.2147/IJN.S420967
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Phong A Tran
Xin Qiao,1– 3,* Jie Tang,1– 3,* Lei Dou,1– 3 Shiyao Yang,1,3,4 Yuting Sun,1,3,4 Hongchen Mao,1– 3 Deqin Yang1– 3
1Department of Endodontics, Stomatological Hospital of Chongqing Medical University, Chongqing, 401147, People’s Republic of China; 2Stomatological Hospital of Chongqing Medical University, Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences, Chongqing, 401147, People’s Republic of China; 3Chongqing Municipal Key Laboratory of Oral Biomedical Engineering of Higher Education, Chongqing, 401147, People’s Republic of China; 4Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences, Chongqing, 401147, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Deqin Yang, Department of Endodontics, Stomatological Hospital of Chongqing Medical University, No. 426, Songshi North Road, Yubei District, Chongqing, People’s Republic of China, Tel +86-23-8886-0060, Email [email protected]
Purpose: Dental pulp stem cell-derived exosomes (DPSC-EXO), which have biological characteristics similar to those of metrocytes, have been found to be closely associated with tissue regeneration. Periodontitis is an immune inflammation and tissue destructive disease caused by plaque, resulting in alveolar bone loss and periodontal epithelial destruction. It is not clear whether DPSC-EXO can be used as an effective therapy for periodontal regeneration. The purpose of this study was not only to verify the effect of DPSC-EXO on reducing periodontitis and promoting periodontal tissue regeneration, but also to reveal the possible mechanism.
Methods: DPSC-EXO was isolated by ultracentrifugation. Then it characterized by transmission electron microscope (TEM), nanoparticle tracking analysis (NTA) and Western Blot. In vitro, periodontal ligament stem cells (PDLSCs) were treated with DPSC-EXO, the abilities of cell proliferation, migration and osteogenic potential were evaluated. Furthermore, we detected the expression of IL-1β, TNF-αand key proteins in the IL-6/JAK2/STAT3 signaling pathway after simulating the inflammatory environment by LPS. In addition, the effect of DPSC-EXO on the polarization phenotype of macrophages was detected. In vivo, the experimental periodontitis in rats was established and treated with DPSC-EXO or PBS. After 4 weeks, the maxillae were collected and detected by micro-CT and histological staining.
Results: DPSC-EXO promoted the proliferation, migration and osteogenesis of PDLSCs in vitro. DPSC-EXO also regulated inflammation by inhibiting the IL-6/JAK2/STAT3 signaling pathway during acute inflammatory stress. In addition, the results showed that DPSC-EXO could polarize macrophages from the M1 phenotype to the M2 phenotype. In vivo, we found that DPSC-EXO could effectively reduce alveolar bone loss and promote the healing of the periodontal epithelium in rats with experimental periodontitis.
Conclusion: DPSC-EXO plays an important role in inhibiting periodontitis and promoting tissue regeneration. This study provides a promising acellular therapy for periodontitis.
Keywords: exosome, anti-inflammatory, osteogenesis, periodontitis, macrophages, IL-6/JAK2/STAT3 signaling pathway
Introduction
As one of the main causes of tooth loss in adults, periodontitis is a destructive periodontal inflammatory disease caused by dental plaque, which affects approximately 90% of the world’s population.1 In clinical practice, current treatments for periodontitis are mechanical debridement techniques (removing subgingival biofilm and dental calculus) supplemented with antibiotics and other drugs.2,3 However, autogenous bone is not easy to form, and damaged periodontal tissue cannot be completely repaired when periodontitis causes severe bone resorption even after treatment.4,5 Reducing periodontitis and reconstructing defective periodontal bone tissue are still ongoing challenges.
In recent years, cell tissue engineering (CTE) has become a promising tissue repair strategy in which stem cells are one of the basic elements.6–8 Current studies have shown that the therapeutic effect of stem cells is mainly due to the release of paracrine factors.9–11 As one of the most important paracrine mediators, exosomes derived from stem cells play a therapeutic role through immune regulation.12 Exosomes, lipid membrane particles (30–150 nm) secreted by all types of cells, act as intercellular communicators by transferring cell proteins, nucleic acids, lipids, metabolites and so on,13 which can be secreted under both normal and pathological conditions.14 Exosomes are released into the extracellular matrix after fusion of the polyvesicular outer membrane and cell membrane,14,15 so the biological function of exosomes is determined by their source cells. They participate in almost all cell interactions, especially in information transmission, antigen presentation, immune and target cell regulation, tissue regeneration and so on.15 Therefore, the use of exosomes derived from MSCs in the treatment of periodontitis seems to have great prospects.
Some studies have shown that exosomes derived from bone marrow mesenchymal stem cells (BMSCs) and stem cells from human exfoliated deciduous teeth (SHEDs) can promote the repair of alveolar bone defects and periodontal tissue regeneration in rats.16–20 However, dental stem cells not only express bone marrow mesenchymal stem cell markers but also show multipotential regeneration (osteogenesis, cartilage, nerve) and immunomodulatory ability.21–23 Meanwhile, tissue-specific mesenchymal stem cells (MSCs) provide more possibilities for the success of tissue engineering.24,25 Among all dental stem cells, DPSCs and PDLSCs are the most easily obtained.26 Ma L found that DPSCs and PDLSCs had similar abilities in proliferation and osteogenesis, but PDLSCs were more likely to differentiate after repeated passage in vitro.27 In general, DPSCs had a higher proliferation rate, lower cell senescence rate and stronger osteogenic maintenance and even improved periodontal regeneration after injection in a miniature pig periodontitis model.27 Other studies confirmed that DPSC-EXO combined with chitosan hydrogel could reduce periodontal inflammation and regulate the immune response.28 Except for this, previous studies on DPSC-EXO have paid more attention to its role in promoting dental pulp regeneration, especially angiogenesis.29,30 We noticed that DPSC-EXO has great potential in the treatment of periodontitis. In addition to the mechanism of regulating inflammation, the effects of DPSC-EXO on the reconstruction of periodontal bone defects are still unclear and need to be further verified.
Generally, DPSCs are prospective candidate stem cells with abundant sources to achieve periodontal regeneration. We suspect that its exosomes have a similar function in promoting tissue regeneration and regulating immunity. To verify whether DPSC-EXO can be used as a cell replacement therapy for periodontal regeneration, this study will explore the effect of DPSC-EXO on reducing periodontitis and promoting periodontal tissue regeneration and further explore its possible mechanism of action to provide a reference for the treatment of periodontitis.
Materials and Methods
Cell Culture and Identification
DPSCs and PDLSCs were obtained from exfoliated third molars of healthy donors in the Surgical Outpatient Department of Stomatological Hospital affiliated to Chongqing Medical University. We also obtained the informed consent of the donor for this study. This study was approved by the Ethics Committee of the affiliated Stomatological Hospital of Chongqing Medical University (CQHS-REC-2020 (LSNo.68)). Pulp tissue and periodontal tissue were collected from freshly extracted teeth and subjected to enzymatic digestion by collagenase type I(3 mg/mL; Sigma‒Aldrich, St. Louis, USA) at 37 °C for 40 minutes. The tissue suspension was inoculated in a 25 cm2 culture flask and cultured with α-minimum essential medium (α-MEM, HyClone, Logan, Utah, USA) containing 10% fetal bovine serum (FBS, Lonsera, Suzhou, China) and 100 U/mL penicillin‒streptomycin (HyClone, Logan, Utah, USA) to maintain growth at 37 °C and 5% CO2. Primary cells were induced to undergo osteogenic and adipogenic differentiation at the third or fourth passage. Alizarin red and oil red staining were performed on the 21st day of induction, and their multidirectional differentiation ability was observed by inverted phase contrast microscopy (Primo vert monitor, Carl Zeiss, Oberkochen, Germany). The cell surface antibody markers were identified by sorted flow cytometry (Influx, BD, New Jersey, USA). DPSCs were incubated with CD73-APC, CD90-FITC, CD31-FITC, CD45-PE, and Nestin-PE (1:100, Invitrogen, California, USA) antibodies. PDLSCs were incubated with CD90-FITC, CD31-FITC, CD29-FITC, CD45-PE, and CD105-APC (1:100, Invitrogen, California, USA) antibodies.
RAW264.7 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (SCSP-5036, Shanghai, China), cultured in Dulbecco’s modified Eagle medium (DMEM, HyClone, Logan, Utah, USA) supplemented with 10% fetal bovine serum (FBS, Lonsera, Suzhou, China) and 100 U/mL penicillin‒streptomycin (HyClone, Logan, Utah, USA) and maintained in 5% CO2 at 37 °C.
Exosomes Isolation and Identification
When the DPSCs reached 60% or 80% confluence, serum-free medium (α-minimum essential medium (α-MEM, HyClone, Logan, Utah, USA), 100 U/mL penicillin‒streptomycin (HyClone, Logan, Utah, USA)) was used instead of conventional medium and cultured for 48 hours. Then, the conditioned medium was collected and centrifuged at 4 °C: centrifuged at 300×g for 10 minutes, 2000×g for 10 minutes and 10,000 × g for 30 minutes by freezing centrifugation (Beckman Microfuge22R, Beckman Coulter, California, USA) to remove dead cells and debris; centrifuged at 100,000 × g for 70 minutes by ultracentrifugation (Optima™ XPN, Beckman Coulter, California, USA); the supernatant was discarded, and the sediment at the bottom of the tube was retained and resuspended in PBS. The previous step was repeated to obtain the exosome suspension. The total concentration of exosomes was determined by a bicinchoninic acid (BCA) kit (Beyotime, Shanghai, China). DPSC-EXO was stored at −80 °C for follow-up experiments.
The morphology of DPSC-EXO was identified by transmission electron microscopy (H-800, Hitachi, Tokyo, Japan). The sizes of these exosomes were analyzed by nanoparticle tracking analysis (Flow NanoAnalyzer, Fuliu Biological, Xiamen, China). In addition, surface marker proteins were detected by Western blot using specific antibodies against CD9 (1:500, Abcam, Cambridge, UK) and CD63 (1:500, Abcam, Cambridge, UK).
Exosomes Labeling and Uptake
According to the manufacturer’s instructions, the exosomes were labeled with PKH26 (Sigma‒Aldrich, St. Louis, USA) and re-extracted by ultracentrifugation. The exosomes labeled with PKH26 in the above steps were added to PDLSCs and RAW264.7 cells for 2 hours. Cells were washed with PBS three times, Calcein AM (Beyotime, Shanghai, China, 1:10) was added to the cells, and the cells were cultured for 20 minutes. Cells were washed with PBS and fixed with 4% paraformaldehyde (PFA, Solarbio, Beijing, China) for 15 minutes. DAPI (Solarbio, Beijing, China) was added for 5 minutes, rinsed with PBS three times, and photographed under a fluorescence inverted microscope (EVOS, Thermo Fisher, Massachusetts, USA).
Cell Proliferation and Migration
Exosomes at different concentrations (0, 1, 5, 10 μg/mL) were used to evaluate cell proliferation by Cell Counting Kit-8 (CCK-8, Baoguang Biotechnology Co. Ltd., Chongqing, China). Specifically, PDLSCs (5000 cells per well) were inoculated on a 96-well plate and incubated for 1, 2 and 3 days. 10% (v/v) CCK-8 solution was added to the cell and treated in the dark for 1 hour. Then, a filter-type enzyme labeling instrument (Elx800, BioTEK, Vermont, USA) measures optical density (OD). The cell viability value was calculated according to the following formula: Cell survival rate = [(experimental hole - blank hole) / (control hole - blank hole)] × 100%.
A wound healing assay was used to evaluate the effect of exosomes on cell migration. In the wound healing assay, PDLSCs were cultured in a 6-well plate with a fusion degree of 90%, the original culture medium was discarded, and the cells were starved overnight in serum-free medium. Each well was scraped with a 200 μL pipette tip to create a linear region without cells. The cell fragments were removed by washing with PBS. After that, the cells were incubated in medium containing different concentrations of exosomes (0, 1, 5, 10 μg/mL). A digital camera (Olympus, Tokyo, Japan) connected to an inverted phase contrast microscope (Primo vert monitor, Carl Zeiss, Oberkochen, Germany) was used to take pictures at 0 and 12 hours. Then, the migration of cells to acellular areas was monitored. The area of cells that moved into the scratch area was calculated by ImageJ analysis software (NIH, Bethesda, USA) for quantitative analysis. The specific formula is: cell migration rate (wound healing rate) = (0 h scratch area - 12 h scratch area)/0 h scratch area. We repeated this process three times for each group of samples.
Osteogenic Differentiation
PDLSCs were cultured in a 6-well plate with growth medium. When the cells reached 80% confluence, the cells were cultured in osteogenic induction medium (α-MEM containing 10% FBS, 0.1 μM dexamethasone, 50 μM ascorbate-2-phosphate and 10 mM β-glycerophosphate; Sigma‒Aldrich, St. Louis, USA) and replaced every three days. After 7 days of induction, total RNA was extracted, and quantitative real time-polymerase chain reaction (qRT‒PCR) was performed to detect the expression of osteogenesis-related genes. The activity of alkaline phosphatase (ALP) was detected by an ALP kit (Jiancheng, Nanjing, China) and stained with a BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime, Shanghai, China). Then, the sections were scanned, and pictures were taken. After 21 days of induction, alizarin red S staining was used to evaluate cell mineralization in vitro (Solarbio, Beijing, China). We repeated this process three times for each group of samples.
Quantitative Real Time-Polymerase Chain Reaction (qRT‒PCR)
To analyze the expression of osteogenesis-related genes, PDLSCs were cultured in 6-well plates, and the total RNA of cells was extracted after 7 days of osteogenesis induction. To analyze the expression of genes related to inflammation and macrophage polarization, PDLSCs/RAW 264.7 cells were cultured in 6-well plates, lipopolysaccharide (LPS) was prestimulated for 24 hours, and the total RNA of cells was extracted after 24 hours of exocrine treatment. Total RNA was extracted using TRIzol reagent (Takara, Tokyo, Japan). After the cells were homogenized in 800 μL of TRIzol reagent per well, 160 μL of chloroform was added to the homogenate solution, mixed well, and then centrifuged to separate the solution into three layers, the top layer of which was a clear liquid containing RNA. The top liquid layer was removed and pipetted into a new tube, followed by precipitation of total RNA by isopropanol. The purity (A260/A280) and concentration of RNA were determined using a NanoDrop2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). According to the manufacturer’s instructions, cDNA was reverse transcribed using the PrimeScript TM RT kit (Takara, Tokyo, Japan). RT‒qPCR was carried out with a Bio-Rad Applied Biosystems ABI 7500 System (Bio-Rad Laboratories, Hercules, CA, USA) using the PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara, Tokyo, Japan). The reaction mixture, 20 μL for each well, contained 10 μL TB Green Premix Ex Taq II (Takara, Tokyo, Japan), 2.0 μL template cDNA, 1 μL 10 μM PCR Forward Primer, 1 μL 10 μM PCR Reverse Primer and 6 μL Sterile purified water. The recommended protocol was as follows: 95 °C for 30 seconds (s) (initial denaturation), followed by 40 cycles of 95 °C for 5 s (denaturation), 60 °C for 31s (primer annealing), 95 °C for 15s, 60 °C for 1 minute (min) and 95 °C for 15s (dissociation). The PCR primers were amplified (Table 1), with GAPDH as the internal control. All assays were performed in triplicate from at least three different experiments.
 |
Table 1 List of Specific Primers |
Western Blot
PDLSCs were cultured in 6-well plates, and 200 μL RIPA lysate (Beyotime, Shanghai, China) was added to each well to extract cell protein, which was split on ice for 30 min, 14,000 g was separated for 10 min, and the supernatant was collected in a new centrifuge tube. BCA kits (Beyotime, Shanghai, China) were used to detect the protein concentration. The protein was boiled with SDS‒PAGE Sample Loading Buffer (Bio-Rad, California, USA) for 10 min and stored at −20 °C. The protein sample (20 μg) was electrophoretically separated and transferred to a 0.2 μm PVDF membrane (Millipore, Massachusetts, USA). The PVDF membrane was blocked with 5% (w/v) skim milk for 2 hours. The primary antibody was then diluted to 1:1000 and incubated with the PVDF membrane overnight at 4 °C. Then, the membranes were washed in Tris-buffered saline containing 0.1% Tween 20, pH 7.5, and incubated with goat anti-rabbit secondary antibody (Thermo Fisher Scientific, Massachusetts, USA) at room temperature for 2 hours. Protein signals were detected with the Immobilon Western Chemiluminescent HRP substrate kit (Beyotime, Shanghai, China), and the signal density was determined with ChemiDoc Touch (ChemiDOCTMXRS+ imaging system, BIO-RAD, California, USA). The primary antibodies used in this study were as follows: CD9 (Abcam, Cambridge, UK), CD63 (Abcam, Cambridge, UK), IL-6 (Abcam, Cambridge, UK), IL-1β (Abcam, Cambridge, UK), TNF-α (Bioworld, Minnesota, USA), STAT3 (Affinity, San Francisco, USA), p-STAT3 (Affinity, San Francisco, USA), JAK2 (Affinity, San Francisco, USA), p-JAK2 (Affinity, San Francisco, USA), and GAPDH (Abcam, Cambridge, UK). ImageJ software was used to analyze the gray value of the strip.
Flow Cytometry
Fourth-generation PDLSCs were collected and washed twice with PBS containing 1% FBS. After incubation in the dark with anti-CD45, anti-CD31, anti-CD29, anti-CD90 and anti-CD105 antibodies (Invitrogen, California, USA) at 4 °C for 30 minutes. The cells were centrifuged and washed three times, and the suspension was analyzed by sorted flow cytometry (Influx, BD, New Jersey, USA). The detection of DPSC surface antibodies was the same as above, and the markers included anti-CD73, anti-CD90, anti-CD45, anti-CD31 and Nestin (Invitrogen, California, USA).
Macrophage polarization markers were detected as follows. After washing with PBS, centrifuged cells with anti-F4/80 and anti-CD86 or anti-CD206 (Invitrogen, California, USA) were incubated in the dark at 4 °C for 30 minutes. The cells were then washed once, suspended in PBS containing 1% FBS and analyzed by flow cytometry. Flow cytometry data were analyzed by FlowJo software v10.2 (BD, New Jersey, USA).
Animal Experiment
Animals used in this study were maintained in accordance with the Guideline for ethical review of animal welfare of laboratory animals published by the China National Standardization Management Committee (publication No. GB/T 35892–2018) and were approved by the Ethics Committee of the Stomatological Hospital Affiliated to Chongqing Medical University (CQHS-REC-2020 (LSNo.68)). 24 healthy male Sprague‒Dawley (SD) rats (8 weeks old) were obtained from the CQMU Animal Experimental Center (G * POWER software to calculate the sample size). In this study, all rats weighed approximately 200–250 g. Rats were fed standard food regularly without any disease. All animals were housed in the Experimental Animal Center of Chongqing Medical University Stomatology Hospital and maintained in a specific-pathogen-free (SPF) environment at 25 °C, 40% humidity, and a 12-h light/dark cycle. The animals had free access to food and water and were randomly divided in the follow-up experiment.
We established a rat model of periodontitis based on previous studies.31 Briefly, a rat was placed in a sealed container with a 4% (v/v) isoflurane flow until it was completely anesthetized. A stainless-steel wire with a diameter of 0.20 mm was fastened around the bilateral maxillary first molars of isoflurane-intoxicated rats. LPS from Porphyromonas gingivalis (10 μg/mL) was injected into the periodontal pocket every other day for 5 times. The ligature remained for 30 days to induce periodontitis. Then, 24 rats were randomly divided into 4 groups (n = 6): healthy group without any treatment; periodontitis group, only stainless-steel ligature wire was removed after periodontitis was induced; and in the PBS group, PBS was injected into the periodontal pocket every other day after periodontitis was induced; in the EXO group, stainless steel ligature wire was removed after periodontitis was induced, and EXO (50 μg/mL) was injected into the periodontal pocket every other day.32 Specifically, 1 mL syringes were used to inject PBS/EXO in the buccal mesial, buccal distal, palatal proximal, and palatal distal regions of the first molars, approximately 10 μL total per sample. After 30 days, the experimental rats were sacrificed. The maxillary alveolar bones were dissected and fixed with 4% paraformaldehyde.
Microcomputed Tomography (Micro-CT) Analysis
To evaluate bone regeneration, a micro-CT scanner (vivaCT 40, SCANCO Medical AG, Bassersdorf, Zurich, Switzerland) was used to scan the maxillary bones with the defects. Dissected rat maxillae were secured in foam plates and placed into the machine, and relevant parameters were set: 45 kVp, 177 mA, and 10 μm resolution to start the scan. Three-dimensional (3D) pictures were created and analyzed in accordance with the software instructions. Volumetric measurement was performed after the selection of a region of interest (ROI), which was defined as an 18×18 pixel2 area on the alveolar bone between the first and the second molar, and the height was 50 slices. The ratio of new bone volume to tissue volume (BV/TV) was calculated.33 The distance between the cementoenamel junction and the alveolar bone crest (ABC-CEJ) was measured and analyzed.
Histological Analysis
After micro-CT analysis, the maxillae were decalcified in 10% disodium ethylenediamine tetraacetate (EDTA), dehydrated by a gradient ethanol series, and embedded in paraffin. Then, the samples were cut into 5 μm thick slices by a pathological slicer (LEICABM2135, Leica, Weizler, Hessen, Germany) and stored at room temperature.
For histological analysis, the maxillary sections were stained with a hematoxylin and eosin staining kit (Solarbio, Beijing, China) and TRAP kit (Solarbio, Beijing, China) according to the manufacturer’s instructions to show periodontal tissue defect healing and bone formation. Specifically, for HE staining, paraffin sections were conventionally dewaxed in water, stained for 2 min with hematoxylin staining solution and rinsed in distilled water. Then, the cells were differentiated for 60s, rinsed in distilled water, placed in eosin stain for 1 min, dehydrated quickly, cleared by xylene and sealed with resinene. For details of the TRAP staining scheme, the trap fixative solution was fixed at 2–8 °C for 30–60 s, slightly dried, put into the trap incubation solution at 37 °C for 50 minutes, dyed with hematoxylin staining solution for 3 minutes, dehydrated quickly, cleared with xylene and sealed with resinene. The TRAP-positive (+) cells, as osteoclast cells (clusters of more than three nuclei were counted as one osteoclast), in the alveolar bone between the maxillary first molar and the second molar were photographed and counted. After staining, the sections were analyzed by a full slide scanning system (VS200, OLYMPUS, Tokyo, Japan). At least four samples were analyzed per group, five fields of view per sample (500 × 500μm2 square areas) were randomly picked between the first and second molars, and the group mean was calculated using the mean of the individual measurements.
Immunostaining was performed on paraffin sections from each group. Paraffin sections were deparaffinized in water, and antigen retrieval was performed (the sections were immersed in citrate buffer (pH 6.0), repairer 80 °C, 20 min; After cooling, PBS was washed 3 times for 5 min each); The sections were incubated in 3% H2O2 for 10 min at room temperature; PBS was washed three times for 5 min each to block endogenous peroxidase; Goat serum blocking solution was added dropwise for 20 min at room temperature; TNF-α primary antibodies (1:200, BOSTER, Wuhan, China), OCN (1:200, Servicebio, Wuhan, China) and iNOS (1:200, Servicebio, Wuhan, China) were added dropwise, respectively, overnight at 4 °C; PBS wash 3 times for 5 min each; Goat anti-rabbit secondary antibody (1:100, Servicebio, Wuhan, China) was added dropwise for 30 min at 37 °C; PBS wash 3 times for 5 min each; Make up fresh DAB solution (Solelybio, Beijing, China) add dropwise onto the tissue at room temperature, control time under microscope until positive was brownish yellow, distilled water washed sections; Hematoxylin counterstained nuclei and dehydrated and sealed with resinene. The sections were analyzed by a full slide scanning system (VS200, OLYMPUS, Tokyo, Japan). Based in the staining intensity, positive area occupancy (area %) was counted by ImageJ. At least three samples were analyzed per group, five fields of view per sample (100 × 100μm2 square areas) were randomly picked between the first and second molars, and the group mean was calculated using the mean of the individual measurements.
Statistical Analysis
Data are presented as the mean ± standard deviation of three independently repeated experiments. T-test was used to compare between the two groups. Comparisons among three or four groups were evaluated by one-way ANOVA followed by Tukey’s post hoc test. For all analyses, P < 0.05 was used to specify statistical significance.
Results
Characterization of DPSCs and PDLSCs
Fresh extracted teeth were collected, and DPSCs and PDLSCs were extracted from pulp tissue and periodontal tissue by enzyme digestion. Alizarin red and oil red O staining showed that the extracted cells could differentiate into osteogenic and adipogenic cells (Figure 1A and B). According to the flow cytometry data, DPSCs exhibited intense expression of mesenchymal surface molecular markers (CD73 (97.1%), CD90 (99.9%)), whereas they exhibited weak expression of hematopoietic surface markers (CD45 (0.13%), CD31 (0.42%), Nestin (0.14%)). PDLSCs exhibited intense expression of mesenchymal surface molecular markers (CD29 (95.9%), CD90 (99.7%), CD105 (94.5%)), whereas they exhibited weak expression of hematopoietic surface markers (CD45 (0.3%), CD31 (0.37%)) (Figure 1C and D). These results were in line with the definition of mesenchymal stem cells of the International Association for Cell Therapy. The above results showed that the characteristics of the cells we extracted were consistent with that of the dental pulp MSCs and periodontal ligament MSCs.
Characterization of DPSC-EXO
The exosomes were isolated from DPSC-conditioned medium by ultracentrifugation. Transmission electron microscopy images showed that DPSCs exhibited a bilayer membrane and cup-plate-shaped structures with a diameter of approximately 100–150 nm (Figure 1E). Western blotting demonstrated that the DPSC-EXOs were positive for characteristic exosomal markers, such as CD9 and CD63 (Figure 1F). NTA analysis showed that 91.59% of the particles were distributed between 30 and 150 nm, and the average particle size was 84.06 nm (Figure 1G). In summary, the above results showed that we successfully isolated DPSC-EXOs.
Uptake Assay of DPSC-EXO
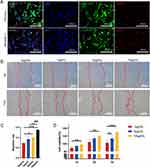
To confirm the uptake of exosomes by cells, the uptake assay of DPSC-EXOs was conducted. PKH26 labeled exosomes as red fluorescence. Calcein AM labeled the cytoplasm as green fluorescence. DAPI was used to label nuclei as blue fluorescence. After coincubating PKH26-labeled exosomes with PDLSCs and RAW264.7 cells, a large number of exosomes were scattered (red fluorescence) in the cytoplasm (green fluorescence) around the nucleus (blue fluorescence) (Figure 2A), which indicated that exosomes could be taken up by PDLSCs and RAW264.7 cells.
DPSC-EXO Promoted the Proliferation and Cell Migration of PDLSCs
To evaluate the effect of DPSC-EXO on cell migration, PDLSCs were cultured with DPSC-EXO at different concentrations (0, 1, 5, and 10 μg/mL). After 12 hours of culture, the wound healing assay showed that DPSC-EXO promoted the migration of PDLSCs in a concentration-dependent manner. However, we observed that when the exosome concentration was low (1 μg/mL), even if the migration effect was enhanced, the difference was not statistically significant. When the exosome concentrations increased (5 and 10 μg/mL), the difference was statistically significant (P<0.01, Figure 2B and C). At the same concentration gradient, we evaluated the effect of DPSC-EXO on the proliferation of PDLSCs by CCK8 assay. The group with exosome culture (5, 10 μg/mL) showed a significant effect on the proliferation of PDLSCs, and the difference was statistically significant (Figure 2D). The above results showed that DPSC-EXOs can effectively promote the migration and proliferation of PDLSCs.
DPSC-EXO Promoted the Osteogenic Differentiation of PDLSCs
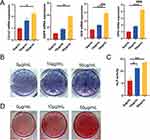
To study the effect of DPSC-EXO on the osteogenesis of PDLSCs, we performed alizarin red staining, ALP staining, quantitative analysis and qRT‒PCR. During osteogenic induction, exosomes at different concentrations (0, 10, and 50 μg/mL) were added. Total RNA was extracted, and qRT‒PCR was used to detect the expression of osteogenic genes. The results showed that DPSC-EXOs significantly improved the gene expression of Col1a1, DSPP, OCN and OPN with increasing exosome concentration, especially at a concentration of 50 μg/mL (Figure 3A). ALP staining showed that the expression of alkaline phosphatase improved significantly with increasing exosome concentration (Figure 3B). ALP quantitative analysis showed similar results, and the expression of alkaline phosphatase increased more than 2-fold in the exosome group (Figure 3C). Alizarin red staining showed that calcium deposition improved significantly with increasing exosome concentration, which indicated that exosomes promoted PDLSCs mineralization and calcium deposition in a concentration-dependent manner (Figure 3D). The above results showed that DPSC-EXO can effectively promote the osteogenic differentiation of PDLSCs.
DPSC-EXO Reduced Inflammation in PDLSCs Induced by LPS
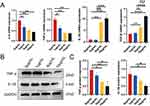
To study the effect of DPSC-EXO on the inflammation of PDLSCs, we first treated PDLSCs with LPS (10 μg/mL) and then cocultured them with exosomes at different concentrations (0, 1, 5, 10 μg/mL). Inflammation-related genes and proteins were measured by qRT‒PCR and Western Blot. The results of qRT‒PCR showed that under exosome coculture conditions, the gene expression of IL-6 and TNF-α was significantly downregulated. Moreover, different concentrations of exosomes showed similar effects, while the gene expression of IL-10 and TGF-β was significantly upregulated in a concentration-dependent manner (Figure 4A). The Western blot results showed that the protein levels of IL-1β and TNF-α also showed a decreasing trend, and the decreasing trend was most obvious when the exosome concentration was 10 μg/mL (Figure 4B and C). These results suggested that DPSC-EXO can inhibit PDLSCs inflammation induced by LPS.
DPSC-EXO Promoted the Polarization of Macrophages from the M1 Phenotype to the M2 Phenotype
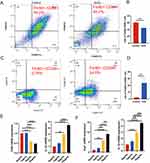
To study the effect of DPSC-EXO on inflammatory macrophages, we used LPS to stimulate RAW264.7 cells M1 polarization. Then, we analyzed the macrophage polarization markers of M1 (CD86) and M2 (CD206, Arg1) with or without EXOs. We chose an exosome concentration of 10 μg/mL to treat RAW264.7 cells. Flow cytometry analysis showed that compared with the control group (59.0%), the number of F4/80+CD86+ cells in the EXO group (46.7%) decreased significantly (Figure 5A). After statistical analysis, the difference was statistically significant (Figure 5B). However, in the EXO group, F4/80+CD206+ cells (24.6%) were significantly higher than those in the control group (0.76%) (Figure 5C). After statistical analysis, the difference was statistically significant (Figure 5D). The qRT‒PCR results showed that the mRNA expression of CD86 and IL-1β, polarizing markers of M1 macrophages, decreased with increasing exosome concentration (Figure 5E). The mRNA expression of Arg1 and IL-10, polarizing markers of M2 macrophages, significantly increased in a concentration-dependent manner, and the differences were statistically significant (Figure 5F). In summary, our results showed that DPSC-EXO could regulate the immune microenvironment by promoting the polarization of macrophages from the M1 phenotype to the M2 phenotype.
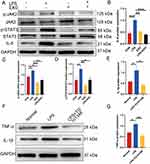
DPSC-EXO Inhibited the IL-6/JAK2/STAT3 Signaling Pathway in PDLSCs Stimulated by LPS
Western blot was used to study the effect of exosomes on the IL-6/JAK2/STAT3 signaling pathway. When PDLSCs were not stimulated or only EXO were used, the expression levels of IL-6, p-JAK2 and p-STAT3 were low. However, the expression of those proteins increased significantly when stimulated by LPS. This indicated that the IL-6/JAK2/STAT3 signaling pathway was activated by acute inflammation stimulated by LPS. The addition of EXO significantly reduced the protein levels of IL-6, p-JAK2 and p-STAT3. The differences were statistically significant (Figure 6A–D). To further verify whether DPSC-EXO can inhibit inflammation by regulating the IL-6/JAK2/STAT3 signaling pathway, we used a JAK2 inhibitor (TG101348, Beyotime, Shanghai, China) to treat PDLSCs stimulated by LPS. The results showed that the protein expression levels of IL-1β and TNF-α were downregulated (Figure 6E–G), which was consistent with the results of DPSC-EXO treatment. Therefore, we believe that DPSC-EXO exert their anti-inflammatory effect by inhibiting the IL-6/JAK2/STAT3 signaling pathway.
DPSC-EXO Inhibited Periodontitis in Rats and Promoted Alveolar Bone Repair
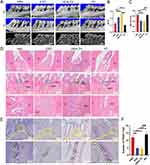
We studied the therapeutic effect of DPSC-EXO on periodontitis in rats and collected periodontal tissues from experimental rats for micro-CT and histological analysis. The main manifestation of periodontitis is alveolar bone destruction and resorption. Micro-CT analysis showed that the periodontitis group (PD) had obvious alveolar bone resorption on the buccal and lingual surfaces compared with the healthy group. The difference was also confirmed by two-dimensional X-ray images. Besides, the BV/TV decreased significantly in the PD group. Especially on buccal side, root furcation was obviously exposed, and the distance from alveolar bone crest to cementoenamel junction (ABC-CEJ) increased significantly. The results indicated that experimental periodontitis of rats was constructed successfully. Compared with that in the PBS group, the BV/TV in the EXO group was significantly higher and slightly lower than that in the healthy group. In addition, compared with the PBS group, ABC-CEJ in the EXO group was significantly decreased. These results indicated that EXO treatment could significantly reduce alveolar bone loss caused by periodontitis in rats (Figure 7A–C).
H&E staining analysis revealed that the DPSC-EXO treatment group had a thicker periodontal epithelial fibrous layer and better alveolar bone height and morphology than the PBS treatment group. In addition to the conditions of connective tissue attachment and alveolar bone, fewer infiltrating inflammatory cells could be seen in gingival tissues and periodontal ligaments between the distal root of the first molar and the mesial root of the second molar. (Figure 7D). Osteoclasts participate in bone resorption. Excessive osteoclast cells destroy the balance of bone resorption and affect new bone formation. TRAP staining analysis showed that there were fewer osteoclasts in periodontal tissue in the DPSC-EXO group than in the PBS group (Figure 7E and F).
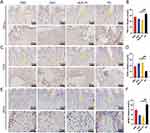
To further confirm the therapeutic effect of DPSC-EXO on experimental periodontitis in rats, we performed immunohistochemical staining with inflammatory markers (TNF-α) and osteogenic markers (OCN). Semiquantitative analysis showed that the EXO group had significantly lower TNF-α and higher OCN expression than the PD group. However, no obvious statistical significance was observed in the PBS group compared with the other groups (Figure 8A–D). These results suggested that DPSC-EXO can reduce experimental periodontitis and save alveolar bone loss in rats. To verify the effect of macrophages on the development of periodontitis, we performed immunohistochemical staining of iNOS (M1 marker). We observed more iNOS positive cell infiltration in the PD combined PBS group, while fewer in the EXO and healthy groups (Figure 8E and F). This indicated that M1 macrophage infiltration affects periodontal healing and alveolar bone repair.
Discussion
DPSCs are Excellent Seed Cell Sources for Periodontal Repair
Mesenchymal stem cells (MSCs) have been widely used in tissue engineering and regenerative medicine, and tissue-specific MSCs are superior to MSCs from other sources in repairing specific tissue injuries.24,25 Based on the above points, we strongly believe that odontogenic stem cells hold promise in promoting periodontal repair. Of all dental stem cells, DPSCs and PDLSCs are the most easily obtained.26 Kotova evaluated proteome characteristics of differences between DPSCs and PDLSCs in pluripotency, transcription of neuroepithelial markers, morphological and functional characteristics, proteome characteristics of osteoblast/odontoblast differentiation and osteoblast differentiation.34 The results showed that DPSCs proliferated more slowly, and their shape was more similar to a polygon. However, compared with PDLSCs, DPSCs responded better to osteogenic stimulation.34 In addition, Ma L found that DPSCs and PDLSCs had similar abilities in proliferation and osteogenesis, but PDLSCs were more likely to differentiate after repeated passage in vitro.27 This result suggested that DPSCs were a good candidate for osteogenesis or odontogenic bone substitute cell implantation. However, other studies have shown that compared with DPSCs, PDLSCs have formed more new bone in the treatment of periodontal defects in animals.35 Therefore, the differences in the osteogenic potential of the two kinds of cells need to be further verified. It is worth noting that PDLSCs are easily affected by the periodontal microenvironment,36 while DPSCs are located in a more closed dental pulp cavity and less affected by the external environment. In addition, the amount of dental pulp tissue in isolated teeth is usually much higher than that in periodontal tissue. Based on the above, DPSCs were chosen as the stem cell source of exosomes in our study.
The Extraction, Identification and Internalization of Exosomes Based on Their Biological Functions
Compared with stem cells, exosome infusion rarely induces an immune response. Exosomes do not contain MHC I or II molecules.37,38 As the isolation and identification of exosomes is still a challenge, continuous technological progress will help to reveal the heterogeneity and biological function of exosomes and improve the efficacy of exosomes in the treatment of periodontitis. It is worth noting that different separation techniques could affect the purity, yield and even biological activity of the exosomes.39 Exosome extraction methods usually include ultracentrifugation (UC), ExoQuick, and size exclusion chromatography.40–42 In particular, compared with other methods, UC has higher purity and less protein contamination.43 In this study, we used UC to extract DPSC-EXO and confirmed the existence of exosomes by TEM, NTA and Western blot. The results showed that the isolated particles had the same morphological characteristics as exosomes, which were a bilayer membrane vesicle-like structure with a diameter of 30–150 nm. Tang found that the exosomes obtained from UC showed high expression of CD9 and CD63, as identified by Western blot,43 which was consistent with our results. UC does not deposit all the bioactive factors of DPSCs, so other bioactive factors of DPSCs that are helpful to the treatment of periodontitis need to be further studied. Exosomes can directly fuse with the plasma membrane of receptor cells, transmit specified information, and internalize through endocytosis or phagocytosis.44 Obviously, endocytosis of exosomes by cells is fundamental for exosomes to exert biological functions. In this experiment, we used exosome tracers and fluorescence microscope to detect exosome uptake by PDLSCs and RAW264.7 cells. We confirmed that exosome labeled PKH26 could be taken up by PDLSCs and macrophages.
DPSC-EXO Has Significant Advantages in Promoting the Proliferation, Migration and Osteogenesis of PDLSCs
In tissue engineering, tissue repair and regeneration mainly depend on the proliferation and migration of endogenous cells, which is called the cell homing effect.45,46 Previous studies on DPSC-EXO have paid more attention to its role in promoting dental pulp regeneration, especially angiogenesis. Xian’s study showed that DPSC-EXO promoted human umbilical vein endothelial cells (HUVEC) proliferation, proangiogenic factors expression and tube formation, which played an important role in the regulation of angiogenesis.30 Swanson also demonstrated that DPSC-EXO promoted the migration of dental pulp cells in a dose-dependent manner.29 Our study focused on the effect of DPSC-EXO on PDLSCs. The results showed that DPSC-EXO could promote the proliferation, cell migration and osteogenesis of PDLSCs in a dose-dependent manner. This further confirmed the great potential value of DPSC-EXO in periodontal regeneration and widened the clinical application of DPSC-EXO.
Periodontitis mainly manifests as inflammatory infiltration of periodontal connective tissue and absorption of alveolar bone.47 Therefore, it is very important for the treatment of periodontitis to reduce inflammatory bone loss and repair bone defects. In this study, we used DPSC-EXO to promote the osteogenic differentiation of PDLSCs. It avoided the disturbance of immune regulation and potential risk of tumorigenesis caused by direct application of stem cells. At the same time, our results confirmed that DPSC-EXO showed obvious characteristics of promoting osteogenic differentiation of PDLSCs in vitro from the aspects of mRNA expression, alkaline phosphatase expression and calcium deposition. In vivo experiments also confirmed that DPSC-EXO can significantly reduce alveolar bone loss in experimental periodontitis rats. All these results suggest that DPSC-EXO has great prospects in the treatment of periodontitis.
DPSC-EXO Act on Periodontitis by Both Improving the Immune Microenvironment and Directly Inhibiting Inflammation in PDLSCs
Bacterial infection is an initial factor in the progression of periodontitis, but what ultimately induces periodontium destruction is dysregulation of the host immune-inflammatory response.48,49 Periodontal pathogens can stimulate strong innate immune responses, including the mobilization of macrophages, blood cells and lymphocytes into periodontal tissue. This can lead to periodontal soft tissue inflammation and alveolar bone resorption.50 Macrophages play a key role in defending periodontal pathogens. For different local microenvironments, primordial macrophages (M0) could be polarized into two opposite phenotypes: pro-inflammatory (M1) and anti-inflammatory (M2). These two phenotypes could play distinct roles under different physiological or pathological conditions.51 Pro-inflammatory macrophages can elevate local expression of RANKL, which promotes osteoclast differentiation in the periodontium.52,53 In contrast, anti-inflammatory macrophages are involved in inflammation resolution and tissue regeneration via the secretion of anti-inflammatory mediators.54 In addition, anti-inflammatory macrophages contribute to the exocytosis of apoptotic osteoblasts and mediate bone formation.55,56 As the imbalance of proinflammatory/anti-inflammatory macrophages is one of the causes of periodontal destruction, the transformation of the macrophage phenotype from proinflammatory to anti-inflammatory can effectively improve the periodontal inflammatory microenvironment. Shen showed that DPSC-EXO combined with chitosan hydrogel promoted the transformation of macrophages from proinflammatory phenotype to an anti-inflammatory phenotype in periodontitis mice, and the mechanism may be related to miR-1246 in DPSC-EXO.28 The results of this study also confirmed that DPSC-EXO could independently regulate the transformation of macrophages from the M1 to M2 phenotype without hydrogel material. In addition, DPSC-EXO also showed a significant anti-inflammatory effect on LPS-induced PDLSCs at both the mRNA and protein levels. These finding suggested that in the inflammatory microenvironment, DPSC-EXO could not only regulate the macrophage phenotype to improve the inflammatory microenvironment to further regulate the regression of inflammation but also directly inhibit the inflammation of PDLSCs. Considering that PDLSCs in the inflammatory microenvironment are different from those in the normal state, the effect of DPSC-EXO on PDLSCs in patients with periodontitis needs to be further explored.
DPSC-EXO Inhibit Inflammation via the IL-6/JAK2/STAT3 Signaling Pathway
STAT3 is a member of the signal transducer and activator of transcription (STAT) protein family.57 Recently, it has been found that STAT, as a downstream gene activated by transcriptome factors, is involved in the key genes of apoptosis, the cell cycle and other pathways.58,59 In the inflammatory state, IL-6 activates the receptor-bound Jak protein, and the receptor-bound Stat protein is further phosphorylated by Jak to form a dimer, which is transferred into the nucleus to regulate the expression of target genes.58 Hu showed that the STAT3 signaling pathway was activated in the periodontal tissue and cortex of periodontitis rats. It is speculated that activation of the STAT3 signaling pathway may play an important role by increasing the inflammatory load and promoting neuroinflammation.60 In this study, we found that DPSC-EXO simultaneously downregulated the expression of IL-6, p-JAK2 and p-STAT3 proteins in PDLSCs induced by LPS. After further use of JAK2 inhibitors, we also observed a downregulation of PDLSCs inflammation-related proteins. This result suggests that the mechanism of the role of DPSC-EXO is related to the inhibition of the IL-6/JAK2/STAT3 signaling pathway. However, the STAT3 protein is regulated by many factors. In addition to the classical and simple signal transduction of the receptor-JAK-STAT- downstream pathway, the activation of some receptor tyrosine kinases (RTKs) can phosphorylate STAT through Src kinase, including epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR).61,62 It can also activate mitogen-activated protein kinase (MAPK) through activation of the RTK/Ras pathway. MAPK can specifically phosphorylate the serine site (Ser727) at the C-terminus of STAT to enhance the transcriptional activity of STAT.63 Therefore, the other mechanisms by which DPSC-EXO regulate periodontitis and improve the immune microenvironment need to be further clarified.
Prospect and Deficiency of DPSC-EXO in Treating Experimental Periodontitis in Rats
In our study, DPSC-EXO was used to treat experimental periodontitis in rats and achieved satisfactory results. We observed less inflammatory cell infiltration, more complete epithelial healing and less alveolar bone loss. This finding is consistent with the research results of Shen et al, who demonstrated that DPSC-Exo combined with chitosan hydrogel (DPSC-Exo/CS) can accelerate the healing of alveolar bone and periodontal epithelium in periodontitis mice.28 In addition, Huang et al found that LPS-preconditioned dental follicle stem cells-derived small extracellular vesicles (DFC-sEV) loaded with hyaluronic acid (HA) injectable system could sustainably release sEV and enhance the therapeutic efficacy for periodontitis in canines.64 Wu et al also confirmed that SHED-derived exosomes contribute to periodontal bone regeneration by promoting neovascularization and new bone formation.20 Lei et al also demonstrated that PDLSC-derived exosomes lead to more bone formation in periodontitis rats.65 Therefore, exosomes derived from odontogenic stem cells are a promising new target for the treatment of periodontitis. The choice of a more appropriate cell model and optimization of conditions for exosome acquisition resulting in better therapeutic outcomes are now primary concerns. As a limitation of experiments in vivo, we did not perform animal experiments with a large sample size, although we observed satisfactory therapeutic outcomes. At the same time, we are concerned that most of the current studies, including our research, adopt only one time point for all analyses in animal experiments. More detailed information can be obtained by analysis at different time points to facilitate a more accurate analysis of the therapeutic effect of exosomes.
Conclusion
This study showed that DPSC-EXO could directly inhibit the gene and protein expression of PDLSCs inflammatory factors in vitro. It also showed that DPSC-EXO could promote the proliferation, cell migration and osteogenesis of PDLSCs. In addition, DPSC-EXO regulated the phenotype of macrophages and improved the microenvironment of periodontitis. It promoted the healing of alveolar bone and periodontal epithelium in periodontitis rats in vivo. Our preliminary study found that the mechanism may be related to the inhibition of IL-6/JAK2/STAT3 signaling pathway. However, due to the limitations of the study, the in-depth mechanism still needs further study. In short, DPSC-EXO played an important role in inhibiting periodontitis and promoting tissue regeneration. Our study provides a promising acellular therapy for periodontitis. DPSC-EXO is expected to be a new target for the treatment of periodontitis.
Data Sharing Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The reported work was supported in part by research grants from the Project Supported by the National Natural Science Foundation of China (No.31970783 to DY, NO.32270888 to DY), Program for Top talent Distinguished Professor from Chongqing Medical University to DY (NO.[2021]215), Program for Youth Innovation in Future Medicine from Chongqing Medical University (No. W0060 to DY).
Disclosure
None of the authors declare any conflicts of interest.
References
1. Kinane D, Stathopoulou P, Papapanou P. Periodontal diseases. Nature Rev Dis Primers. 2017;3:17038. doi:10.1038/nrdp.2017.38
2. Meire M, Bronzato J, Bomfim R, et al. Effectiveness of adjunct therapy for the treatment of apical periodontitis: a systematic review and meta-analysis. Int Endod J. 2022. doi:10.1111/iej.13838
3. Tavelli L, Barootchi S, Rasperini G, et al. Clinical and patient-reported outcomes of tissue engineering strategies for periodontal and peri-implant reconstruction. Periodontol 2000. 2022; 91(1):217.
4. Dragana R, Jelena M, Jovan M, et al. Antibacterial efficiency of adjuvant photodynamic therapy and high-power diode laser in the treatment of young permanent teeth with chronic periapical periodontitis. A prospective clinical study. Photodiagnosis Photodyn Ther. 2022;41:103129. doi:10.1016/j.pdpdt.2022.103129
5. Ray R. Periodontitis: an oral disease with severe consequences. Appl Biochem Biotechnol. 2022;195:17.
6. Hagelaars M, Rijns L, Dankers P, et al. Engineering strategies to move from understanding to steering renal tubulogenesis. Tissue Eng Part B Rev. 2022;29:203–216. doi:10.1089/ten.TEB.2022.0120
7. Liu G, Zhou Y, Zhang X, et al. Advances in hydrogels for stem cell therapy: regulation mechanisms and tissue engineering applications. J Materials Chem B. 2022;10:5520–5536. doi:10.1039/D2TB01044E
8. Wen Y, Dai N, Hsu S. Biodegradable water-based polyurethane scaffolds with a sequential release function for cell-free cartilage tissue engineering. Acta Biomaterialia. 2019;88:301–313. doi:10.1016/j.actbio.2019.02.044
9. Park J, Lee J, Yoon B, et al. Additive effect of bFGF and selenium on expansion and paracrine action of human amniotic fluid-derived mesenchymal stem cells. Stem Cell Res Ther. 2018;9:293. doi:10.1186/s13287-018-1058-z
10. Park J, Park G, Hong H. Age affects the paracrine activity and differentiation potential of human adipose-derived stem cells. Mol Med Rep. 2021;23:1.
11. Sikora B, Skubis-Sikora A, Prusek A, et al. Paracrine activity of adipose derived stem cells on limbal epithelial stem cells. Sci Rep. 2021;11:19956. doi:10.1038/s41598-021-99435-1
12. Liu Y, Zhuang X, Yu S, et al. Exosomes derived from stem cells from apical papilla promote craniofacial soft tissue regeneration by enhancing Cdc42-mediated vascularization. Stem Cell Res Ther. 2021;12:76. doi:10.1186/s13287-021-02151-w
13. Chamberlain C, Kink J, Wildenauer L, et al. Exosome-educated macrophages and exosomes differentially improve ligament healing. Stem Cells. 2021;39(1):55–61. doi:10.1002/stem.3291
14. Jafari D, Malih S, Eini M, et al. Improvement, scaling-up, and downstream analysis of exosome production. Crit Rev Biotechnol. 2020;40:1098–1112. doi:10.1080/07388551.2020.1805406
15. Pu X, Ma S, Gao Y, et al. Mesenchymal stem cell-derived exosomes: biological function and their therapeutic potential in radiation damage. Cells. 2020;10:10. doi:10.3390/cells10010010
16. Liu L, Guo S, Shi W, et al. Bone marrow mesenchymal stem cell-derived small extracellular vesicles promote periodontal regeneration. Tissue Eng Part A. 2021;27:962–976. doi:10.1089/ten.tea.2020.0141
17. Song X, Xue Y, Fan S, et al. Lipopolysaccharide-activated macrophages regulate the osteogenic differentiation of bone marrow mesenchymal stem cells through exosomes. PeerJ. 2022;10:e13442. doi:10.7717/peerj.13442
18. Wang M, Li J, Ye Y, et al. SHED-derived conditioned exosomes enhance the osteogenic differentiation of PDLSCs via Wnt and BMP signaling in vitro. Res Biol Diversity. 2020;111:1–11. doi:10.1016/j.diff.2019.10.003
19. Wei J, Song Y, Du Z, et al. Exosomes derived from human exfoliated deciduous teeth ameliorate adult bone loss in mice through promoting osteogenesis. J Mol Histol. 2020;51:455–466. doi:10.1007/s10735-020-09896-3
20. Wu J, Chen L, Wang R, et al. Exosomes secreted by stem cells from human exfoliated deciduous teeth promote alveolar bone defect repair through the regulation of angiogenesis and osteogenesis. ACS biomaterials sci eng. 2019;5:3561–3571. doi:10.1021/acsbiomaterials.9b00607
21. Cui D, Li H, Wan M, et al. The origin and identification of mesenchymal stem cells in teeth: from odontogenic to non-odontogenic. Curr Stem Cell Res Ther. 2018;13:39–45.
22. Morsczeck C, Reichert T. Dental stem cells in tooth regeneration and repair in the future. Expert Opin Biol Ther. 2018;18:187–196. doi:10.1080/14712598.2018.1402004
23. Ziauddin S, Nakashima M, Watanabe H, et al. Biological characteristics and pulp regeneration potential of stem cells from canine deciduous teeth compared with those of permanent teeth. Stem Cell Res Ther. 2022;13:439. doi:10.1186/s13287-022-03124-3
24. Gupta S, Rawat S, Krishnakumar V, et al. Hypoxia preconditioning elicit differential response in tissue-specific MSCs via immunomodulation and exosomal secretion. Cell Tissue Res. 2022;388:535–548. doi:10.1007/s00441-022-03615-y
25. Paliwal S, Chaudhuri R, Agrawal A, et al. Human tissue-specific MSCs demonstrate differential mitochondria transfer abilities that may determine their regenerative abilities. Stem Cell Res Ther. 2018;9:298. doi:10.1186/s13287-018-1012-0
26. Shoushrah S, Transfeld J, Tonk C, et al. Sinking our teeth in getting dental stem cells to clinics for bone regeneration. Int J Mol Sci;2021. 22. doi:10.3390/ijms23010022
27. Ma L, Hu J, Cao Y, et al. Maintained properties of aged dental pulp stem cells for superior periodontal tissue regeneration. Aging Dis. 2019;10:793–806. doi:10.14336/AD.2018.0729
28. Shen Z, Kuang S, Zhang Y, et al. Chitosan hydrogel incorporated with dental pulp stem cell-derived exosomes alleviates periodontitis in mice via a macrophage-dependent mechanism. Bioactive Materials. 2020;5:1113–1126. doi:10.1016/j.bioactmat.2020.07.002
29. Swanson W, Gong T, Zhang Z, et al. Controlled release of odontogenic exosomes from a biodegradable vehicle mediates dentinogenesis as a novel biomimetic pulp capping therapy. J Control Release. 2020;324:679–694. doi:10.1016/j.jconrel.2020.06.006
30. Xian X, Gong Q, Li C, et al. Exosomes with highly angiogenic potential for possible use in pulp regeneration. J Endod. 2018;44:751–758. doi:10.1016/j.joen.2017.12.024
31. Li H, Deng Y, Tan M, et al. Low-intensity pulsed ultrasound upregulates osteogenesis under inflammatory conditions in periodontal ligament stem cells through unfolded protein response. Stem Cell Res Ther. 2020;11:215. doi:10.1186/s13287-020-01732-5
32. Shi J, Zhang Y, Zhang X, et al. Remodeling immune microenvironment in periodontitis using resveratrol liposomes as an antibiotic-free therapeutic strategy. J Nanobiotechnology. 2021;19:429. doi:10.1186/s12951-021-01175-x
33. Zhang X, Xu M, Xue Q, et al. A modified method for constructing experimental rat periodontitis model. Front bioeng biotechnol. 2022;10:1098015. doi:10.3389/fbioe.2022.1098015
34. Kotova A, Lobov A, Dombrovskaya J, et al. Comparative analysis of dental pulp and periodontal stem cells: differences in morphology, functionality, osteogenic differentiation and proteome. Biomedicines. 2021;9(11):1606. doi:10.3390/biomedicines9111606
35. Amghar-Maach S, Gay-Escoda C, Sánchez-Garcés M. Regeneration of periodontal bone defects with dental pulp stem cells grafting: systematic Review. J Clin Exp Dentistry. 2019;11:e373–e381. doi:10.4317/jced.55574
36. Chen Q, Liu X, Wang D, et al. Periodontal inflammation-triggered by periodontal ligament stem cell pyroptosis exacerbates periodontitis. Front Cell Dev Biol. 2021;9:663037. doi:10.3389/fcell.2021.663037
37. Buschow S, van Balkom B, Aalberts M, et al. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol Cell Biol. 2010;88:851–856. doi:10.1038/icb.2010.64
38. Lu M, Peng L, Ming X, et al. Enhanced wound healing promotion by immune response-free monkey autologous iPSCs and exosomes vs. their allogeneic counterparts. EBioMedicine. 2019;42:443–457. doi:10.1016/j.ebiom.2019.03.011
39. Singh K, Nalabotala R, Koo K, et al. Separation of distinct exosome subpopulations: isolation and characterization approaches and their associated challenges. Analyst. 2021;146:3731–3749. doi:10.1039/D1AN00024A
40. Ding L, Yang X, Gao Z, et al. A holistic review of the state-of-the-art microfluidics for exosome separation: an overview of the current status, existing obstacles, and future outlook. Small. 2021;17:e2007174. doi:10.1002/smll.202007174
41. Huang S, Ji X, Jackson K, et al. Rapid separation of blood plasma exosomes from low-density lipoproteins via a hydrophobic interaction chromatography method on a polyester capillary-channeled polymer fiber phase. Anal Chim Acta. 2021;1167:338578. doi:10.1016/j.aca.2021.338578
42. Yang D, Zhang W, Zhang H, et al. Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics. 2020;10:3684–3707. doi:10.7150/thno.41580
43. Tang Y, Huang Y, Zheng L, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med. 2017;40:834–844. doi:10.3892/ijmm.2017.3080
44. Zhao X, Wu D, Ma X, et al. Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomed Pharmacother. 2020;128:110237. doi:10.1016/j.biopha.2020.110237
45. Liesveld J, Sharma N, Aljitawi O. Stem cell homing: from physiology to therapeutics. Stem Cells. 2020;38:1241–1253. doi:10.1002/stem.3242
46. Wang S, Niu Y, Jia P, et al. Alkaline activation of endogenous latent TGFβ1 by an injectable hydrogel directs cell homing for in situ complex tissue regeneration. Bioactive Materials. 2022;15:316–329. doi:10.1016/j.bioactmat.2021.12.015
47. Chen H, Wang Z, He Y, et al. Pyroptosis may play a crucial role in modifications of the immune microenvironment in periodontitis. J Periodontal Res. 2022;57:977–990. doi:10.1111/jre.13035
48. Bartold P, Van Dyke T. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. J Clin Periodontol. 2019;46:6–11. doi:10.1111/jcpe.13046
49. Ma J, Kageyama S, Takeshita T, et al. Clinical utility of subgingival plaque-specific bacteria in salivary microbiota for detecting periodontitis. PLoS One. 2021;16:e0253502. doi:10.1371/journal.pone.0253502
50. Almubarak A, Tanagala K, Papapanou P, et al. Disruption of Monocyte and Macrophage Homeostasis in Periodontitis. Front Immunol. 2020;11:330. doi:10.3389/fimmu.2020.00330
51. Zhang B, Yang Y, Yi J, et al. Hyperglycemia modulates M1/M2 macrophage polarization via reactive oxygen species overproduction in ligature-induced periodontitis. J Periodontal Res. 2021;56:991–1005. doi:10.1111/jre.12912
52. Alam M, Mae M, Farhana F, et al. NLRP3 inflammasome negatively regulates rankl-induced osteoclastogenesis of mouse bone marrow macrophages but positively regulates it in the presence of lipopolysaccharides. Int J Mol Sci. 2022;23(11):6096. doi:10.3390/ijms23116096
53. Zhu H, Tamura A, Zhang S, et al. Mitigating RANKL-induced cholesterol overload in macrophages with β-cyclodextrin-threaded polyrotaxanes suppresses osteoclastogenesis. Biomaterials Science. 2022;10(18):5230–5242. doi:10.1039/D2BM00833E
54. Lundahl M, Mitermite M, Ryan D, et al. Macrophage innate training induced by IL-4 and IL-13 activation enhances OXPHOS driven anti-mycobacterial responses. eLife. 2022;11.
55. Shang N, Wu J. Egg-Derived Tripeptide IRW Attenuates LPS-Induced Osteoclastogenesis in RAW 264.7 Macrophages via Inhibition of Inflammatory Responses and NF-κB/MAPK Activation. J Agric Food Chem. 2020;68:6132–6141. doi:10.1021/acs.jafc.0c01159
56. Zhu K, Yang C, Dai H, et al. Crocin inhibits titanium particle-induced inflammation and promotes osteogenesis by regulating macrophage polarization. Int Immunopharmacol. 2019;76:105865. doi:10.1016/j.intimp.2019.105865
57. Menon P, Doudin A, Gregus A, et al. The anti-parallel dimer binding interface in STAT3 transcription factor is required for the inactivation of cytokine-mediated signal transduction. Biochimica et biophysica acta Mol cell res. 2021;1868:119118. doi:10.1016/j.bbamcr.2021.119118
58. Mohassab A, Hassan H, Abdelhamid D, et al. STAT3 transcription factor as target for anti-cancer therapy. Pharmacol Rep. 2020;72:1101–1124. doi:10.1007/s43440-020-00156-5
59. Nikolskii A, Shilovskiy I, Barvinskaia E, et al. Role of STAT3 Transcription Factor in Pathogenesis of Bronchial Asthma. Biochemistry (Mosc). 2021;86:1489–1501. doi:10.1134/S0006297921110122
60. Hu Y, Zhang X, Zhang J, et al. Activated STAT3 signaling pathway by ligature-induced periodontitis could contribute to neuroinflammation and cognitive impairment in rats. J Neuroinflammation. 2021;18:80. doi:10.1186/s12974-021-02071-9
61. de Los Fayos Alonso I G, Zujo L, Wiest I, et al. PDGFRβ promotes oncogenic progression via STAT3/STAT5 hyperactivation in anaplastic large cell lymphoma. Mol Cancer. 2022;21:172. doi:10.1186/s12943-022-01640-7
62. Yan M, Han M, Yang X, et al. Dual inhibition of EGFR and IL-6-STAT3 signalling by miR-146b: a potential targeted therapy for epithelial ovarian cancer. J Enzyme Inhib Med Chem. 2021;36:1905–1915. doi:10.1080/14756366.2021.1963240
63. Zhao K, Dai Q, Wu J, et al. Morusin enhances the antitumor activity of MAPK pathway inhibitors in BRAF-mutant melanoma by inhibiting the feedback activation of STAT3. Eur j Cancer. 2022;165:58–70. doi:10.1016/j.ejca.2022.01.004
64. Huang Y, Liu Q, Liu L, et al. Lipopolysaccharide-preconditioned dental follicle stem cells derived small extracellular vesicles treating periodontitis via reactive oxygen species/mitogen-activated protein kinase signaling-mediated antioxidant effect. Int J Nanomedicine. 2022;17:799–819. doi:10.2147/IJN.S350869
65. Lei F, Li M, Lin T, et al. Treatment of inflammatory bone loss in periodontitis by stem cell-derived exosomes. Acta Biomater. 2022;141:333–343. doi:10.1016/j.actbio.2021.12.035
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.