Back to Journals » Nature and Science of Sleep » Volume 13
Deep Learning for Diagnosis and Classification of Obstructive Sleep Apnea: A Nasal Airflow-Based Multi-Resolution Residual Network
Authors Yue H , Lin Y, Wu Y, Wang Y, Li Y, Guo X, Huang Y, Wen W, Zhao G, Pang X, Lei W
Received 22 December 2020
Accepted for publication 19 February 2021
Published 12 March 2021 Volume 2021:13 Pages 361—373
DOI https://doi.org/10.2147/NSS.S297856
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ahmed BaHammam
Huijun Yue,1,* Yu Lin,1,* Yitao Wu,2,* Yongquan Wang,1 Yun Li,1 Xueqin Guo,1 Ying Huang,3 Weiping Wen,1 Gansen Zhao,2 Xiongwen Pang,2 Wenbin Lei1
1Otorhinolaryngology Hospital, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, 510080, People’s Republic of China; 2School of Computer Science, South China Normal University, Guangzhou, 510631, People’s Republic of China; 3Guangdong Province Traditional Chinese Medical Hospital, Guangzhou, 510000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenbin Lei
Otorhinolaryngology Hospital, The First Affiliated Hospital, Sun Yat-sen University, 58 Zhongshan Road II, Guangzhou, Guangdong, 510080, People’s Republic of China
Tel +86-13922113299
Email [email protected]
Xiongwen Pang
School of Computer Science, South China Normal University, 55 West Zhongshan Avenue, Guangzhou, Guangdong, 510631, People’s Republic of China
Tel +86-18620638848
Email [email protected]
Purpose: This study evaluated a novel approach for diagnosis and classification of obstructive sleep apnea (OSA), called Obstructive Sleep Apnea Smart System (OSASS), using residual networks and single-channel nasal pressure airflow signals.
Methods: Data were collected from the sleep center of the First Affiliated Hospital, Sun Yat-sen University, and the Integrative Department of Guangdong Province Traditional Chinese Medical Hospital. We developed a new model called the multi-resolution residual network (Mr-ResNet) based on a residual network to detect nasal pressure airflow signals recorded by polysomnography (PSG) automatically. The performance of the model was assessed by its sensitivity, specificity, accuracy, and F1-score. We built OSASS based on Mr-ResNet to estimate the apnea‒hypopnea index (AHI) and to classify the severity of OSA, and compared the agreement between OSASS output and the registered polysomnographic technologist (RPSGT) score, assessed by two technologists.
Results: In the primary test set, the sensitivity, specificity, accuracy, and F1-score of Mr-ResNet were 90.8%, 90.5%, 91.2%, and 90.5%, respectively. In the independent test set, the Spearman correlation for AHI between OSASS and the RPSGT score determined by two technologists was 0.94 (p < 0.001) and 0.96 (p < 0.001), respectively. Cohen’s Kappa scores for classification between OSASS and the two technologists’ scores were 0.81 and 0.84, respectively.
Conclusion: Our results indicated that OSASS can automatically diagnose and classify OSA using signals from a single-channel nasal pressure airflow, which is consistent with polysomnographic technologists’ findings. Thus, OSASS holds promise for clinical application.
Keywords: deep learning, nasal airflow, obstructive sleep apnea, residual network
Introduction
Undiagnosed and untreated obstructive sleep apnea (OSA) is a major health burden worldwide,1,2 and is associated with several severe health consequences, such as cardiac arrhythmia, heart attacks, stroke, and even sudden death,3,4 and can increase the risk of traffic accidents.5 Polysomnography (PSG) is regarded as the gold standard for the accurate diagnosis of OSA, and is performed at a specialized sleep center or laboratory.6 Multiple physiological signals pertaining to electroencephalogram (EEG), electrooculograms (EOG), electromyogram (EMG), electrocardiogram (ECG), respiration, and pulse oxygen saturation (SpO2) are recorded during overnight PSG.7 The output requires manual scoring by sleep specialists, which is time-consuming, labor-intensive, and error-prone, making it a complex and expensive process, which markedly affects the availability and accessibility of OSA diagnostic resources.
Although unattended abbreviated testing for routine screening of OSA at home, based on a reduced number of physiological signals, has been encouraged during the last few years,8 the accuracy of most analysis software has been shown to be relatively poor, underestimating the condition.9,10 Therefore, there is a need for an intelligent OSA diagnostic system that uses fewer signals, analyzes more patients more rapidly, and eliminates the high intra- and inter-scorer variability.11
Deep learning (DL)12 has achieved some success in the field of automated OSA diagnosis using single-channel signals. A convolutional neural network (CNN) is a popular structure in DL.12 Urtnasan et al13 showed the performance of a six-layer CNN model for OSA detection from a single-lead ECG signal, with a sensitivity, specificity, and accuracy of 96% each. Jiang et al14 proposed a multi-scale parallel CNN for automatic sleep apnea detection using single-channel EEG signals, with an accuracy of 89.1%. Haidar et al15 compared two classification methods based on nasal airflow signals and proved that CNN (with an accuracy of 74.7%) was superior to a support vector machine (SVM), a traditional machine-learning algorithm (which had an accuracy of 72.0%). Van Steenkiste et al16 identified sleep apnea from chest and abdominal breathing data using a long short-term memory network (LSTM),17,18 another deep learning algorithm called a recurrent neural network (RNN),12,18 with an average accuracy of 77.2% and 75.0%, respectively. However, most methods only classify apnea, ignoring hypopnea,19 which is also important for individualized diagnosis20,21 and treatment.22,23
Using airflow signals to estimate the apnea‒hypopnea index (AHI) is a suitable method, as it is completely in accordance with the rules of manual scoring, but models based on one-channel respiratory signals exhibit inferior performance. The residual network (ResNet) approach24 is an outstanding means of implementing a CNN and can accelerate the training of ultra-deep neural networks, yielding improved accuracy. To the best of our knowledge, there have been no reports using ResNet and airflow signals to detect OSA. In this study, we propose a DL architecture called multi-resolution ResNet (Mr-ResNet) based on ResNet, that uses only raw nasal pressure airflow signals to learn and extract relevant features automatically, and achieves high sensitivity (90.8%) and specificity (90.5%), with an accuracy of 91.2%. The architecture of Mr-ResNet was presented at 2020 International Conference on Neural Computing for Advanced Applications (Shenzhen, China).25 Based on Mr-ResNet, we built an OSA Smart System (OSASS) to estimate AHI and classify OSA severity in a manner that was consistent with scoring by polysomnographic technologists in a real clinical setting. These results demonstrate that the novel concept has good prospects for clinical application, which can be widely used for large-scale OSA screening, rapid diagnosis, and remote diagnosis systems.
Methods
Subjects and Database
The Clinical Research Ethics Committee of the First Affiliated Hospital, Sun Yat-sen University approved the research protocol (approval No. 2,019,119). Given the retrospective nature of the study and considering that we ensured participant privacy by confirming that the study was conducted in compliance with the Declaration of Helsinki, the Institutional Review Board approved our application for an exemption regarding the requirement to obtain written informed consent from the patients.
In this study, two datasets were used to develop and verify the model, including one database of adult patients with suspected OSA collected from the sleep center of the First Affiliated Hospital, Sun Yat-sen University (FAH database), and another one collected from the Integrative Department of Guangdong Province Traditional Chinese Medical Hospital (CMH database). The FAH database contains 405 PSG records, which were collected using an Embla device (Embla N7000, San Diego, CA, USA), which recorded seven EEGs, right and left EOGs, one chin muscle EMG, ECG, SpO2, nasal pressure airflow, thoracic and abdominal respiratory movements, body position, snoring sound, and video monitoring, between January 2018 and December 2019. The CMH database information was collected from 45 patients using the same device. All records were manually analyzed by RemLogic 3.4.1 software (Embla) using the 2012 American Academy of Sleep Medicine (AASM) scoring rules.19 According to these rules, an obstructive apnea event was defined as a ≥ 90% drop in respiratory flow for at least 10 s, associated with breathing effort. A hypopnea event was defined as a ≥ 30% drop in respiratory flow for at least 10 s, associated with ≥ 3% oxygen desaturation or arousal; and the AHI was defined as the number of apneas and hypopneas per hour of total sleep time (TST).
Subjects who met the following criteria were excluded: patients (1) aged < 18 years; (2) with TST < 240 min; (3) with central sleep apnea events > 50%; (4) with other coexisting sleep disorders; and (5) with pulmonary dysfunction (Figure 1). The subjects from the FAH database were divided into two groups to make up the training and primary test sets, by first sorting all patients based on their AHI and randomly selecting about 10% of every severity category for the primary test set. The CMH test set included a similar proportion of subjects for the primary test set. The demographic and polysomnographic characteristics of all subjects in the whole FAH dataset, training set, primary test set, and the CMH test set are presented in Table 1.
 |
Table 1 Demographic and Polysomnographic Characteristics of Subjects |
Preprocessing and Datasets
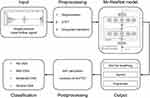
The database contained time-series and one-channel nasal pressure airflow signals obtained with a sampling frequency of 200 Hz. To take full advantage of classic neural networks, we needed to convert the original time series data into spectrograms. Therefore, the preprocessing procedures for the raw signal included signal segmentation, short-time Fourier transform (STFT),26 and grayscale transform. The 6-h original airflow data of each subject was split into 30-s non-overlapping segments, and each segment had a corresponding label. We labeled a segment A or H if an apnea or hypopnea waveform lasted longer than 10 seconds, respectively, while other segments were labeled N. Following this rule, the numbers of A, H, and N events were obtained; the statistics of the event distribution are shown in Figure 1. Then, 30-s segments were converted into corresponding spectrograms by STFT to generate our datasets (Figure 2). The size of the sliding windows was 64 and the overlap of the windows was 32 for the STFT calculation in each 30-s non-overlapping segment.
Deep Learning Algorithm
In this study, we proposed an intelligent system called OSASS for OSA diagnosis and classification. The core of OSASS is Mr-ResNet, a DL architecture based on ResNet. A schematic representation of the proposed approach is shown in Figure 3. First, Mr-ResNet was used to determine normal breathing, apnea, and hypopnea events from spectrograms. Next, post-processing was used to calculate the AHI. Finally, the estimated AHI values were used to classify subjects into the standard OSA severity categories: no OSA (AHI < 5), mild (5 ≤ AHI < 15), moderate (15 ≤ AHI < 30), and severe OSA (30 ≤ AHI).
We implemented the proposed Mr-ResNet using PyTorch, and trained it using the stochastic gradient descent algorithm with a learning rate of 0.0001, batch size of 64, and iterations of 20. All the training and validating work was performed on a CentOS 7 server with 2 Tesla K80 graphics cards each of whose running memory has 11,439 MiB.
Statistical Analysis
All statistical analyses were performed using IBM SPSS version 25.0 (SPSS, Chicago, IL, USA) and R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria). The performance of the Mr-ResNet model was quantified by several accepted measurements, including sensitivity, specificity, accuracy, and F1-score. PSG records of the CMH database were manually reanalyzed in accordance with the same standard of the 2012 AASM by two registered polysomnographic technologists (RPSGT). Spearman correlation coefficient and Bland‒Altman plots were evaluated to measure the agreement of OSASS and both manual RPSGT scores. A p-value < 0.05 was considered statistically significant. Finally, to assess the OSA severity classification accuracy, confusion matrices and Cohen’s Kappa were calculated. The Kappa scores were interpreted as follows: values ≤ 0, indicated no agreement, values of 0.01‒0.20 indicated none to slight agreement, 0.21‒0.40 indicated fair agreement, 0.41‒0.60 indicated moderate agreement, 0.61‒0.80 indicated substantial agreement, and 0.81‒1.00 indicated almost perfect agreement.27
Results
Performance of Mr-ResNet
Table 2 shows the performance of the proposed Mr-ResNet model for the automated detection of OSA from a single-channel airflow signal in the primary test set. The overall sensitivity, specificity, accuracy, and F1-score were 90.8%, 90.5%, 91.2%, and 90.5%, respectively. While these values slightly decreased with increasing AHI, the performance did not significantly change among the different severity groups.
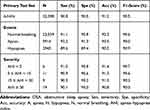 |
Table 2 Performance of Mr-ResNet for OSA Detection in the Primary Test Set |
Additionally, we compared the performance of the proposed method with previous studies using single-channel signals. As shown in Table 3, three conclusions can be drawn: the CNN model based on ECG signals performed better than other single-channel signal methods; the DL algorithm performed better than the machine-learning algorithm; the model for only apnea detection performed better than one using apnea and hypopnea detection. Choi et al28 proposed estimating apnea‒hypopnea (AH) events using CNN from airflow signals, and achieved a satisfactory specificity (98.5%) and accuracy (96.6%) in detection, but not high sensitivity (81.1%). It is thus apparent that the foremost advantage of the Mr-ResNet model is that it can distinguish apnea and hypopnea events with high sensitivity (90.8%), while maintaining high specificity (90.5%) and accuracy (91.2%), which implies that our results provide more important guidance for diagnosing OSA, which is a complex and heterogeneous disorder.29
 |
Table 3 Comparison with Previous Studies |
Estimation of AHI
The proposed OSASS performed well in the independent test set. The sleep respiratory events for all patients in the CMH test set were calculated by OSASS, and strong significant correlations were observed between OSASS and RPSGT scores. Spearman correlation coefficients for AI, HI, and AHI between OSASS and the scores of technologist 1 were 0.96, 0.71, and 0.94, respectively. Spearman correlation coefficients for AI, HI, and AHI between OSASS and scores of technologist 2 were 0.97, 0.59, and 0.96, respectively. The correlation and Bland‒Altman plots are shown in Figures 4 and 5. In the Bland–Altman plot, the x-axis depicts the mean values of AI, HI, and AHI measured with the OSASS and RPSGT and the y-axis depicts the difference between the estimated and reference values. The black dashed lines denote the bias and 95% upper and lower limits of agreement.
Classification of OSA
The estimated AHI values were used to classify the subjects from the CMH test set into the standard OSA severity categories (no, mild, moderate, and severe OSA). Table 4 shows the confusion matrices for OSA severity classification for each technologist’s scores vs OSASS. The Kappa scores between OSASS and the individual technologist’s scores were both above 0.8 (Table 5), which indicates almost perfect agreement (0.8 < Kappa < 1).
 |
Table 4 Confusion Matrices for OSA Classification in CMH Test Set |
 |
Table 5 Kappa Values for OSA Classification Among OSASS and Individual Technologists |
Discussion
In this study, we developed an intelligent OSA diagnosis and classification system for respiratory event detection using a DL algorithm based on nasal pressure airflow signals. The main findings of our work are as follows: (1) We improved a neural network from ResNet to capture multi-resolution imaging features of airflow signals without any handcrafted engineering to generate high-accuracy AH estimates, which has not been presented previously. (2) In comparison with other research, the proposed model offers competitive performance with only one-channel airflow signals, which can detect both apnea and hypopnea events overnight with a sensitivity of 90.8%, a specificity of 90.5%, an accuracy of 91.2%, and F1-score of 90.5%. (3) Our proposed method corresponds well with RPSGT scores for OSA diagnosis and classification, in which performance was validated using an independent hospital database.
Owing to the non-stationary and nonlinear characteristics of sleep data, traditional machine-learning methods require researchers to expend much energy on feature extraction, while neural networks can solve such problems well. Therefore, neural network technology is increasingly applied to automated OSA detection. As a popular structure in DL, the accuracy of a CNN increases with increasing network depth. However, when the network depth increases to a certain level, the accuracy decreases rapidly, as the deeper network becomes more difficult to optimize. ResNet solves this problem with a simple structure, and its short connection structure breaks the traditional layer-by-layer transmission of neural networks, which can accelerate the training of ultra-deep neural networks, yielding improved accuracy.34 In this study, we proposed the DL architecture called MR-ResNet, based on ResNet, in which the 3 × 3 filters were replaced by a new block containing multi-level group convolution. The structure we used makes full use of group convolution and residual-like connections at both levels, to maximize feature extraction capabilities and reduce the number of network parameters, which greatly improves the training speed of our network.
To simplify the diagnostic technique, various alternative methods have been proposed to replace PSG and minimize the number of sensors required in recent decades, such as ECG,13,30,35 EEG,14,36 SpO2,31,37 respiratory,16,38 and snoring signals.39,40 However, the most appropriate physiological signals for the development of simple and accurate screening tests for OSA remain unknown. Overall, single-channel systems have been reported, and the results were similar to those obtained with algorithms using multiple data channels. These approaches place fewer limitations on patient movement and can be more convenient for use in the ambulatory environment than systems employing multiple channels.
Previous single-channel studies for the automated detection of OSA were mostly based on ECG.41,42 Dey et al30 reported that their framework employing a CNN can achieve a very high degree of sensitivity, specificity, and accuracy using single-lead ECG signals, ie, 98.9%, 97.8%, and 99.2%, respectively. However, they did not classify hypopnea events. Yet, this information is important, because hypopnea is the main respiratory event in some OSA patients, which has significance for guiding individualized treatment.19,22,23 According to the AASM criteria of sleep apnea and hypopnea, many studies also use airflow and SpO2 signals to detect respiratory events. In the study of Álvarez et al,32 airflow signals were analyzed based on the machine-learning algorithm, OSA detection with a sensitivity of more than 90%, but a specificity of less than 70%. Choi et al28 proposed estimating AH events using CNN from airflow signals and achieved a satisfactory specificity (98.5%) and accuracy (96.6%) in detection, but the sensitivity was only 81.1%. Vaquerizo-Villar et al31 proposed using CNN to detect AH events from SpO2 raw data. The experiment achieved a classification sensitivity, specificity, and accuracy of 95.4%, 96.7%, and 93.6%, respectively, but only in pediatric patients. The above results show that the performance of the model based on a single-channel airflow signal can still be improved. This was due to the non-standard airflow waveforms, especially the hypopnea waveform, which challenged the learning algorithms. In this study, to solve this problem, we converted the original time series data into spectrograms by STFT and make full use of the advantage of Mr-ResNet to maximize feature extraction capabilities. Based on this approach, we were able to achieve high sensitivity (90.8%) and specificity (90.5%), with an accuracy of 91.2%.
Airflow signals have been used to estimate AHI and determine OSA severity categories.15,28,33 However, these studies were applied to a limited number of segments and performance was not assessed on over-night data or on an OSA population. One novelty of this study is that the performance of OSASS in OSA diagnosis and classification was compared with the scores of two sleep specialists using an independent hospital database. Although the OSASS performed slightly worse in hypopnea index (HI) estimation (Spearman correlation coefficient for HI between OSASS and technologist 1, and technologist 2, was 0.71 and 0.59, respectively), the results are still impressive considering that the dataset was completely new and scored by different people. With our study design, the Bland–Altman plot, as a measure of agreement, was appropriate to perform within-subject comparisons for the two methods.43 The Kappa agreement between OSASS and technologist 1 and technologist 2 was 0.81 and 0.84, respectively, which suggests that OSASS is capable of reasonably estimating OSA severity. These results show that OSASS generalizes reasonably well.
There were some limitations to this study. First, OSASS is likely to underestimate the severity of hypopnea when compared to a full PSG scoring, due to a lack of SpO2 and EEG data; this is particularly true for OSA patients in whom hypopnea events are dominant. Second, OSASS only classifies obstructive apnea, hypopnea, and normal breathing, but not central and mixed sleep apnea, as this would require information about the breathing effort, which is not present in the airflow signal. Although central and mixed sleep apneas occur very rarely, it affects the results partly. Third, OSASS is not able to auto-extract TST. Yet, accurate estimation of sleeping time can increase the accuracy of the estimated AHI, particularly in individuals with low sleep efficiency.44 Finally, a small population was investigated in this study, additional evidence on the effectiveness of OSASS for OSA screening in the presence of comorbidities, particularly cardiovascular and pulmonary diseases, is required. Thus, further studies should be conducted to address these limitations to increase the clinical usefulness of the present model.
Conclusion
This paper proposed the Mr-ResNet framework for AHI estimation using only one-channel nasal pressure airflow signals, and validated the model against two datasets. Our findings imply that this new diagnosis and classification system of OSA demonstrated good feasibility for clinical application as well as scalability.
Abbreviations
AASM, American Academy of Sleep Medicine; AB‒CART, AdaBoost‒classification and regression trees; AF, airflow; AH, apnea‒hypopnea; AHI, apnea‒hypopnea index; AI, apnea index; BMI, body mass index; CHAT, Childhood Adenotonsillectomy Trial; CMH, Chinese Medical Hospital; CNN, convolutional neural network; CSA, central sleep apnea; DL, deep learning; ECG, electrocardiogram; EEG, electroencephalogram; EMG, electromyogram; EOG, electrooculogram; FAH, First Affiliated Hospital; HI, hypopnea index; LSTM, long short-term memory network; MESA, Multi-Ethnic Study of Atherosclerosis; MIT-BIH, Massachusetts Institute of Technology-Beth Israel Hospital; Mr-ResNet, multi-resolution residual network; OSA, obstructive sleep apnea; OSASS, Obstructive Sleep Apnea Smart System; PSG, polysomnography; ResNet, residual network; RNN, recurrent neural network; RPSGT, registered polysomnographic technologist; SpO2, pulse oxygen saturation; STFT, short-time Fourier transform; SVM, support vector machine; TST, total sleep time.
Data Sharing Statement
The code and corresponding model used during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors thank their department for support and would like to thank Editage for the English language editing.
Funding
This work was supported by the National Key R&D Program of China (No. 2020YFC1316903, No. 2018YFB1404402), the Science and Technology Program of Guangzhou of China (No. 201704020092, No. 20180203004, No. 201804010314), the 5010 Clinical Research Program of Sun Yat-sen University (No. 2017004), the General Program of National Natural Science Foundation of China (No. 81972528), and the Major Program of National Social Science Fund of China (No. 19ZDA041).
Disclosure
Dr. Huijun Yue, Professor Gansen Zhao, Professor Xiongwen Pang and Professor Wenbin Lei report a patent a processing method, system and storage media for sleep respiratory events detection pending. The authors report no other conflicts of interest in this work.
References
1. Lévy P, Kohler M, McNicholas WT, et al. Obstructive sleep apnoea syndrome. Nat Rev Dis Primers. 2015;1:15015. doi:10.1038/nrdp.2015.15
2. Jordan AS, McSharry DG, Malhotra A. Adult obstructive sleep apnoea. Lancet. 2014;383(9918):736–747. doi:10.1016/S0140-6736(13)60734-5
3. Mokhlesi B, Varga AW. Obstructive sleep apnea and cardiovascular disease. REM sleep matters! Am J Respir Crit Care Med. 2018;197(5):554–556. doi:10.1164/rccm.201710-2147ED
4. Zinchuk AV, Jeon S, Koo BB, et al. Polysomnographic phenotypes and their cardiovascular implications in obstructive sleep apnoea. Thorax. 2018;73(5):472–480. doi:10.1136/thoraxjnl-2017-210431
5. Karimi M, Hedner J, Häbel H, Nerman O, Grote L. Sleep apnea-related risk of motor vehicle accidents is reduced by continuous positive airway pressure: Swedish traffic accident registry data. Sleep. 2015;38(3):341–349. doi:10.5665/sleep.4486
6. de Chazal P, Sutherland K, Cistulli PA. Advanced polysomnographic analysis for OSA: a pathway to personalized management? Respirology. 2020;25(3):251–258. doi:10.1111/resp.13564
7. Hirshkowitz M. Polysomnography challenges. Sleep Med Clin. 2016;11(4):403–411. doi:10.1016/j.jsmc.2016.07.002
8. Mendonça F, Mostafa SS, Ravelo-García AG, Morgado-Dias F, Penzel T. Devices for home detection of obstructive sleep apnea: a review. Sleep Med Rev. 2018;41:149–160. doi:10.1016/j.smrv.2018.02.004
9. BaHammam AS, Sharif M, Gacuan DE, George S. Evaluation of the accuracy of manual and automatic scoring of a single airflow channel in patients with a high probability of obstructive sleep apnea. Med Sci Monit. 2011;17(2):MT13–MT19. doi:10.12659/msm.881379
10. Aurora RN, Swartz R, Punjabi NM. Misclassification of OSA severity with automated scoring of home sleep recordings. Chest. 2015;147(3):719–727. doi:10.1378/chest.14-0929
11. Kuna ST, Benca R, Kushida CA, et al. Agreement in computer-assisted manual scoring of polysomnograms across sleep centers. Sleep. 2013;36(4):583–589. doi:10.5665/sleep.2550
12. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi:10.1038/nature14539
13. Urtnasan E, Park JU, Joo EY, Lee KJ. Automated detection of obstructive sleep apnea events from a single-lead electrocardiogram using a convolutional neural network. J Med Syst. 2018;42(6):104. doi:10.1007/s10916-018-0963-0
14. Jiang D, Ma Y, Wang Y. A multi-scale parallel convolutional neural network for automatic sleep apnea detection using single-channel EEG signals.
15. Haidar R, Koprinska I, Jeffries B. Sleep apnea event detection from nasal airflow using convolutional neural networks. In: Liu D, Xie S, Li Y, Zhao D, El-Alfy ES, editors. Neural Information Processing. ICONIP 2017. Lecture Notes in Computer Science. Cham: Springer; 2017:10638. doi:10.1007/978-3-319-70139-4_83
16. Van Steenkiste T, Groenendaal W, Deschrijver D, Dhaene T. Automated sleep apnea detection in raw respiratory signals using long short-term memory neural networks. IEEE J Biomed Health Inform. 2019;23(6):2354–2364. doi:10.1109/JBHI.2018.2886064
17. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi:10.1162/neco.1997.9.8.1735
18. Yu Y, Si X, Hu C, Zhang JA. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 2019;31(7):1235–1270. doi:10.1162/neco_a_01199
19. Berry RB, Brooks R, Gamaldo CE, Harding SM, Marcus C, Vaughn B. The AASM Manual for the Scoring of Sleep and Associated Events. Rules, Terminology and Technical Specifications. Illinois, Darien: American Academy of Sleep Medicine; 2012.
20. Duce B, Milosavljevic J, Hukins C. The 2012 AASM respiratory event criteria increase the incidence of hypopneas in an adult sleep center population. J Clin Sleep Med. 2015;11(12):1425–1431. doi:10.5664/jcsm.5280
21. Casey KR, Tiwari R. The (still) elusive definition of hypopnea. J Clin Sleep Med. 2018;14(12):1971–1972. doi:10.5664/jcsm.7512
22. Korotinsky A, Assefa SZ, Diaz-Abad M, Wickwire EM, Scharf SM. Comparison of American Academy of Sleep Medicine (AASM) versus Center for Medicare and Medicaid Services (CMS) polysomnography (PSG) scoring rules on AHI and eligibility for continuous positive airway pressure (CPAP) treatment. Sleep Breath. 2016;20(4):1169–1174. doi:10.1007/s11325-016-1327-y
23. Arora S, Collen JF. What are we missing? Hypertension and hypopneas. J Clin Sleep Med. 2020;16(10):1653–1654. doi:10.5664/jcsm.8764
24. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition.
25. Chen Q, Yue H, Pang X, et al. Mr-ResNeXt: a multi-resolution network architecture for detection of obstructive sleep Apnea.
26. Kiymik MK, Güler I, Dizibüyük A, Akin M. Comparison of STFT and wavelet transform methods in determining epileptic seizure activity in EEG signals for real-time application. Comput Biol Med. 2005;35(7):603–616. doi:10.1016/j.compbiomed.2004.05.001
27. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22(3):276–282. doi:10.11613/BM.2012.031
28. Choi SH, Yoon H, Kim HS, et al. Real-time apnea-hypopnea event detection during sleep by convolutional neural networks. Comput Biol Med. 2018;100:123–131. doi:10.1016/j.compbiomed.2018.06.028
29. Mazzotti DR, Lim DC, Sutherland K, et al. Opportunities for utilizing polysomnography signals to characterize obstructive sleep apnea subtypes and severity. Physiol Meas. 2018;39(9):09TR01. doi:10.1088/1361-6579/aad5fe
30. Dey D, Chaudhuri S, Munshi S. Obstructive sleep apnoea detection using convolutional neural network based deep learning framework. Biomed Eng Lett. 2018;8:95–100. doi:10.1007/s13534-017-0055-y
31. Vaquerizo-Villar F, Alvarez D, Kheirandish-Gozal L, et al. Convolutional neural networks to detect pediatric apnea-hypopnea events from oximetry. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:3555–3558. doi:10.1109/EMBC.2019.8857934
32. Álvarez D, Cerezo-Hernández A, Crespo A, et al. A machine learning-based test for adult sleep apnoea screening at home using oximetry and airflow. Sci Rep. 2020;10(1):5332. doi:10.1038/s41598-020-62223-4
33. Gutiérrez-Tobal GC, Álvarez D, Del Campo F, Hornero R. Utility of AdaBoost to detect sleep apnea-hypopnea syndrome from single-channel airflow. IEEE Trans Biomed Eng. 2016;63(3):636–646. doi:10.1109/TBME.2015.2467188
34. Miao F, Wen B, Hu Z, et al. Continuous blood pressure measurement from one-channel electrocardiogram signal using deep-learning techniques. Artif Intell Med. 2020;108:101919. doi:10.1016/j.artmed.2020.101919
35. Cheng M, Sori WJ, Jiang F, Khan A, Liu S. Recurrent neural network based classification of ECG signal features for obstruction of sleep apnea detection.
36. Almuhammadi WS, Aboalayon KAI, Faezipour M. Efficient obstructive sleep apnea classification based on EEG signals.
37. Nikkonen S, Afara IO, Leppänen T, Töyräs J. Artificial neural network analysis of the oxygen saturation signal enables accurate diagnostics of sleep apnea. Sci Rep. 2019;9(1):13200. doi:10.1038/s41598-019-49330-7
38. Hafezi M, Montazeri N, Zhu K, Alshaer H, Yadollahi A, Taati B. Sleep apnea severity estimation from respiratory related movements using deep learning. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:1601–1604. doi:10.1109/EMBC.2019.8857524
39. Solà-Soler J, Fiz JA, Morera J, Jané R. Multiclass classification of subjects with sleep apnoea-hypopnoea syndrome through snoring analysis. Med Eng Phys. 2012;34(9):1213–1220. doi:10.1016/j.medengphy.2011.12.008
40. Perero-Codosero JM, Espinoza-Cuadros F, Antón-Martín J, Barbero-álvarez MA, Hernández-Gómez LA. Modeling obstructive sleep apnea voices using deep neural network embeddings and domain-adversarial training. IEEE J Sel Top Signal Process. 2020;14(2):240–250. doi:10.1109/JSTSP.2019.2957977
41. Penzel T, McNames J, Murray A, de Chazal P, Moody G, Raymond B. Systematic comparison of different algorithms for apnoea detection based on electrocardiogram recordings. Med Biol Eng Comput. 2002;40(4):402–407. doi:10.1007/BF02345072
42. Erdenebayar U, Kim YJ, Park JU, Joo EY, Lee KJ. Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram. Comput Methods Programs Biomed. 2019;180:105001. doi:10.1016/j.cmpb.2019.105001
43. Haghayegh S, Kang HA, Khoshnevis S, Smolensky MH, Diller KR. A comprehensive guideline for bland-altman and intra class correlation calculations to properly compare two methods of measurement and interpret findings. Physiol Meas. 2020;41(5):055012. doi:10.1088/1361-6579/ab86d6
44. Montazeri Ghahjaverestan N, Akbarian S, Hafezi M, et al. Sleep/wakefulness detection using tracheal sounds and movements. Nat Sci Sleep. 2020;12:1009–1021. doi:10.2147/NSS.S276107
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.