Back to Journals » OncoTargets and Therapy » Volume 10
Decrease of RBM4 indicates poor prognosis in patients with hepatocellular carcinoma after hepatectomy
Received 20 October 2016
Accepted for publication 19 November 2016
Published 11 January 2017 Volume 2017:10 Pages 339—345
DOI https://doi.org/10.2147/OTT.S125250
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Jian-yao Chen,1 Li-ping Liu,2 Jiang-feng Xu3
1Department of Hepatobiliary Surgery, Shaoxing Second Hospital, Shaoxing, 2Department of Surgery, Zhuzhou Clinical Institute, Central South University School of Medicine, Zhuzhou, 3Department of Surgery, The Fourth Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang, People’s Republic of China
Abstract: RNA-binding motif 4 (RBM4) has been reported to play an important role in many human tumors such as lung cancer, breast cancer, ovarian cancer and gastric cancer by regulating alternative splicing and messenger RNA (mRNA) translation. However, little is known about the role of RBM4 in the development of hepatocellular carcinoma (HCC). This study aimed to investigate the expression of RBM4 in HCC tissues. Expression of RBM4 was detected by immunohistochemistry in 95 cases of HCC. Correlations of RBM4 expression with the overall survival and disease-free survival of HCC were also studied. Patients with high RBM4 expression had better overall survival rate and disease-free survival rate than those with low RBM4 expression (P<0.001, P=0.007, respectively). RBM4 expression, together with tumor numbers, capsular formation, vascular invasion and Barcelona clinic liver cancer (BCLC) stage, was an independent prognostic factor for overall survival rate and disease-free survival rate of HCC. Our data implicate RBM4 as a novel prognostic marker and a potential therapeutic target for HCC.
Keywords: RBM4, hepatocellular carcinoma, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers.1 The overall survival rate of patients with advanced HCC remains unsatisfactory as a high incidence of recurrence and metastasis after liver resection has been noted.2,3 Therefore, it is very important to elucidate the underlying mechanisms and find novel biological markers for prognosis of HCC.
Recently, Wang et al4 reported that RNA-binding motif 4 (RBM4) suppresses proliferation and migration in various cancers by specifically controlling cancer-related splicing. The study indicated that RBM4, as a tumor suppressor, had therapeutic potential and clinical value. However, there is no report on the study of RBM4 expression in HCC. Roles of RBM4 in development and prognosis of HCC remain unknown. This study aimed to study the relationship between RBM4 expression and the prognosis of HCC after liver resection.
Methods
Specimens
HCC and the adjacent non-tumor liver tissue specimens were obtained from 95 HCC patients (without any preoperative treatment) during tumor resection surgery at Department of Hepatobiliary Surgery, Shaoxing Second Hospital and Department of Surgery, Zhuzhou Clinical Institute, Central South University School of Medicine, from January 2010 to December 2015. Preoperative evaluation protocol included blood biochemistry, chest radiography, liver and renal function tests, ultrasonography and computed tomography scan. Liver resection was performed in the patients with good cardiopulmonary and renal function and Pugh–Child’s grades A and B. All patients were followed up for tumor recurrence by clinical examination, alpha feto protein and ultrasonography.
Participants
Our research was in compliance with the Helsinki Declaration. The ethics approval and consent in study were approved by the Ethics Committee of Shaoxing Second Hospital, Zhuzhou Clinical Institute, Central South University School of Medicine and the Fourth Affiliated Hospital, Zhejiang University School of Medicine. Prior written informed consent was obtained from all patients for this study.
Immunohistochemistry
Formalin-fixed paraffin sections were stained for RBM4 using anti-human RBM4 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and the streptavidin–peroxidase system (Zhongshan Goldenbridge Biotechnology, Beijing, People’s Republic of China). Negative control slides were probed with goat serum followed by the secondary antibody (Zhongshan Goldenbridge Biotechnology) under the same conditions. The expression levels of RBM4 were scored using a 4-point scale according to the percentage of positive hepatocytes: 0, the percentage of positive hepatocytes was no more than 10%; 1, the percentage of positive hepatocytes was 10%–25%; 2, the percentage of positive hepatocytes was 25%–50%; 3, the percentage of positive hepatocytes was more than 50%. The protein expression of RBM4 was thus considered as low expression if the score was 0 or 1; a score of 2 or 3 was considered as high expression. RBM4 expression in HCC specimens was also divided into low-expression group (score 0 or 1) and high-expression group (score 2 or 3).
Statistical analysis
All data were analyzed using the statistical software Statistical Package for the Social Sciences (SPSS) 19 for Windows (SPSS Inc., Chicago, IL, USA). Continuous variables were expressed as mean ± standard deviation and compared using the independent-samples t-test. Survival analysis was estimated by the Kaplan–Meier survival method and compared using the log-rank test. The log-rank was compared between the survival of patients with low and high RBM4 expression. Multivariate analysis by the Cox proportional hazard regression model was used to identify independent prognostic factors. P<0.05 was considered to be statistically significant.
Results
Clinicopathologic features of patients
Ninety-five HCC patients without any preoperative treatment received liver resection from January 2010 to December 2015. Of the 95 patients (Table 1), there were 75 patients with hepatitis B infection. Sixty-one patients had liver cirrhosis, while 34 patients were without liver cirrhosis. Forty-two patients were with solitary tumors, while 53 patients were with multiple tumors. Thirty-five patients were with capsular formation, while 60 patients were without capsular formation. Twenty-five patients had vascular invasion. Forty-one patients belonged to UICC stage I, while 54 patients belonged to UICC stage II or III. Thirty-two patients belonged to Barcelona clinic liver cancer (BCLC) stages 0–A while, 63 patients belonged to BCLC stages B–C.
Association between RBM4 expression and the clinicopathologic characteristics of HCC
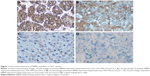
Immunohistochemistry showed that RBM4 expression was significantly low (25.3%) in HCC tissues (Table 1). According to the immunohistochemistry results, all HCC patients were divided into two groups: the high-expression group in which RBM4 expression scored 2 or 3 (n=24) (Figure 1) and the low-expression group in which RBM4 expression scored 1 or 0 (n=71). We found that RBM4 expression negatively correlates with hepatitis B infection (P=0.039). The RBM4 expression in patients with liver cirrhosis was significantly lower than in those without liver cirrhosis (P=0.033). The RBM4 expression also has significant relationship with tumor number (P=0.047). The patients with capsular formation had higher RBM4 expression than those without capsular formation (P=0.041). The expression of RBM4 in patients with vascular invasion was lower than those without vascular invasion (P=0.003). The patients in UICC stage I had higher RBM4 expression than the patients in stages II–III (P=0.046). The RBM4 expression level also had significant relationship with BCLC stage (P=0.001) (Table 1).
Association between RBM4 expression and the overall survival rate of HCC
Overall survival rate was analyzed according to the expression of RBM4 in 95 cases of HCC. RBM4 high-expression group had better overall survival rate than the low-expression group (P<0.001) (Figure 2A). In addition, the results from the univariate Cox regression analysis indicated that RBM4 expression (relative risk, 2.016; P=0.022), together with tumor numbers (relative risk, 1.962; P=0.033), absence of capsular formation (relative risk, 1.898; P=0.040), vascular invasion (relative risk, 2.468; P=0.001) and BCLC stages B–C (relative risk, 2.125; P=0.041), was an important prognostic factor for overall survival rate in HCC patients (Table 2). The multivariate Cox regression analysis showed similar results. RBM4 expression (relative risk, 1.693; P=0.033), together with tumor numbers (relative risk, 1.723; P=0.041), capsular formation (relative risk, 1.696; P=0.038), vascular invasion (relative risk, 2.698; P=0.003) and BCLC stage (relative risk, 2.058; P=0.045), was an independent prognostic factor for overall survival of HCC patients.
Association between RBM4 expression and the disease-free survival of HCC
Disease-free survival was also analyzed according to the expression of RBM4 in 95 cases of HCC (Table 3). RBM4 high-expression group had better disease-free survival rates than the low-expression group (P=0.007) (Figure 2B). In addition, the results from the univariate Cox regression analysis indicated that RBM4 expression (relative risk, 2.694; P=0.026) together with tumor numbers (relative risk, 2.168; P=0.037), capsular formation (relative risk, 1.683; P=0.031), vascular invasion (relative risk, 2.073; P=0.018) and BCLC stage (relative risk, 2.567; P=0.033) was an important prognostic factor for disease-free survival of HCC patients (Table 3). The multivariate Cox regression analysis showed the same result. RBM4 expression (relative risk, 1.683; P=0.038) together with tumor numbers (relative risk, 1.783; P=0.041), capsular formation (relative risk, 1.62; P=0.037), vascular invasion (relative risk, 2.284; P=0.021) and BCLC stage (relative risk, 2.382; P=0.041) was an independent prognostic factor for disease-free survival of HCC patients.
Discussion
The splicing factor RBM4 suppresses proliferation and migration in various cancers by specifically controlling cancer-related splicing.5–9 Wang et al4 reported that RBM4 regulates Bcl-x splicing to induce apoptosis and RBM4 expression is decreased dramatically in human cancer such as lung, breast and ovarian cancer. Recently, Yong et al10 found that RBM4 expression is also decreased dramatically in gastric cancer and a reduced RBM4 level is correlated with poor survival rate. However, it is still unknown about the RBM4 expression in HCC. In this study, our result showed that the RBM4 expression was low in HCC tissues. RBM4 high-expression group had better overall survival rate and disease-free survival rate than the low-expression group (P<0.001, P=0.007, respectively). Furthermore, RBM4 expression, together with tumor numbers, capsular formation, vascular invasion and BCLC stage, was as independent prognostic factor for overall and disease-free survival of HCC. These implicated RBM4 as a novel prognostic marker for HCC after liver resection. Interestingly, we found that RBM4 expression negatively correlates with hepatitis B infection and liver cirrhosis. As most HCC cases in People’s Republic China were caused by the HBV infection,11–14 RBM4 may play an important role in the development of HCC by regulating HBV-related cirrhosis. This part of work remains to be done in our future research.
Subsequently, we found that the RBM4 expression also had significant relationship with tumor number (P=0.047), capsular formation (P=0.041) and vascular invasion (P=0.003). Tumor number together with capsular formation and vascular invasion is related to the invasion and metastasis of HCC.15–17 Accordingly, we speculate that RBM4 may participant in the invasion and metastasis of HCC. There may remain new factors such as epithelial–mesenchymal transition. Future studies are needed to find out the exact mechanism.
Conclusion
In conclusion, our study showed for the first time that high expression of RBM4 correlated with better overall survival rate and disease-free survival rate in HCC patients after liver resection. Our data suggested RBM4 as a novel biomarker for HCC.
Acknowledgments
This work was supported by the grants from Natural Science Foundation of Zhejiang Province (LQ15H160007), Science and Technology Innovation Project of Shaoxing (2016CX017) and Scientific Research Project of Department of Education of Zhejiang Province (Y20141279).
Author contributions
Jian-yao Chen, Li-ping Liu and Jiang-feng Xu carried out the data collection and analysis and drafted the manuscript. All the authors read and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM. Cancer statistics. CA Cancer J Clin. 2011;57:43–66. | ||
Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512.e2. | ||
Mayer J, Auriol J, Muscari F, et al. Worst prognosis of hypovascular hepatocellular carcinoma. J Hepatol. 2010;52:227. | ||
Wang Y, Chen D, Qian H, et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26(3):374–389. | ||
Qi Y, Yu J, Han W, et al. A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tumour proliferation. Nat Commun. 2016;7:ncomms11840. | ||
Markus MA, Yang YH, Morris BJ. Transcriptome-wide targets of alternative splicing by RBM4 and possible role in cancer. Genomics. 2016;107(4):138–144. | ||
Gowen BG, Chim B, Marceau CD, et al. A forward genetic screen reveals novel independent regulators of ULBP1, an activating ligand for natural killer cells. Elife. 2015;4.pii:e08474. | ||
Liang YC, Lin WC, Lin YJ, Lin JC. The impact of RNA binding motif protein 4-regulated splicing cascade on the progression and metabolism of colorectal cancer cells. Oncotarget. 2015;6(35):38046–38060. | ||
Lin JC, Lin CY, Tarn WY, Li FY. Elevated SRPK1 lessens apoptosis in breast cancer cells through RBM4-regulated splicing events. RNA. 2014;20(10):1621–1631. | ||
Yong H, Zhu H, Zhang S, et al. Prognostic value of decreased expression of RBM4 in human gastric cancer. Sci Rep. 2016;6:28222. | ||
Tajiri K, Shimizu Y. New horizon for radical cure of chronic hepatitis B virus infection. World J Hepatol. 2016;8(21):863–873. | ||
Lin XJ, Lao XM, Shi M, et al. Changes of HBV DNA after chemoembolization for hepatocellular carcinoma and the efficacy of antiviral treatment. Dig Dis Sci. 2016;61(9):2465–2476. | ||
Höner Zu Siederdissen C, Cornberg M. Management of HBV and HBV/HDV-associated liver cirrhosis. Visc Med. 2016;32(2):86–94. | ||
Zhang YQ, Peng LJ, Cao YR, et al. Risk factors for hepatocellular carcinoma in cirrhotic patients with chronic hepatitis B. Genet Test Mol Biomarkers. 2016;20(9):535–543. | ||
Xu JF, Liu XY, Wang S, Wen HX. Surgical treatment for hepatocellular carcinoma with portal vein tumor thrombus: a novel classification. World J Surg Oncol. 2015;13:86. | ||
Xu JF, Liu XY. PIVKA-II is an independent prognostic factor for overall survival of HCC patients and maybe associated with epithelial-mesenchymal transition. J Hepatol. 2015;63(4):1040–1041. | ||
Liu XY, Xu JF. Liver resection for young patients with large hepatocellular carcinoma: a single center experience from China. World J Surg Oncol. 2014;12:175. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.