Back to Journals » Journal of Pain Research » Volume 13
Decreased MiR-485-5p Contributes to Inflammatory Pain Through Post-Transcriptional Upregulation of ASIC1 in Rat Dorsal Root Ganglion
Authors Xu M, Wu R, Zhang L, Zhu HY, Xu GY , Qian W, Zhang PA
Received 4 September 2020
Accepted for publication 22 October 2020
Published 19 November 2020 Volume 2020:13 Pages 3013—3022
DOI https://doi.org/10.2147/JPR.S279902
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor E Alfonso Romero-Sandoval
Meijie Xu,1 Rui Wu,1,2 Ling Zhang,1 Hong-Yan Zhu,1 Guang-Yin Xu,1,2 Wenxia Qian,1 Ping-An Zhang1,2
1Department of Respiratory and Critical Care Medicine, Affiliated Zhangjiagang Hospital of Soochow University, Zhangjiagang 215600, People’s Republic of China; 2Center for Translational Pain Medicine, Institute of Neuroscience, Soochow University, Suzhou 215123, People’s Republic of China
Correspondence: Ping-An Zhang
Center for Translational Pain Medicine, Institute of Neuroscience, Soochow University, Suzhou 215123, People’s Republic of China
Tel +86-15006281769
Email [email protected]
Wenxia Qian
Department Of Respiratory and Critical Care Medicine, Affiliated Zhangjiagang Hospital of Soochow University, Zhangjiagang 215600, People’s Republic of China
Tel +86-13952440813
Email [email protected]
Background: Inflammatory pain is the most common type of pain treated clinically. However, the currently available treatments for inflammatory pain have limited effects and can cause severe side effects. The aim of this study is to describe the effect of miRNA-485-5p on osteoarthritis-related inflammatory pain.
Methods: Paw withdrawal threshold (PWT) of rats was measured by von Frey filaments. The expressions of miRNA-485-5p and acid-sensing ion channel 1 (ASIC1) in the dorsal root ganglion (DRG) were measured with real-time quantitative PCR and Western blotting analysis. Fluorescent in situ hybridization and fluorescent immunohistochemistry were employed to detect expression of miRNA-485-5p, acid-sensing ion channelASIC1 and co-location of miRNA-485-5p with ASIC1.
Results: The PWT of rats was significantly reduced after complete Freund’s adjuvant (CFA) injection. The miRNA-485-5p expression level clearly decreased while the ASIC1 expression level was upregulated in the L4-6 dorsal root ganglion (DRG) of CFA rats. MiRNA-485-5p and ASIC1 were co-expressed in the same DRG cells of CFA rats. Amiloride, an inhibitor of ASIC1, clearly increased the PWT of CFA rats. Further, miRNA-485-5p agomir reversed the upregulation of ASICI1 and alleviated CFA-induced mechanical hypersensitivity of CFA rats.
Conclusion: These results suggest that reduced expression of miRNA-485-5p contributes to inflammatory pain through upregulating ASIC1 expression, implying a promising strategy for pain therapy.
Keywords: miR-485-5p, ASIC1, dorsal root ganglion (DRG), inflammatory pain
Introduction
Chronic pain is the predominant symptom for people with inflammatory arthritis and osteoarthritis (OA).1 The treatment for OA-related inflammatory pain is still very limited due to the complicated pathological mechanism. Therefore, the identification of specific inflammatory mediators and characterization of their mechanism of action is of utmost importance for the development of selective and effective therapeutic approaches to inflammatory pain.2 MiRNAs, endogenous small non-coding RNA molecules, contain about 22 nucleotides.3,4 They function in RNA silencing and post-transcriptional regulation of gene expression.5 The dysregulation of miRNAs has been associated with many diseases such as cancer and obesity, as well as diseases of chronic pain. It is reported that dysregulation of miR-1224, miR-46 and miR-124 participated in CFA-induced inflammatory pain.6–9 Further, MiR-485-5p is reported to be involved in OA development and progression,10,11 however, the effect of miR-485-5p on OA-related inflammatory pain remains unclear.
Acid-sensing ion channels (ASICs) are voltage-insensitive, proton-gated cation channels activated by extracellular acidosis.12 In rodents, four genes encode at least six different subunits and three identical or different subunits are required to form a functional channel.12,13 Acid-sensing ion channels are expressed in various cells of the body, especially in the central and peripheral nervous systems.14 Importantly, ASIC1 is one of the targets of miR-485-5p predicted by bioinformatics, and recent studies have demonstrated the influence of ASIC channels on the symptoms of inflammatory pain,15,16 whose pathophysiological mechanism is related to changes in pH.1 However, the regulation of ASIC1 in inflammatory pain remains unknown.
Here, we assume that inflammation induced by CFA suppresses miR-485-5p expression and subsequently leads to an increase in ASIC1 expression in dorsal root ganglions (DRGs), which contributes to pain syndromes of rats. In the present study, persistent inflammation was induced by injecting complete Freund’s adjuvant (CFA) subcutaneously to the plantar surface of the left hind paw. The results of the present study suggest a miR-485-5p/ASIC1-mediated crucial target for the treatment of inflammatory pain in patients with osteoarthritis.
Materials and Methods
Animals
The experiments were performed on male Sprague Dawley rats weighing 180–200 g, which were provided by Shanghai SLAC Laboratory Animal Company Ltd. Rats were kept under controlled conditions (24 ± 2°C, 06:00–18:00 lighting) and had free access to both food and water. For behavioral studies, 1 h before recording, the animals were placed in a wire-bottom cage. All experiments were performed according to the approved guidelines of the Institutional Association for the Study of Pain.
Inflammatory Pain Rat Model and Nociceptive Testing
Persistent inflammation was induced by injecting 100 ul of complete Freund’s adjuvant (CFA) subcutaneously to the plantar surface of the left hind paw, while the rats were briefly anesthetized with isoflurane. Control rats received an injection of normal saline (NS). The rats were placed on an elevated mesh grid that completely exposed the middle of the hind paw. Prior to each testing session, the animals were habituated to the testing environment for at least 30 min. Mechanical hypersensitivity was assessed using calibrated von Frey filaments by those blinded to the group assignments. The behavioral responses were used to calculate the paw-withdrawal threshold, by fitting a Gaussian integral psychometric function using a maximum-likelihood fitting method. This fitting method allowed parametric statistical analysis. The filaments had the following log-stiffness values: Positive signs of withdrawal included pulling back rapidly, biting and shaking the hind limb within 5 sof the hind limb being pricked by one of the von Frey filaments. The interval between trials was at least 3 min. For each trial, the same hind limb was stimulated 5 times by a single von Frey filament before being stimulated by the next larger filament. The smallest value of filament that induced positive signs after pricking the rat hind limb was recorded. The rats were tested before CFA injection to determine a baseline value for each animal and were then tested at the time of 8 h and at 1, 3, 7 days after CFA injection.
Drug Administration
For behavioral experiments, miR-485-5p and NC agomir/antagomir (purchased from GenePharma) were delivered by intrathecal injection. Amiloride was diluted with normal saline (NS). Inflammatory pain threshold was measured after drug microinjection and tested until the effect of the drug disappeared. The doses of the drug used in the study were chosen according to previous reports.
Western Blotting
The animals were euthanized with isoflurane and then total extracts from ipsilateral L4-6 DRG tissues was immediately isolated and frozen in liquid. Protein levels were assayed by Western blotting. Briefly, a 10% separating gel and stacking gel were prepared the day prior to running the Western blot. An equal volume of loading buffer with 5% β-mercaptoethanol to each tissue sample was prepared and boiled for 10 min. An equal dose of protein from each sample was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, PVDF membrane was used to transfer protein. The membranes were then probed with monoclonal rabbit anti-ASIC1 (YM3487, 1/500, Immunoway), or polyclonal rabbit anti-GAPDH (sc-25,778, 1/1000, Santa). Densitometric analyses of proteins were performed using ImageJ software. The ASIC1 protein (~72 KD) expression was normalized to GAPDH (~37 KD).
Real-Time Quantitative PCR (qPCR)
Total RNA used for the present study was extracted from DRG of control and inflammatory rats with TRIzol (15,596,026, Ambion, Shanghai, China). Reverse transcription of miR-485-5p and U6 from total RNA was carried out using a miRNA selective reverse transcription kit (Applied Biosystems by Thermo Fisher Scientific), other cDNA was synthesized by total RNA using a Reverse transcription kit (AE301-03, Transgen Biotech, Beijing, China) following the supplier’s instructions. The primer sequences used here are listed in Table 1. The PCR reaction were performed in triplicate for each experiment. QPCR assays were implemented in a Quantstudio Dx Real-Time PCR instrument (Applied Biosystems, USA). The relative results were calculated using the 2−ΔΔCT equation.
 |
Table 1 Primer Sequences |
Fluorescence in situ Hybridization (FISH) and Immunofluorescence Assay
First, rats were deeply anesthetized with isoflurane. L4-L6 DRGs were then removed from the rats and postfixed for 0.5 h with paraformaldehyde and dehydrated in 10%, 20% and 30% sucrose (Sinopharm Chemical Reagent Co. Ltd, dissolved with PBS) in succession until the DRGs sank to the bottom of the 30% sucrose solution. DRGs were then cut into slices about 15 μm with a freezing microtome (Leika).
Probe for miR-485-5p was synthesized by Qiagen. The specific sequences of miR-485-5p were as follows: 5ʹ-AGAGGCUGGCCGUGAUGAAUUC-3ʹ. FISH was performed using a FISH kit (Boster, MK1030). Sections were treated with 30% H2O2 in methanol (1:50) for 30 min and washed in DEPC-treated water 3 times. Sections were pretreated with proteinase K at 37°C for 2 min and pre-hybridized with pre-hybridize buffer at 37°C for 2–4 h. Then sections were hybridized with probes (100 nM) in hybridization buffer at 40°C overnight. Sections were rinsed with 2× SSC, and subsequently washed with 0.5× SSC, then washed with 0.2× SSC. Sections were incubated with blocking solution at 37°C for 30 min, and then with a mouse anti-digoxigenin antibody at 37°C for 1 hour. Then sections were washed in 0.5 M PBS for 3 times and incubated with SABC-CY3 (POD) at 37°C for 30 mins. Sections were then stained using immunofluorescence staining method.
To perform immunofluorescence, the sections were rinsed in PBS three times (5 min each), and blocked for 1 h at 37°C then incubated with primary antibodies overnight at 4°C and then incubated with secondary antibody for 2 h at room temperature. Negative controls were performed without the primary antibody. The primary antibody used in our experiment is ASIC1 (YM3487,1/500, Immunoway). The slides were imaged using fluorescent microscopy, and the pictures were trimmed with AxioVision.
Quantification and Statistics
In all experiments, animals were randomly assigned to groups. In behavioral experiments, measurements were performed with the experimenter blind to the drug condition. All values are expressed as mean ± SEM, and no samples/animals were excluded from analysis. All reported statistical analyses were justified based on sample size, homogeneity and variance, and normal distribution of the data. The p-values reported for simple main effects were the adjusted p-values for multiple comparisons. The differences with a p value of 0.05 or less were considered significant.
Results
MiR-485-5p Expression Was Decreased in L4-6 DRGs of Rats with Persistent Inflammatory Pain
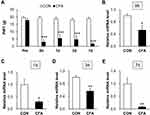
Inflammatory pain was induced in adult rats by single intra-plantar injection with CFA. Firstly, we assessed baseline values for the paw withdrawal threshold (PWT) of rats before CFA injection. The values were 17.73 ± 1.33 (n = 7 rats) and 19.54 ± 0.76 (n = 6 rats) for CFA-treated (CFA) and normal saline-treated (CON) rats, statistical analysis showed that they did not significant differ. Then we determined PWT of CFA and CON rats from 8 hours to 7 days following injection. The values of 8 h, 1 d, 3 d and 7 d after injection were 3.01 ± 1.40 (n = 7 rats), 5.53 ± 2.51 (n = 7 rats), 4.70 ± 2.60 (n = 7 rats), 1.77 ± 0.16 (n = 7 rats) for CFA rats, respectively. The relative values of 8 h, 1 d, 3 d and 7 d were 19.54 ± 0.76 (n = 6 rats), 18.79 ± 0.96 (n = 6 rats), 18.03 ± 1.02 (n =6 rats), 18.79 ± 0.96 (n = 6 rats) for CON rats. Statistical analysis showed that the PWT of CFA rats in response to mechanical stimulation was significantly decreased at these time points compared with CON rats (Figure 1A, ***p < 0.001 vs CON, two-way ANOVA).
To determine whether microRNAs are involved in this setting, we measured the expression of miR-485-5p in L4-L6 DRGs by qPCR. The relative miRNA level of miR-485-5p at the times of 8 h, 1 d, 3 d and 7 d after injection were 0.53 ± 0.15 (n = 3 rats), 0.30 ± 0.05 (n = 4 rats), 0.47 ± 0.11 (n = 4 rats), 0.08 ± 0.02 (n = 4 rats) in CFA rats. The relative miRNA level of miR-485-5p at the times of 8 h, 1 d, 3 d and 7 d after injection were 1.00 ± 0.05 (n = 3 rats), 1.00 ± 0.22 (n = 4 rats), 1.00 ± 0.08 (n = 5 rats), 1.00 ± 0.22 (n = 4 rats) in CON rats. There was a significant downregulation of miR-485-5p in L4-L6 DRGs when compared with CON rats at 8 h (*p < 0.05 vs CON, two sample t-test), 1 d (*p < 0.05 vs CON, two sample t-test), 3 d (**p < 0.01 vs CON, two sample t-test) and 7 d (**p < 0.01 vs CON, two sample t-test) (Figure 1B–E).
The Expression of MiR-7a, 23a, 31, 106, 125b and 181-5p Was Not Changed
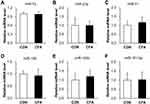
To further identify the involvement of miRNAs in inflammatory pain, qPCR was performed to check the expression levels of several pain-related miRNAs in L4-6 DRG at day 3 following injection. As shown in Figure 2, the expressions of miR-7a, miR-23a, miR-31, miR-106, miR-125b and miR-181-5p were not significantly changed (p > 0.05 vs CON, two sample t-test). The relative values of miR-7a, miR-23a, miR-31, miR-106, miR-125b and miR-181-5p in CFA rats were 0.98 ± 0.09, 1.00 ± 0.27, 1.16 ± 0.36, 0.93 ± 0.28, 1.19 ± 0.41 and 1.02 ± 0.43, respectively. The relative values of miR-7a, miR-23a, miR-31, miR-106, miR-125b and miR-181-5p in CON rats were 1 ± 0.05, 1 ± 0.43, 1 ± 0.22, 1 ± 0.12, 1 ± 0.36 and 1 ± 0.40. The above results indicate that these miRNAs may not be involved in CFA-induced inflammatory pain.
Targeted Genes of MiR-485-5p Enriched in Sensory Perception of Pain
To explore the mechanisms underlying the effects of miR-485-5p on CFA-induced inflammatory pain, we first predicted the potential target genes of miR-485-5p from miRWalk, miRanda, miRNAMap and Targetscan databases (Figure 3A). Of them, 189 genes were picked out to be further analyzed by GO. We found that 189 target genes were mainly involved in sensory perception of pain (Figure 3B, red bar) and protein phosphorylation (Figure 3B). We also found that acid-sensing ion channel 1 (ASIC1) is one of the genes connected with the sensory perception of pain. Furthermore, ASIC1 has been widely reported to be involved in regulating neuroinflammation in various pathological conditions.1,16 As neuroinflammation is a characteristic pathological change and an important cause of inflammatory pain, we focused our interest on studying the roles of ASIC1 under the regulation of miR-485-5p in the following experiments.
Inhibition of ASIC1 Attenuated Inflammatory Pain of CFA Rats
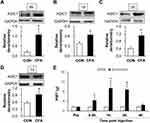
To verify the role of ASIC1 on inflammatory pain of CFA rats, the protein level of ASIC1 in L4-L6 DRGs at the times of 8 h, 1 d, 3 d and 7 d after CFA injection was detected by Western blotting. The relative densitometry of ASIC1 in inflammatory rats at the times of 8 h, 1 d, 3 d and 7 d after CFA injection were 1.08 ± 0.34 (n = 4 rats), 0.64 ± 0.09 (n = 3 rats), 1.30 ± 0.39 (n = 4 rats) and 0.93 ± 0.29 (n = 3 rats), the relative densitometry of ASIC1 in CON rats at the times of 8 h, 1 d, 3 d and 7 d after CFA injection were 0.20 ± 0.17 (n = 4 rats), 0.38 ± 0.10 (n = 3 rats), 0.69 ± 0.29 (n = 4 rats) and 0.58 ± 0.11 (n = 3 rats). Statistical analysis showed the protein expression of ASIC1 in L4-L6 DRGs was increased significantly at the times of 8 h, 1 d, 3 d and 7 d after CFA injection (Figure 4A–D, *p < 0.05 vs CON, two sample t-test). These results demonstrate that upregulation of ASIC1 may contribute to the mechanical pain of CFA rats.
To confirm the role of ASIC1 in inflammatory pain, we tested paw-withdrawal threshold before and after intrathecal injection of 10 μL amiloride, an inhibitor of ASIC1, or NS. Paw-withdrawal thresholds of inflammatory rats before the administration were 1.67 ± 0.14 (amiloride, n = 7 rats), 1.67 ± 0.15 (NS, n = 6 rats), respectively. At 0.5, 1, 2 and 4 h after treatment with amiloride, the thresholds were 4.34 ± 2.67 (n = 7 rats), 9.34 ± 3.08 (n = 7 rats), 8.78 ± 2.17 (n = 7 rats) and 2.45 ± 0.45 (n = 7 rats), respectively. At 0.5, 1, 2 and 4 h after delivery of NS, the thresholds were 1.61 ± 0 (n = 6 rats), 1.61 ± 0 (n = 6 rats), 1.74 ± 0.08 (n = 6 rats) and 1.67 ± 0.06 (n = 6 rats), respectively. Statistical analysis showed that there was a significant increase of threshold at 1 h and 2 h after injection of amiloride (Figure 4E, *p < 0.05 vs CON, two-way ANOVA). These results indicate that increased ASIC1 in L4-6 DRGs contributes to the inflammatory pain of CFA rats.
MiR-485-5p Overexpression Decreased ASIC1 Expression
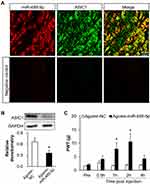
To confirm the regulation of miR-485-5p and ASIC1, immunofluorescence assay was conducted to detect their location in L4-6 DRG. As shown in Figure 5A, miR-485-5p (red) was mostly localized with ASIC1 (green). Additionally, we overexpressed miR-485-5p by intrathecal injection of its agomir and detected the protein expression of ASIC1 in L4-L6 DRGs. Western blot assays showed the relative densitometry of ASIC1 in inflammatory rats treated with miR- 485-5p agomir was 0.36 ± 0.20 (n = 7) and 0.66 ± 0.26 (n = 7) with agomir NC. Statistical analyses showed that ASIC1 protein expression was significantly reduced after intrathecal injection of miR-485-5p agomir when compared with NC group (Figure 5B, *p < 0.05 vs Agomir-NC, two sample t-test).
Overexpression of MiR-485-5p Alleviated Inflammatory Pain
To confirm the role of miR-485-5p in inflammatory pain, we tested paw-withdrawal threshold before and after intrathecal injection of 10 μL miR-485-5p agomir or negative control (NC). We first injected CFA and assessed baseline values at 3 days. The values were 2.07 ± 0.40 (n = 8 rats) and 1.67 ± 0.06 (n = 6 rats), statistical analysis showed that there was no significant difference (p > 0.05 vs agomir-NC, two-way ANOVA). Then the paw-withdrawal threshold was measured 30 min after intrathecal injection of miR-485-5p agomir or NC. The values of inflammatory rats at 0.5, 1, 2 and 4 hafter injection of miR-485-5p agomir were 4.11 ± 1.28, 7.85 ± 2.38, 10.54 ± 3.05 and 4.25 ± 1.25 (n = 8 rats), respectively. The values at 0.5, 1, 2 and 4 h after injection of agomir NC were 2.70 ± 0.70, 1.74 ± 0.08, 1.86 ± 0.19 and 1.99 ± 0.22 (n = 6 rats), respectively. Statistical analysis showed that there was a significant increase of paw-withdrawal threshold of inflammatory rats injected with miR-485-5p agomir when compared with agomir NC group at 1 and 2 h after injection (Figure 5C, *p < 0.05 vs agomir-NC, two-way ANOVA).
Discussion
Our present study adds new evidence to support the hypothesis that microRNAs are critical in the development of pain associated with inflammatory disorders.17 We demonstrated that miR-485-5p was involved in inflammatory pain in the present study based on the following observed results. Firstly, we showed that miR-485-5p was downregulated in L4-6 DRGs, and this decrease appeared at the times of 8 h, 1 d, 3 d and 7 d after CFA injection, which was well matched with the pain response of rats, suggesting a potential role of miR-485-5p in inflammatory pain. This result was then further confirmed by application of miR-485-5p agomir which significantly reverses paw withdrawal response threshold of CFA rats. We did not measure the miR-485-5p expression at a longer timepoint, but our data strongly suggested a potential involvement of miR-485-5p in early development of inflammatory pain. Additionally, the regulation mechanism of miR-485-5p in inflammatory pain remains unclear. It was reported that noncoding RNA such as circRNA and lncRNA target and affect miR-485-5p expression.18,19 Therefore, whether epigenetic regulation including these or other noncoding RNAs affect the function of miR-485-5p in inflammation needs to be investigated in future study.
MicroRNAs are involved in the regulation of cellular activity by targeting specific mRNAs and result in translation repression or degradation of the mRNAs.20–22 By bioinformatics analysis, we found 189 target genes of miR-485-5p. Further, GO analysis showed that the sensation of pain was one of the primary biological processes of these genes, and there were six genes involved this process, including ASIC1. There are findings suggesting that ASIC1 probably participates in both the direct activation of primary afferent fibers and indirect central sensitization.23 ASIC1 can perceive the acidic microenvironment induced by cell damage, immune response, or hypoxic metabolism in the inflammatory state,7,24 and ASICs can directly respond to acidic pH (below pH 7.0) in the DRG.23,25 Consistent with this, we proved here that ASIC1 in L4-L6 DRGs was involved in inflammatory pain based on the following observations. First of all, ASIC1 was upregulated at protein level in L4-L6 DRGs of rats at the times of 8 h, 1 d, 3 d after injection of CFA. Secondly and importantly, inhibition of ASIC1 by amiloride significantly reduced the paw withdrawal response threshold of inflammatory rats.
Additionally, we further established how miR-485-5p modulates ASIC1 expression. We showed by FISH experiments that miR-485-5p was co-localized with ASIC1, indicating an anatomical possibility of miR-485-5p in regulation of ASIC1 expression. Then, application of miR-485-5p agomir clearly reversed the upregulation of ASIC1 in L4-L6 DRGs at protein level. While we had not excluded the regulation effect of other miRNAs on ASIC1 in inflammatory pain, at least, the present study strongly suggested that miR-485-5p regulates ASIC1 expression through a post-transcriptional process. Further, we did not detect the expression of other targets of miR-485-5p in DRGs, so we cannot determine whether miR-485-5p also regulates the expression of other molecules affecting inflammatory pain.
Conclusions
In summary, miR-485-5p was downregulated in L4-L6 DRGs of inflammatory pain rats which was negatively correlated with the protein expression of ASIC1. MiR-485-5p may inhibit inflammatory pain by targeting ASIC1. The restoration of miR-485-5p expression may be an attractive strategy for inflammatory pain therapy.
Ethics Approval
This work was performed in accordance with the recommendations of the International Association for the Study of Pain (IASP). The protocol was approved by the Institutional Animal Care and Use Committee of Soochow University, P.R. China.
Acknowledgments
The present work was supported by grants from the National Natural Science Foundation of China (31730040, 81801115, 81801109, 81920108016), China Postdoctoral Science Foundation Grant (2018M642304), and from the Priority Academic Program Development of Jiangsu Higher Education Institutions of China. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
No conflicts of interest, financial or otherwise, are declared by the authors.
References
1. Geenen R, Overman CL, Christensen R, et al. EULAR recommendations for the health professional’s approach to pain management in inflammatory arthritis and osteoarthritis. Ann Rheum Dis. 2018;77:797–807. doi:10.1136/annrheumdis-2017-212662.
2. Barkai O, Puig S, Lev S, et al. Platelet-derived growth factor activates nociceptive neurons by inhibiting M-current and contributes to inflammatory pain. Pain. 2019;160(6):1281–1296. doi:10.1097/j.pain.0000000000001523.
3. Mo JL, Pan ZG, Chen X, et al. MicroRNA-365 knockdown prevents ischemic neuronal injury by activating oxidation resistance 1-mediated antioxidant signals. Neurosci Bull. 2019;35:815–825. doi:10.1007/s12264-019-00371-y.
4. Gao F, Wu H, Wang R, et al. MicroRNA-485-5p suppresses the proliferation, migration and invasion of small cell lung cancer cells by targeting flotillin-2. Bioengineered. 2019;10(1):1–12. doi:10.1080/21655979.2019.1586056.
5. Wang ZH, Liu T. MicroRNA21 meets neuronal TLR8: non-canonical functions of MicroRNA in neuropathic pain. Neurosci Bull. 2019;35(5):949–952. doi:10.1007/s12264-019-00366-9.
6. Sun W, Ma M, Yu H, Yu H. Inhibition of lncRNA X inactivate-specific transcript ameliorates inflammatory pain by suppressing satellite glial cell activation and inflammation by acting as a sponge of miR-146a to inhibit Nav1.7. J Cell Biochem. 2018;119(12):9888–9898. doi:10.1002/jcb.27310.
7. Pan Z, Li GF, Sun ML, et al. MicroRNA-1224 splicing circularRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. J Neurosci. 2019;39(11):2125–2143. doi:10.1523/JNEUROSCI.1631-18.2018.
8. Chen O, Donnelly CR, Ji RR. Regulation of pain by neuro-immune interactions between macrophages and nociceptor sensory neurons. Curr Opin Neurobiol. 2020;62:17–25. doi:10.1016/j.conb.2019.11.006.
9. Huang S, Ge X, Yu J, et al. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018;32(1):512–528. doi:10.1096/fj.201700673R.
10. Chen HO, Zhang L, Tang ZY, Gong ZM. MiR-485-5p promotes the development of osteoarthritis by inhibiting cartilage differentiation in BMSCs. Eur Rev Med Pharmacol Sci. 2018;22:3294–3302. doi:10.26355/eurrev_201806_15148.
11. Tsai CH, Liu SC, Chung WH, Wang SW, Wu MH, Tang CH. Visfatin increases VEGF-dependent angiogenesis of endothelial progenitor cells during osteoarthritis progression. Cells. 2020;9(5):1315. doi:10.3390/cells9051315.
12. Lee CH, Chen CC. Roles of ASICs in nociception and proprioception. Adv Exp Med Biol. 2018;1099:37–47. doi:10.1007/978-981-13-1756-9_4.
13. Grunder S, Pusch M. Biophysical properties of acid-sensing ion channels (ASICs). Neuropharmacology. 2015;94:9–18. doi:10.1016/j.neuropharm.2014.12.016.
14. Rhoades JL, Nelson JC, Nwabudike I, et al. ASICs mediate food responses in an enteric serotonergic neuron that controls foraging behaviors. Cell. 2019;176(1–2):85–97e14. doi:10.1016/j.cell.2018.11.023.
15. Guo L, Zhao L, Ming P, Hong L, Liu A, Li R. Sumatriptan inhibits the electrophysiological activity of ASICs in rat trigeminal ganglion neurons. Eur J Pharmacol. 2018;841:98–103. doi:10.1016/j.ejphar.2018.10.013.
16. Lynagh T, Romero-Rojo JL, Lund C, Pless SA. Molecular basis for allosteric inhibition of acid-sensing ion channel 1a by ibuprofen. J Med Chem. 2017;60(19):8192–8200. doi:10.1021/acs.jmedchem.7b01072.
17. Lopez-Gonzalez MJ, Landry M, Favereaux A. MicroRNA and chronic pain: from mechanisms to therapeutic potential. Pharmacol Ther. 2017;180:1–15. doi:10.1016/j.pharmthera.2017.06.001.
18. Zhang Z, Li X, Li A, Wu G. miR-485-5p suppresses schwann cell proliferation and myelination by targeting cdc42 and Rac1. Exp Cell Res. 2020;388(1):111803. doi:10.1016/j.yexcr.2019.111803.
19. Lai J, Xin J, Fu C, Zhang W. CircHIPK3 promotes proliferation and metastasis and inhibits apoptosis of renal cancer cells by inhibiting MiR-485-3p. Cancer Cell Int. 2020;20(1):248. doi:10.1186/s12935-020-01319-3.
20. Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019;20(24):6249. doi:10.3390/ijms20246249.
21. Cao T, Zhen XC. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci Ther. 2018;24(7):586–597. doi:10.1111/cns.12840.
22. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034.
23. Fu H, Fang P, Zhou HY, et al. Acid-sensing ion channels in trigeminal ganglion neurons innervating the orofacial region contribute to orofacial inflammatory pain. Clin Exp Pharmacol Physiol. 2016;43(2):193–202. doi:10.1111/1440-1681.12510.
24. Lee JYP, Saez NJ, Cristofori-Armstrong B, et al. Inhibition of acid-sensing ion channels by diminazene and APETx2 evoke partial and highly variable antihyperalgesia in a rat model of inflammatory pain. Br J Pharmacol. 2018;175(12):2204–2218. doi:10.1111/bph.14089.
25. Cai Q, Qiu CY, Qiu F, et al. Morphine inhibits acid-sensing ion channel currents in rat dorsal root ganglion neurons. Brain Res. 2014;1554:12–20. doi:10.1016/j.brainres.2014.01.042.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.