Back to Journals » International Journal of Nanomedicine » Volume 16
Cyclam-Modified Polyethyleneimine for Simultaneous TGFβ siRNA Delivery and CXCR4 Inhibition for the Treatment of CCl4-Induced Liver Fibrosis
Authors Ullah A , Chen G, Hussain A, Khan H , Abbas A , Zhou Z, Shafiq M , Ahmad S, Ali U , Usman M , Raza F , Ahmed A, Qiu Z , Zheng M, Liu D
Received 5 April 2021
Accepted for publication 1 June 2021
Published 1 July 2021 Volume 2021:16 Pages 4451—4470
DOI https://doi.org/10.2147/IJN.S314367
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Aftab Ullah, 1 Gang Chen, 2 Abid Hussain, 3, 4 Hanif Khan, 1 Azar Abbas, 5 Zhanwei Zhou, 5 Muhammad Shafiq, 6 Saleem Ahmad, 7 Usman Ali, 8 Muhammad Usman, 6 Faisal Raza, 8 Abrar Ahmed, 8 Zijie Qiu, 5 Maochao Zheng, 1 Daojun Liu 1
1Department of Pharmacy, Shantou University Medical College, Shantou, 515041, Guangdong, People’s Republic of China; 2Institute of Comparative Medicine, College of Veterinary Medicine, Yangzhou University, Yangzhou, 225009, Jiangsu, People’s Republic of China; 3School of Life Science, Key Laboratory of Molecular Medicine and Biotherapy, Beijing Institute of Technology, Beijing, 100081, People’s Republic of China; 4Chinese Academy of Sciences (CAS) Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety, CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology of China, Beijing, 100190, People’s Republic of China; 5School of Pharmacy, China Pharmaceutical University, Nanjing, 210028, Jiangsu, People’s Republic of China; 6Department of Cell Biology and Genetics, Shantou University Medical College, Shantou, Guangdong, 515041, people’s Republic of China; 7Department of Medicine, Shantou University Medical College Cancer Hospital, Shantou, People’s Republic of China; 8School of Pharmacy, Shanghai Jiaotong University, Shanghai, 200240, Shanghai, People’s Republic of China
Correspondence: Aftab Ullah; Daojun Liu Email [email protected]; [email protected]
Background: Liver fibrosis is a chronic liver disease with excessive production of extracellular matrix proteins, leading to cirrhosis, hepatocellular carcinoma, and death.
Purpose: This study aimed at the development of a novel derivative of polyethyleneimine (PEI) that can effectively deliver transforming growth factor β (TGFβ) siRNA and inhibit chemokine receptor 4 (CXCR4) for TGFβ silencing and CXCR4 Inhibition, respectively, to treat CCl 4-induced liver fibrosis in a mouse model.
Methods: Cyclam-modified PEI (PEI-Cyclam) was synthesized by incorporating cyclam moiety into PEI by nucleophilic substitution reaction. Gel electrophoresis confirmed the PEI-Cyclam polyplex formation and stability against RNAase and serum degradation. Transmission electron microscopy and zeta sizer were employed for the morphology, particle size, and zeta potential, respectively. The gene silencing and CXCR4 targeting abilities of PEI-Cyclam polyplex were evaluated by luciferase and CXCR4 redistribution assays, respectively. The histological and immunohistochemical staining determined the anti-fibrotic activity of PEI-Cyclam polyplex. The TGFβ silencing of PEI-Cyclam polyplex was authenticated by Western blotting.
Results: The 1H NMR of PEI-Cyclam exhibited successful incorporation of cyclam content onto PEI. The PEI-Cyclam polyplex displayed spherical morphology, positive surface charge, and stability against RNAse and serum degradation. Cyclam modification decreased the cytotoxicity and demonstrated CXCR4 antagonistic and luciferase gene silencing efficiency. PEI-Cyclam/siTGFβ polyplexes decreased inflammation, collagen deposition, apoptosis, and cell proliferation, thus ameliorating liver fibrosis. Also, PEI-Cyclam/siTGFβ polyplex significantly downregulated α-smooth muscle actin, TGFβ, and collagen type III.
Conclusion: Our findings validate the feasibility of using PEI-Cyclam as a siRNA delivery vector for simultaneous TGFβ siRNA delivery and CXCR4 inhibition for the combined anti-fibrotic effects in a setting of CCl 4-induced liver fibrosis.
Keywords: polyethyleneimine, cyclam, gene delivery, liver fibrosis, CXCR4
Corrigendum for this paper has been published
Introduction
Liver fibrosis is an essential characteristic of chronic liver diseases, which boosts the disease progression by damaging normal hepatic parenchyma.1 Liver fibrosis involves multiple cell types and signaling pathways. If the disease is not treated in time, it will lead to liver cirrhosis and cancer,2 which are the leading causes of mortalities globally.3 Unfortunately, because of lack of symptoms in the early onset of liver fibrosis and inadequate therapeutic options, liver replacement is the only treatment option. Thus, we need to develop an effective anti-fibrotic strategy to inhibit the fibrotic process and disease progression.4–8 Among current conventional treatment options, ursodeoxycholic acid, vitamin E, caffeine, chenodeoxycholic acid, and silymarin revealed improved therapeutic outcomes in liver fibrosis, but none of them displayed a potential survival benefit.9
Chemokines and chemokine receptors lead to a complex network in a variety of pathological conditions. Among the family of chemokines and their receptors, stromal-derived growth factor-1 (SDF-1) along with its CXCR4 causes cell proliferation, survival, angiogenesis, chronic diseases like cancer, and eventually death.10,11 Hepatic stellate cells (HSC) are key players involved in liver fibrosis.1,12 During liver damage, the upregulated SDF-1/CXCR4 axis activates HSC.13,14 The activated HSC results in overexpression of TGFβ, α-smooth muscle actin (α-SMA), extracellular matrix (ECM; mainly collagen), and liver fibrosis.15 Previous studies revealed promising results using cyclam like AMD3100 (a CXCR4 antagonist) to inhibit the SDF-1/CXCR4 axis for ameliorating liver fibrosis.14,16–19 Thus, inhibiting the upregulated SDF-1/CXCR4 axis prevents HSC activation and could be a treatment of choice for liver fibrosis.6,14,20
TGFβ is a major cytokine released by infiltrating mononuclear cells, hepatocytes, and HSC.21 The released TGFβ further activates HSC and results in ECM overproduction.22,23 Studies show that the regulation of TGFβ significantly ameliorates liver fibrosis.24,25 Silencing TGFβ expression can be an excellent treatment option to prevent the activation of HSC, ECM overproduction, and liver fibrosis.
Recently, RNA interference (RNAi)26,27 has emerged as a powerful therapeutic tool for treating numerous diseases28–30 like liver fibrosis, cirrhosis, and liver cancer by silencing the target genes.31–35 Targeted siRNA delivery (for gene silencing) is a complicated and challenging process and the lack of efficient and safe nanocarriers remains a major obstacle for its clinical application.36 Currently, cationic polymers are extensively being employed as siRNA delivery vectors because of their efficient complexation ability with siRNA (to form polyplexes) and resistance against enzymatic and serum degradation, thus promising safe siRNA delivery across cell membranes.37 Polyplexes offer multiple advantages, including versatility in functional modification, minimal immunogenicity, and low toxicity.38,39 Among cationic polymers, PEI is an ideal one with a well-reported transfection efficiency.40,41 Also, PEI efficiently facilitates the endosomal escape of the polyplex, in part because of its high cationic charge density contributing to its membrane-active characteristics. Still, such high cationic charge density leads to higher cytotoxicity, which is a key concern for PEI that limits its clinical applications. Numerous methods are employed to solve this issue, like changing the molecular architecture and molecular weight of the polymer,42,43 modification of amines using hydrophobic moieties,44 and complexing PEI with lipid scaffolds to form hybrid polycation liposomes.45,46
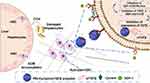
SDF-1/CXCR4 axis regulates chronic diseases such as cancer.10,11,47 The Inhibition of such interactions using a CXCR4 antagonist like cyclam (eg, AMD3100, a type of cyclam) revealed significant improvements.48,49 Previously, PEI-based polymeric CXCR4 antagonists have been employed as a dual functional delivery vector for simultaneous siRNA delivery and CXCR4 inhibition50,51 to constrain tumor angiogenesis and metastasis.52,53 Therapeutic strategies that can silence TGFβ expression and inhibit CXCR4 may be a potential therapeutic choice to treat liver fibrosis.20,25 Similarly, the goal of the present study is to present a novel strategy to modify PEI with a cyclam-based CXCR4 antagonist (to form PEI-Cyclam). The potential of PEI-Cyclam/siTGFβ polyplex as an anti-fibrotic treatment strategy that combines TGFβ silencing with simultaneous CXCR4 inhibition was evaluated in a setting of the CCl4-induced liver fibrosis in a mouse model (Scheme 1).
Materials and Methods
Cyclam was bought from Vesina Industrial company (Tianjin, China). Branched PEI (MW = 10 kDa) was purchased from Polysciences (Warrington, PA, USA). AMD3100 was obtained from Biochempartner (Shanghai, China). TGFβ siRNA (5-GCAACAACGCCAUCUAUGATT-3), FAM siRNA (5-UUCUCCGAACGUGUCACGUTT-3), scrambled siRNA (siScr) (5-UUCUCCGAACGUGUCACGUTT-3), siRNA targeting luciferase (5-GGACGAGGACGA GCACUUCUU-3) were acquired from Gene Pharma (Shanghai, China). Phosphate buffered saline (PBS) and the assay kits to measure aspartate aminotransferase (AST), alanine aminotransferase (ALT), bicinchoninic acid (BCA), and hydroxyproline (HYP) were obtained from Nanjing Jiancheng Bioengineering Institute (Nanjing, Jiangsu, China). CCl4 was from Sinopharm chemical reagent (Shanghai, China). Olive oil was bought from Agric-bemvig (Barcelona, Spain). FBS, trypsin, penicillin/streptomycin (Pen-Strep), and RPMI-1640 medium were purchased from Hyclone (Waltham, MA, USA). DMEM and LysoTracker™ Red were obtained from Gibco (CA, USA) and Beyotime (Shanghai, China). G418 and SDF-1 were bought from Life Technologies (Carlsbad, CA, USA). α-SMA, anti-TGFβ, anti-PCNA, anti-Col-III, and β-actin antibodies were purchased from Abcam (Cambridge, MA, USA). 3,3-Diaminobenzidine (DAB) kit was obtained from Boster biological (Hubei, China).
PEI-Cyclam Synthesis
PEI-Cyclam was synthesized following the previously reported method.53 Briefly, branched PEI (10 kDa), cyclam derivative, and K2CO3 were suspended in acetonitrile (20 mL) and followed by refluxing for 16 h. Then, 20 mL of trifluoroacetic acid (TFA) was added and followed by immediate stirring overnight to achieve deprotection. The obtained product was a hydrochloride salt after dialysis in water (pH 3.0) for 48 h, followed by another dialysis with deionized water for an additional 24 h before lyophilization. The final product was characterized by the technique of 1H NMR.
Preparation and Characterization of Polyplexes
The desired w/w ratios of PEI-Cyclam polyplex were obtained by mixing equal volumes of PEI-Cyclam with either siRNA TGFβ (siTGFβ) or siRNA scrambled (siScr) solution (20µg/mL) prepared in 20 mM HEPES buffer (pH 7.4). Then, the mixture was followed by immediate vortexing for 30 s and incubated for 30 min at room temperature. To determine the formation of polyplexes, the samples at various w/w ratios were loaded in the wells of agarose (2% Wt) gel containing JelRed, and run for 30 min. UV illumination was used for siRNA band detection. The hydrodynamic particle size and zeta potential were analyzed by dynamic light scattering and zeta potential analyzer (ZetaPlus, NY, USA). Briefly, PEI-Cyclam polyplex were obtained by mixing equal volumes of PEI-Cyclam with siRNA scrambled (siScr) solution (20µg/mL) prepared in 20 mM HEPES buffer (pH 7.4) and the obtained polyplexes were diluted with HEPES buffer before measuring the particle size and zeta potential. The morphology of polyplex was determined by using TEM (H-600, Hitachi, Ibaraki-ken, Japan).
In vitro Enzymatic and Serum Stability of Polyplexes
The protection ability of PEI-Cyclam against RNAse and serum degradation was investigated by using gel electrophoresis. For RNAse stability, different w/w ratios of PEI-Cyclam polyplexes were incubated with 2µL of RNAse (5U/µL) at 37 °C for 30 min. Then, a 2% sodium dodecyl sulfate (SDS) solution was added to each sample to extract siRNA from the polyplexes and followed by vortexing. The integrity of siRNA was detected on agarose (2% Wt) gel. For serum stability, PEI-Cyclam polyplexes (w/w ratio 1.5) were incubated with an equal volume of FBS at 37 °C. Then, 20 µL samples were collected at 0 h, 1 h, 2 h, 4 h, 8 h, 16 h, and 24 h, respectively. Then, a 2% SDS solution was added to each sample and vortexed for displacing siRNA from the complexes. Finally, the released siRNA was examined by gel electrophoresis.
Cell Culture
Murine breast 4T1 (stably expressing luciferase) cells and murine melanoma B16 (stably expressing luciferase) cells (ATCC, VA, USA) were cultured in RPMI-1640 (10% FBS and 1% Pen-Strep). Human epithelial osteosarcoma (U2OS) cells expressing EGFP-CXCR4 (Thermo Scientific, Waltham, MA, USA) were cultured in DMEM (10% FBS, 1% Pen-Strep, 2 mM L-Glutamine, and 0.5 mg/mL G418). Immortalized rat HSC-T6 (HSC-T6) cells were obtained from Shanghai FuMeng Gene Biotechnology Co., LTD. (Shanghai, China) and were cultured in DMEM (10% FBS and 1% Pen-Strep). The cells were maintained at 37 °C with 5% CO2 in a humidified incubator.
Cytotoxicity Assay
The toxicity of the synthesized polycations was determined by MTT assay using 4T1, B16, HSC-T6, and U2OS cells. Briefly, 200µL of complete medium (10% FBS) containing cells (8x103 cells per well) were added into 96-well plates and incubated overnight. The old medium was replaced with a complete fresh medium containing PEI, PEI-Cyclam, and PEI-Cyclam polyplexes for 24 h. Then, 20 µL of MTT reagent (5 mg/mL) was subsequently incorporated into the fresh medium and incubated for an additional 4 h. Finally, the medium was removed, and 150 μL of DMSO was added to each well. The cells were maintained at 37 °C with 5% CO2 in a humidified incubator. The absorbance was detected at 490 nm by using a microplate reader (BioTek Synergy, Montpelier, VT, USA).
CXCR4 Antagonism
The CXCR4 antagonism of PEI-Cyclam and polyplex was evaluated by using the CXCR4 redistribution assay. Briefly, 200 μL of U2OS cells were seeded (8x103 cells per well) in a 96 well plate and cultured for 24 h. Cells were washed twice with assay buffer (DMEM containing 2 mM L-glutamine, 1% FBS, 1% Pen-Strep, and 10 mM HEPES), and incubated with AMD3100 (300 nM), PEI (1μg/mL), PEI-Cyclam (2 μg/mL), and polyplexes for 30 min. Then, SDF-1 (final concentration 10 nM) was incorporated. After 1 h incubation, cells were then fixed (4% paraformaldehyde) for 20 min at room temperature, followed by washing and staining with PBS, and 1 μM Hoechst solution, respectively. Finally, cells were imaged using the FL microscope (Thermo Scientific, Waltham, MA, USA).
Cell Uptake and Endosomal Escape
To investigate the cell uptake and endosomal escape, fluorescently labeled FAM siRNA was employed. The cellular uptake of polyplex was studied by confocal laser scanning microscopy and flow cytometry. Briefly, for confocal laser scanning microscopy, HSC-T6 cells were seeded (5x104 cells per glass-bottom dish) and incubated for 24 h. Then, PEI/FAM siRNA polyplexes or PEI-Cyclam/FAM siRNA polyplex (FAM siRNA; 100 nM) was added to each well. After 2 h incubation, cells were washed twice (PBS) and fixed (4% paraformaldehyde) for 20 min at room temperature. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min, and cell uptake was observed under a confocal microscope (Carl Zeiss, Jena, TH, Germany). For flow cytometry analysis, the HSC-T6 cells were cultured and treated as above. After 2 h incubation, cellular uptake was stopped with ice-cold PBS. Then, the cells were collected by trypsinization, and fluorescence intensity for FAM siRNA in the cytoplasm of cells was measured by flow cytometer (FACS-Calibur, BD, Biosciences). For the observation of endosomal escape, cells were treated with polyplex for 2 h or 6 h, washed twice (PBS), and stained (LysoTracker™ Red) for 30 min. Finally, cells were washed thrice (PBS) and imaged using the confocal microscope.
Luciferase (Luc) Gene Silencing Assay
The Luc gene silencing ability of PEI-Cyclam/siLuc polyplex was investigated in 4T1 and B16 cells. Briefly, cells were cultured (5x104 cells per well) in a 48-well plate and incubated for 24 h. Then, cells were incubated with free siLuc, PEI/siLuc polyplexes, or PEI-Cyclam/siLuc polyplexes at various w/w ratios in a serum-free medium. After 4 h incubation, a fresh medium containing 10% FBS was added, and cells were further incubated for 24 h. Cells were then washed and lysed with PBS and lysis buffer, respectively. Finally, cell lysates were incubated with luciferase kit reagents, and the relative luciferase expression was detected by using a microplate reader.
The in vivo Anti-Fibrotic Activity of PEI-Cyclam/siTGFβ Polyplexes
BALB/C mice (20±2 g, 3–4 weeks old), provided by the Laboratory Animal Center Yangzhou University (Jiangsu, China), were maintained in a pathogen-free laboratory environment. All animal procedures were approved by the Animal Ethics Committee of Experimental Animal Center, Shantou University medical college, and carried out following the Regulations on Experimental Animals of Shantou University medical college (SUMC2016-200). The in vivo anti-fibrotic activity of PEI-Cyclam and PEI-Cyclam/siTGFβ polyplexes was evaluated in BALB/C mice. To induce liver fibrosis, BABL/C mice (n=5) received CCl4 (1mL/kg body weight) in olive oil (CCl4: olive oil; 1:9) by intraperitoneal injection (twice a week for 4 weeks). Mice received only olive oil by intraperitoneal route was used as controls. After 48 h of the last injection of CCl4, animals were killed to obtain blood and liver samples. Blood and liver samples were assayed to confirm the successful generation of the liver fibrosis model. To investigate the combined anti-fibrotic effect of TGFβ siRNA and PEI-Cyclam in vivo, mice with induced liver fibrosis were separated into 3 groups, which were intravenously administered with PBS (n=5) or PEI-Cyclam/siScr polyplex (n=5) (siScr 0.5 mg/kg), and PEI-Cyclam/siTGFβ polyplex (siTGFβ 0.5 mg/kg) (n=5) twice a week for four weeks. Mice were killed for blood and liver sample collection.
Effect of PEI-Cyclam/siTGFβ Polyplex on Liver Functions
Liver functions were evaluated by serological assay for serum biomarkers using AST and ALT kits. Briefly, the obtained blood samples were centrifuged (3000 rpm, 10 min) at 4 °C. The serum AST and ALT were detected by using a microplate reader.
Effect of PEI-Cyclam/siTGFβ Polyplex on Liver Collagen Content
Hydroxyproline (HYP) content level is a direct measure of collagen expression.54,55 Liver HYP content was determined by using a HYP testing kit. Briefly, liver tissue (50 mg) was homogenized at 100 °C (pH 6.0–6.8) for 10 min. Then activated carbon was incorporated into the obtained tissue homogenates and centrifuged (3500 rpm, 20 min). Next, the supernatant was obtained, and chloramine-T was incorporated. After 10 min incubation, dimethylaminobenzaldehyde (DMBA) was added to each sample. After 15 min incubation at 60 °C, the absorbance was measured by using a microplate reader. Liver HYP content was expressed as μg/gram.
Effect of PEI-Cyclam/siTGFβ Polyplex on Inflammation and Immunohistochemistry (IHC)
The hepatic morphology (inflammation) was determined by H&E staining, while collagen content was determined by Sirius red and Masson trichrome staining, respectively. For inflammation and collagen deposition, liver tissues were fixed in 10% formalin followed by embedding in paraffin and were cut precisely into 4 μm thick sections. Then, the samples were deparaffinized, hydrated, and finally stained. Images were taken under the FL microscope.
For IHC staining, liver specimens were deparaffinized using xylene, followed by hydration with sequential ethanol (100%, 95%, 85%, and 75%) and deionized water, respectively. To block endogenous peroxidase, the samples were incubated in 3% H2O2. The liver tissues were boiled in sodium citrate buffer (10 mM, pH 6.0). After this, samples were blocked by using the serum for 1 h at room temperature. Then, samples were incubated with primary antibodies anti-TGFβ antibody (Abcam, Cambridge, MA, USA) (Product number: ab190503, diluted 1:500), anti-α-SMA antibody (Abcam, Cambridge, MA, USA) (Product number: ab7817, 1:500), anti-Collagen type III (Col-III) antibody (Abcam, Cambridge, MA, USA) (Product number: ab7778 diluted 1:1000), and anti-proliferating cell nuclear antigen (PCNA) antibody (Abcam, Cambridge, MA, USA) (Product number: ab92552, diluted 1:100) at 4 °C. The liver tissues were incubated with secondary HRP-conjugated antibodies for an additional 1 h. The immune-reactivity was observed using DAB staining, and the brown color displayed the expressions of proteins. The stained areas were quantified by Image J (NIH, MD, USA).
Terminal Deoxyuridine Triphosphate Nick-End Labeling (TUNEL) Staining
To detect cells undergoing apoptosis, the deparaffinized liver sections were exposed to TUNEL staining using an apoptosis detection kit (Roche, Germany), as reported previously.56 The deparaffinized and dehydrated samples were incubated sequentially with cytonin, 3% H2O2, TUNEL solution (200 µL), and TUNEL peroxidase-labeled antibody (TUNEL POD), respectively. Then, the samples were treated with chromogen, washed with PBS, and finally stained with Mayer hematoxylin staining solution. TUNEL-positive cells were identified by 3-amino-9-ethyl carbazole staining. The stained areas were quantified by using Image J software.
Western Blotting
Liver tissue samples were homogenized in RIPA buffer (Beyotime, Shanghai, China) and centrifuged (12,000 rpm, 4°C) for 20 min. The obtained supernatants (protein extracts) were used for the biochemical analysis. Equal amounts of total protein were separated by 10% SDS-PAGE and transferred to PVDF membrane (St. Louis, MO, USA). The membrane was blocked with 5% nonfat milk and incubated with anti-TGFβ antibody at 4°C overnight and washed with Tris-buffered saline. Then, the membrane was again incubated with a secondary IgG-HRP antibody for 2 h at (room temperature). Finally, the protein bands were detected by ECL reagent (Beyotime, Shanghai, China) with an ECL system (Tanon 5200, Beijing, China).
Biodistribution of Polyplexes
The fibrotic mice were administered with a single i.v. injection of the polyplexes prepared with Cy5-labeled siRNA (2.5 mg/kg, w/w 4). The fluorescence signals of the mice were monitored and analyzed at 0.5 h, 3 h, 12 h, and 24 h after injection using the IVIS Lumina imaging system (Xenogen Co., USA). The organs were isolated and excised. The fluorescence was determined using ex-vivo imaging assays.
Statistical Analysis
Graph Pad Prism (San Diego, CA) was used for statistical analysis using a two-tailed Student’s t-test and one-way ANOVA. A p-value of <0.05 was considered statistically significant. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001
Results
Synthesis, Characterization and of PEI-Cyclam
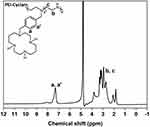
As shown in Scheme 2, the synthesis and characterization of PEI-Cyclam (25 mol%) were carried out according to the previously reported method.53 The cyclam content in PEI-Cyclam was determined using 1H NMR (Figure 1 and Table 1). The signal of the protons on the phenylene ring (δ 6.9–7.8) was compared to the signals of the PEI ethylene groups (δ 2.2–2.8), and the cyclam content was calculated as shown in Table 1.
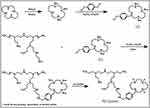 |
Scheme 2 Synthesis of PEI-Cyclam. *Could be any primary, secondary or tertiary amine. |
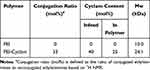 |
Table 1 Composition of PEI-Cyclam |
Preparation and Characterization PEI-Cyclam Polyplexes
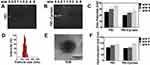
PEI-Cyclam completely condensed siRNA at a w/w ratio of 1.5. The disappearance of the white band was observed, which demonstrates the complete formation of the polyplexes (Figure 2A and B). The polyplexes displayed a positive surface charge, and the zeta potential was increased with increased w/w ratios (Figure 2C). The particle size distribution recorded ranging from 84 to 206 nm (Figure 2D). TEM observations revealed that the polyplexes adopted a spherical morphology (Figure 2E). Hydrodynamic sizes of PEI-Cyclam polyplexes were prepared at various w/w ratios and were measured by DLS. The particle size of PEI-Cyclam polyplexes was the smallest at the lowest w/w around 85 nm (Figure 2F). Polydispersity index (PDI) characterization is vital in nanoparticle applications, as it is problematic to control sample-wide uniformity with surface conjugation chemistry, and frequently aggregation of nanoparticles occurs. The PDI detected for different w/w ratios was less than 0.200 (Figure S1).
Protection Ability of PEI-Cyclam on siRNA Against RNAse and Serum Degradation
Compared to PEI (Figure S2A and B), siRNA remains intact at a w/w ratio of 1.5 in the presence of PEI-Cyclam (Figure S2C and D). Also, compare to free siRNA, the intact siRNA was protected by PEI (Figure S3A and B) and PEI-Cyclam and survived against serum degradation for 24 h (Figure S3C and D). Also, upon incubation with FBS and PBS, a variation in particle size and PDI was observed (Figure S4A and B).
Cytotoxicity of PEI-Cyclam and Polycations
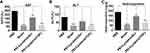
Despite the higher gene delivery efficacy of PEI, it is also cytotoxic.57–60 Biosafety of polycations is a major concern for biomedical applications.61 It’s well reported that cyclam moiety decreases cytotoxicity.53 To prove this, MTT assay was utilized to investigate the cytotoxicity of PEI-Cyclam in U2OS (a model cell line to determine CXCR4 antagonism), B16, breast cancer 4T1, and non-cancerous HSC-T6 cells. Unmodified PEI was added as a control group. The cell viability curves and IC50 values of PEI-Cyclam and PEI are shown in Figures 3 and S5 and summarized in Tables 2 and S1. Compared with PEI, PEI-Cyclam displayed less cytotoxicity in all cell lines, as indicated by the comparatively higher IC50 values. Compared to PEI, the observed IC50 of PEI-Cyclam was ∼ 3.25-fold higher than PEI in U2OS cells, ∼ 4.67-fold higher than PEI in 4T1 ∼ 7.79-folds higher than PEI in B16 cells. Interestingly, in HSC-T6 cells, the cell viability was more than 80% even after treatment with 100 μg/ mL. These findings suggest that PEI-Cyclam is safe in non-cancerous cells, but it may show selective cytotoxicity in cancerous due to their CXCR4-mediated effects by the incorporated cyclam. Also, PEI-Cyclam polyplexes formed with siRNA were safe at increasing w/w ratio (Figures S5–S7). These findings show that the cytotoxicity of PEI-Cyclam is less than unmodified PEI, demonstrating increased biocompatibility.
 |
Table 2 IC50 Values of PEI and PEI-Cyclam |
 |
Figure 3 Cytotoxicity of PEI-Cyclam in U2OS, 4T1, B16, and HSC-T6 cells (A–C) IC50 values were calculated from the figures as shown in Table 2 (D) cell viability assay using HSC-T6 cells. |
CXCR4 Antagonistic Activity of PEI-Cyclam and Polyplex
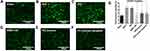
The translocation of CXCR4 on the plasma membrane to endosomes after SDF-1 interaction is a distinctive characteristic of G-protein coupled receptors. Herein, the sham group displayed normal intracellular behavior (Figure 4A). The SDF-1 and PEI showed CXCR4 translocation, as shown by the bright green fluorescence (Figure 4B and C). AMD3100 (a cyclam) was used as a positive control, and it showed a diffused green fluorescence, demonstrating the prevention of CXCR4 translocation (Figure 4D). Also, PEI-Cyclam and PEI-Cyclam polyplexes displayed strong CXCR4 antagonism compared with PEI and SDF-1 (Figure 4E–G).
Cellular Uptake and Endosomal Escape
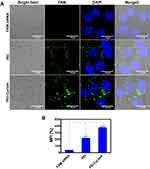
To observe whether cyclam modification can preserve the siRNA delivery efficacy of PEI, we incubated FAM-labeled siRNA polyplexes with HSC-T6 cells. As shown in Figure 5A, compared to PEI and FAM siRNA groups, significant green fluorescence after 2 h was observed in the PEI-Cyclam polyplex group, possibly due to CXCR4 targeting, which is consistent with the flow cytometry results (Figure 5B). These results demonstrate that PEI-Cyclam polyplex exhibits improved cellular uptake and intracellular distribution, confirming that PEI-Cyclam is a suitable siRNA delivery vector. Furthermore, cells treated with increasing concentration of pretreated PEI-Cyclam, the fluorescence intensity was decreased because of the reduced affinity of PEI-Cyclam polyplex for the CXCR4 positive cells due to CXCR4 blockade by the pretreated PEI-Cyclam through competitive inhibition (Figure S8).
Besides the cellular uptake, endosomal escape is another factor that contributes to siRNA delivery.62 The endo-lysosomal escape capacity of PEI-Cyclam polyplex was examined post 2 h and 6 h incubation with HSC-T6 cells. The LysoTracker™ Red (red fluorescence) stained the endo/lysosomes. After 2 h incubation, the green fluorescence of FAM-siRNA overlapped the red fluorescence of LysoTracker™ Red, and both PEI and PEI-Cyclam polyplexes displayed strong yellow fluorescences shown in the merged images, demonstrating that maximum of the endocytosed FAM siRNA was contained in the endosome (Figure S9A). However, 6 h later, compared to PEI, a substantial separation between the green and the red fluorescence was observed, showing that FAM siRNA was effectively escaped from the endosomes (Figure S9-AB). Taken all together, PEI-Cyclam polyplex was capable of entering into HSC-T6 cells by endocytosis and achieved endosomal/lysosomal escape for efficient siRNA delivery.
Luc Gene Silencing Effects
To investigate the PEI-Cyclam polyplexes could achieve gene silencing effects, the Luc gene silencing assay was conducted. 4T1 and B16 cells were transfected with different w/w ratios of PEI-Cyclam polyplexes, but w/w ratios 4 and 8 displayed more than 75% decreased luciferase expression than controls (PEI-Cyclam/siScr polyplex and free siLuc RNA) (Figure 6A and B). Luc expression was determined relative to total proteins (Figure 6C and D). The luciferase gene silencing effect was increased with increasing w/w ratios. These results suggested that cyclam modification can be a promising gene delivery strategy. On the other hand, with prolonged incubation time, the Luc expression was decreased at lower w/w ratios (Figure S10A–D) while increasing the FBS concentration increased the Luc expression (Figure S11A–D).
Restoration of Liver Functions
The restoration effect of PEI-Cyclam/siTGFβ polyplexes on liver functions like serum AST and ALT levels and liver HYP level in fibrotic mice was determined by serological and HYP assays. Compared to Sham mice (without CCl4 treatment), PBS-treated fibrotic mice displayed elevated serum AST, ALT, and liver HYP levels, exhibiting chronic liver damage (Figure 7A–C). PEI-Cyclam/siTGFβ polyplex significantly reduced the levels of AST (73%), ALT (67%), and HYP (58%) compared to the PBS-treated group. Also, PEI-Cyclam/siScr polyplex significantly reduced AST (43%), ALT (38%), and HYP levels (22%) and revealed modest anti-fibrotic activity, which might be ascribed to the CXCR4 antagonistic activity from cyclam moiety. Collectively, these results suggested that the combined TGFβ silencing and CXCR4 Inhibition of PEI-Cyclam/siTGFβ polyplex ameliorated chronic liver damage.
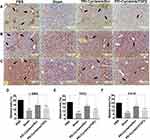
PEI-Cyclam/siTGFβ Polyplex Reverses Liver Fibrosis
H&E staining of liver tissues from Sham mice showed normal liver architecture, PBS-treated fibrotic mice exhibited severe hepatic injuries like centrilobular necrosis, swelling of hepatocytes, extensive fatty changes (an indication of microvesicular steatosis), and extensive immune cells infiltration (inflammation) into the hepatic parenchyma in 4 weeks of CCl4 treatment (Figure 8A and D). PEI-Cyclam/siScr polyplex displayed signs of tissue damage and immune cell infiltration, but to a lesser degree than the PBS-treated group. Those abnormal morphological changes were considerably ameliorated in the PEI-Cyclam/siTGFβ polyplexes treated mice except with only small areas of identifiable atypia, and the overall appearance was similar to the Sham mice. To identify the areas of high collagen deposition as a result of chronic liver damage and to gauge how the polyplexes may impact liver fibrosis, Masson trichrome staining, and Sirius red staining were conducted. Masson trichrome staining and Sirius red staining results showed excessive accumulation of collagen (represented by blue and red color, respectively) in the PBS-treated group, demonstrating liver fibrosis (Figure 8B, C, E and F). Interestingly, fibrotic mice treated with PEI-Cyclam/siScr polyplex also revealed reduced levels of collagen accumulation when compared to PBS-treated mice, additionally supporting the anti-fibrotic effect of the PEI-Cyclam, which might be accredited to the CXCR4 inhibitory activity from the cyclam containing moiety of the polymer. However, mice receiving PEI-Cyclam/siTGFβ polyplex significantly reduced collagen deposition, revealing attenuated liver fibrosis. These findings suggest that PEI-Cyclam/siTGFβ polyplex can efficiently reduce liver fibrosis by combining CXCR4 inhibitory and TGFβ silencing effects.
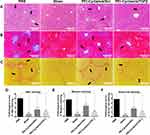
Effect of PEI-Cyclam/siTGFβ Polyplexes on Hepatocytes Apoptosis and Proliferation
PEI-Cyclam/siTGFβ polyplexes might be expected to attenuate liver fibrosis by reducing hepatocyte apoptosis. Therefore, the TUNEL assay was performed. The sectional images are shown in Figure 9A, and the quantitative evaluation is demonstrated in Figure 9C. Increased apoptosis of hepatocytes was observed in the PBS-treated fibrotic mice. However, PEI-Cyclam/siTGFβ polyplex treatment significantly decreased the positively TUNEL-labeled cells, demonstrating attenuated liver fibrosis (Figure 9A and C). PCNA staining was further employed to investigate the effect of PEI-Cyclam/siTGFβ polyplex on the proliferation of hepatocytes.63 Compared to Sham mice, PCNA was highly expressed in the PBS-treated mice, indicating increased hepatocyte proliferation in the fibrotic tissue. However, the treatment of fibrotic mice with PEI-Cyclam/siScr and PEI-Cyclam/siTGFβ polyplexes gave rise to low expression of PCNA. PEI-Cyclam/siTGFβ polyplex decreased the expression of PCNA more significantly, demonstrating inhibition of further disease progression (Figure 9B and D).
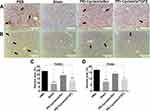
Effect of PEI-Cyclam/siTGFβ Polyplexes on the Expression of Major Fibrotic Proteins
Following histopathological and serological analysis, the therapeutic activity of PEI-Cyclam/siTGFβ polyplexes was further authenticated using IHC staining on the liver. The effect of PEI-Cyclam/siTGFβ polyplexes on the expression of major fibrotic proteins like α-SMA, TGFβ, and Col-III was investigated (Figure 10A–C). These proteins were highly expressed in PBS-treated mice compared to Sham mice, demonstrating liver fibrosis. PEI-Cyclam/siTGFβ polyplex treatment significantly reduced the overexpression of α-SMA (decreased activation of HSC), TGFβ, and Col-III levels (Figure 10A–F). Also, PEI-Cyclam/siScr polyplexes exhibited a limited reduction, which is possibly accredited to inhibiting the SDF-1/CXCR4 axis due to the presence of cyclam moiety. Collectively, these results revealed that, compared to PBS-treated mice, PEI-Cyclam/siTGFβ polyplexes alleviated liver fibrosis by inhibiting the activation of HSC and overexpression of TGFβ.
The PEI-Cyclam/siTGFβ Polyplexes Cause TGFβ Silencing
The effect of PEI-Cyclam/siTGFβ polyplexes on TGFβ expression in fibrotic mice was further investigated by Western blotting. As shown in Figure 11A, TGFβ was highly expressed in the livers isolated from PBS-treated mice compared to Sham control, demonstrating liver fibrosis. Treatment with the PEI-Cyclam/siTGFβ polyplexes significantly reduced the TGFβ expression (Figure 11B). The PEI-Cyclam/siScr polyplexes also showed a limited reduction of TGFβ expression, which is probably ascribed to blocking the SDF-1/CXCR4 axis. The Western blot results agreed well with those from the IHC staining. These findings showed that PEI-Cyclam/siTGFβ polyplexes display TGFβ silencing. Conclusively, PEI-Cyclam could be utilized as a promising siRNA delivery nanocarrier and an anti-fibrotic polymeric drug for combined anti-fibrotic effects.
In vivo Imaging and Tissue Distribution
Effective biodistribution and liver accumulation is an obligatory condition for PEI-Cyclam polyplexes for liver fibrosis therapy. Herein, we compared the biodistribution of PEI-Cyclam polyplexes and PEI- polyplexes prepared with Cy5-labeled siRNA. Compared with PEI-polyplexes, PEI-Cyclam polyplexes exhibited longer biodistribution throughout 24 h (Figure S12A). Also, compare to PEI-polyplexes, the PEI-Cyclam polyplexes showed maximum liver accumulation at 24 h (Figure S12B).
Discussion
Liver fibrosis is a sophisticated process related to the release of numerous cytokines and ECM overproduction. Due to the complex signaling networks involved in liver fibrosis that initiate and control disease progression, selecting a suitable drug and siRNA combination targeting multiple signaling pathways will be an effective treatment strategy for accomplishing the best therapeutic outcomes. The key obstacles for combination therapy are the absence of effective fibrosis-related pathway inhibitors and safe siRNA delivery vectors. Previously polymeric CXCR4 antagonists were employed for simultaneous delivery of siRNA and CXCR4 inhibition. The cyclam moieties provide both the siRNA binding and CXCR4 inhibition, therefore carrying the burden of offering both biological functions and improving the overall activity.50,64,65 Herein, we developed PEI-Cyclam (Polymeric CXCR4 antagonist) by conjugating cyclam (a CXCR4 antagonist) onto PEI for delivering TGFβ siRNA (a therapeutic nucleic acid for TGFβ silencing) and CXCR4 Inhibition. We assumed that the novel polymer design would combine CXCR4 antagonism and TGFβ siRNA delivery in a single molecule, representing a simple tool to present an additional anti-fibrotic function to the existing nanocarrier (Scheme 1).
Compare to PEI, the PEI-Cyclam shows a slightly decreased affinity for siRNA complexation because of its complex and unusual protonation behavior. Out of four amines, only two could be protonated at physiological pH. In PEI-Cyclam, the fraction of protonated amines decreases, resulting in a decreased overall cationic charge density of the polymer. Thus, due to decreased cationic charge density, the affinity for complexation with siRNA also decreases (Figure 2A and B). It is well known that the particle size of drug delivery systems affects pharmacokinetics. Physiological processes like hepatic uptake and accumulation, tissue diffusion, tissue extravasation, and renal excretion of nanoparticles depend on particle size. Only nanocarriers of a specific size (≤150 nm) can enter or exit from the liver.66,67 Herein, the particle size distribution ranged from 84 to 206 nm (Figure 2D), suitable for liver diseases. DLS is a commonly used PDI measurement technique. PDI as an indicator of particle uniformity.68 PDI was employed to evaluate the average uniformity of particles. The larger the PDI values demonstrate larger particle size distribution and particle aggregation, while smaller PDI values indicate particle uniformity.68 The average PDI for nanoparticles ranging from 61–140 nm is between 0.097 and 0.22.69 Our prepared polyplexes showed PDI less than 0.200 indicating particle uniformity (Figure S1).
RNA degradation by nucleases and serum is one of the major obstacles in siRNA delivery, which eventually decreases the gene silencing efficiency.70,71 PEI-Cyclam protected siRNA against RNAse and serum degradation and infers the enhanced siRNA stability in vitro and in vivo (Figures S2, S3C and D).
The biosafety of nanocarriers is one of the crucial factors when investigating any drug or gene delivery systems.61 In the PEI chemical structure, every third atom is a protonizable amine, which gives the PEI a high charge density and significant buffering capacity. However, this high cationic charge density becomes a double-edged sword. At physiological pH, the high protonation PEI offers higher complexation with nucleic acids (negatively charged) via electrostatic interactions, but this may lead to nonspecific interactions with cell membranes, leading to toxicity.72–74 However, Cyclam, has a sophisticated protonation behavior demonstrated by the pKa of the four amines in its chemical structure: 11.29, 10.19, 1.61, and 1.91.75 Interestingly, at physiological pH, only two amines out of the four can be protonated. Thus, after cyclam modification, the fraction of protonated amines in PEI-Cyclam at physiological pH decreases significantly and decreases the polymer overall cationic charge density.53 Thus, cyclam modification significantly reduces protonated amines, resulting in a reduction of the polymer overall cationic charge density, hence decreasing cytotoxicity.53 Also, binding of polycations with nucleic acids typically decreases the toxicity.76 Compared to PEI, cell viability was increased in cells upon cyclam modification several folds (Figures 3A–D, S5 and 6, Tables S1 and 2). Also, PEI-Cyclam polyplex exhibited 100% cell viability in all cell lines (Figure S7). The possible reason for decreased toxicity is cyclam modification of PEI and nucleic acid-binding.
SDF-1 activates HSC and results in proliferation and differentiation of myofibroblasts, causing pro-fibrogenic effects.13 The CXCR4 antagonistic ability of PEI-Cyclam was investigated by using a phenotypic CXCR4 receptor redistribution assay.50 Cyclam moiety functions as a CXCR4 antagonist.64,77 PEI-Cyclam and PEI-Cyclam polyplex displayed significant CXCR4 antagonistic activity, blocking the SDF-1/CXCR4 axis (Figure 4E and F), which may infer the in vivo inhibition of activated HSC, verified by the decreased α-SMA overexpression (Figure 10A and D).
Polyplexes cross cell membranes and enter the cytoplasm for efficient gene delivery.78 Cyclam like AMD3100 binds with CXCR4 at the plasma membrane blocks CXCR4 receptor. However, for polymeric CXCR4 antagonists like PEI-Cyclam to attain dual functions as both CXCR4 antagonists and efficient delivery vectors for siRNA, the polyplexes are essential to be internalized by the cells. The incorporation of cyclam moiety in PEI may alter cellular uptake.53 Previously, polymeric CXCR4 antagonists have been used as effective gene delivery vectors.53 Herein, PEI-Cyclam efficiently delivered siRNA as evidenced by the significant cellular uptake and endosomal escape confirmed by confocal microscopy and flow cytometry (Figures 5A and B, S8, S9A and B). Also, PEI-Cyclam/siLuc polyplex demonstrated robust luciferase gene silencing (Figure 6A and B).
During liver damage, numerous inflammatory mediators like TGFβ and PDGF activate KCs to activate HSC.79 In this study, the potent anti-inflammatory and anti-fibrotic potential of PEI-Cyclam/siTGFβ polyplexes were observed ascribed to the simultaneous CXCR4 inhibition and TGFβ silencing due to CXCR4 antagonistic effects of cyclam moiety and TGFβ siRNA in a single molecule (Figure 8A–F). Elevated levels of TGFβ induce hepatocyte apoptosis and massive cell death contributing to the advancement of the disease and eventually liver cancer.80–82 Numerous mechanisms demonstrate that apoptotic cells lead to fibrosis and increased apoptosis leads to fibrosis progression.83 Apoptotic cells stimulate neutrophils and macrophages, which facilitates fibrotic effects.84 PEI-Cyclam/siTGFβ polyplexes treatment significantly reduced the elevated apoptosis due to TGFβ silencing effects (Figure 9A and C). Additionally, the over-expressed TGFβ facilitates the production of other cytokines that contribute to cell proliferation and eventually leads to cancer.85 Normally, healthy liver tissues display less than 1% PCNA-positive hepatocytes; however, the number of PCNA-positive hepatocytes is markedly increased in fibrosis.86 PEI-Cyclam/siTGFβ polyplexes significantly reduced PCNA-positive hepatocytes, displaying inhibition of disease progression (Figure 9B and D).
The overexpressed α-SMA is considered to be an indicator of activated HSC.22 Activated HSC overexpresses TGFβ,21 which further activates HSC and results in excessive ECM overproduction.22,23 Also, the upregulated SDF-1/CXCR4 axis during liver damage leads to HSC activation,13,14 TGFβ overexpression, and progression of the disease to liver fibrosis.15 TGFβ is reported to activate the expression of many other profibrogenic cytokines like IL-13 and TNF-α, thus further boosting and spreading the fibrotic response.87 PEI-Cyclam/siTGFβ polyplexes, decrease α-SMA, TGFβ, and Col-III expression by combined CXCR4 inhibition (SDF-1/CXCR4 axis blocking) and TGFβ silencing effects (Figures 4E–G, 10A–F, 11A and B). In a nutshell, TGFβ and α-SMA are overexpressed in the fibrotic livers, and polyplexes equipped with cyclam and TGFβ siRNA efficiently inhibited and silenced the CXCR4 (HSC activation: SDF-1/CXCR4 axis) and TGFβ, respectively, thus mitigating liver fibrosis.
Conclusion
This study designed and developed novel polymeric CXCR4 antagonists based on PEI (PEI-Cyclam) that can concurrently deliver TGFβ siRNA and inhibit CXCR4/SDF-1 axis, attaining combined anti-fibrotic effects in CCl4-induced liver fibrosis in a mouse model. Our findings established that conjugation of cyclam moiety to PEI is capable of (1) retain the siRNA delivery efficiency of PEI, (2) alleviate the cytotoxicity of PEI, and (3) exert CXCR4 antagonistic activity. The PEI-Cyclam/siTGFβ polyplexes confirmed the combined anti-fibrotic activity because of the concurrent CXCR4 antagonism/inhibition and TGFβ silencing effects. The reported PEI modification presents a simple tool to introduce a new pharmacological function to existing siRNA delivery vectors to attain an enhanced siRNA delivery and therapeutic efficiency for potential anti-fibrotic applications. Conclusively, The PEI-Cyclam/siTGFβ polyplex treatment demonstrated combined anti-fibrotic effects because of the simultaneous CXCR4 Inhibition (HSC activation) and TGFβ silencing efficiency. In summary, the PEI-Cyclam/siTGFβ polyplex presents a promising delivery system for designing a new treatment strategy for liver fibrosis.
Acknowledgment
This work was financially supported by the 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (2020LKSFG18B, 2020LKSFG02E), and the grant for Key Disciplinary Project of Clinical Medicine under the Guangdong High-Level University Development Program (002-18120314, 002-18120311) and the postdoc fund by Shantou University Medical College, Shantou, Guangdong Province.
Disclosure
The authors declared no conflicts of interest in this work.
References
1. Bataller R, Brennea DA. Liver fibrosis. J Clin Investig. 2005;115(2):209–218. doi:10.1172/JCI24282
2. Garcia-Tsao G, Lim J. Management and treatment of patients with cirrhosis and portal hypertension: recommendations from the department of veterans affairs hepatitis C resource center program and the national hepatitis C program. Am J Gastroenterol. 2009;104(7):1802. doi:10.1038/ajg.2009.191
3. Kim Y, Ejaz A, Tayal A, et al. Temporal trends in population-based death rates associated with chronic liver disease and liver cancer in the United States over the last 30 years. Cancer. 2014;120(19):3058–3065. doi:10.1002/cncr.28843
4. Schuppan D, Kim YO. Evolving therapies for liver fibrosis. J Clin Invest. 2013;123(5):1887–1901. doi:10.1172/JCI66028
5. Popov Y, Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology. 2009;50(4):1294–1306. doi:10.1002/hep.23123
6. Friedman SL. Evolving challenges in hepatic fibrosis. Nat Rev Gastroenterol Hepatol. 2010;7(8):425–436. doi:10.1038/nrgastro.2010.97
7. Friedman SL, Sheppard D, Duffield JS, Violette S. Therapy for fibrotic diseases: nearing the starting line. Sci Transl Med. 2013;5(167):167sr161. doi:10.1126/scitranslmed.3004700
8. Schuppan D, Pinzani M. Anti-fibrotic therapy: lost in translation? J Hepatol. 2012;56:S66–S74. doi:10.1016/S0168-8278(12)60008-7
9. Altamirano-Barrera A, Barranco-Fragoso B, Mendez-Sanchez N. Management strategies for liver fibrosis. Ann Hepatol. 2017;16(1):48–56. doi:10.5604/16652681.1226814
10. Hao L, Zhang C, Qiu Y, et al. Recombination of CXCR4, VEGF, and MMP-9 predicting lymph node metastasis in human breast cancer. Cancer Lett. 2007;253(1):34–42. doi:10.1016/j.canlet.2007.01.005
11. Guo P, You JO, Yang J, Jia D, Moses MA, Auguste DT. Inhibiting metastatic breast cancer cell migration via the synergy of targeted, pH-triggered siRNA delivery and chemokine axis blockade. Mol Pharm. 2014;11(3):755–765. doi:10.1021/mp4004699
12. Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(6):1655–1669. doi:10.1053/j.gastro.2008.03.003
13. Hong F, Tuyama A, Lee TF. Hepatic stellate cells express functional CXCR4: role in stromal cell–derived factor-α–mediated stellate cell activation. Hepatology. 2009;49(6):2055–2067. doi:10.1002/hep.22890
14. Chen Y, Huang Y, Reiberger T, et al. Differential effects of sorafenib on liver versus tumor fibrosis mediated by stromal-derived factor 1 alpha/C-X-C receptor type 4 axis and myeloid differentiation antigen-positive myeloid cell infiltration in mice. Hepatology. 2014;59(4):1435–1447. doi:10.1002/hep.26790
15. Lin CH, Shih CH, Tseng CC, et al. CXCL12 induces connective tissue growth factor expression in human lung fibroblasts through the Rac1/ERK, JNK, and AP-1 pathways. PLoS One. 2014;9(8):e104746. doi:10.1371/journal.pone.0104746
16. Makino H, Aono Y, Azuma M, et al. Antifibrotic effects of CXCR4 antagonist in bleomycin-induced pulmonary fibrosis in mice. J Med Invest. 2013;60(1–2):127–137. doi:10.2152/jmi.60.127
17. Watanabe M, Matsuyama W, Shirahama Y, et al. Dual effect of AMD3100, a CXCR4 antagonist, on bleomycin-induced lung inflammation. J Immunol. 2007;178(9):5888–5898. doi:10.4049/jimmunol.178.9.5888
18. Welschinger R, Liedtke F, Basnett J, et al. Plerixafor (AMD3100) induces prolonged mobilization of acute lymphoblastic leukemia cells and increases the proportion of cycling cells in the blood in mice. Exp Hematol. 2013;41(3):293–302.e291. doi:10.1016/j.exphem.2012.11.004
19. Broxmeyer HE, Orschell CM, Clapp DW, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201(8):1307–1318. doi:10.1084/jem.20041385
20. Lin TT, Gao DY, Liu YC, et al. Development and characterization of sorafenib-loaded PLGA nanoparticles for the systemic treatment of liver fibrosis. J Control Release. 2016;221:62–70. doi:10.1016/j.jconrel.2015.11.003
21. Bauer M, Schuppan D. TGFbeta1 in liver fibrosis: time to change paradigms? FEBS Lett. 2001;502(1–2):1–3. doi:10.1016/S0014-5793(01)02655-2
22. Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123(5):1902–1910. doi:10.1172/JCI66369
23. Moreno M, Bataller R. Cytokines and renin-angiotensin system signaling in hepatic fibrosis. Clin Liver Dis. 2008;12(4):825–852. doi:10.1016/j.cld.2008.07.013
24. Breitkopf K, Haas S, Wiercinska E, Singer MV, Dooley S. Anti-TGF-β strategies for the treatment of chronic liver disease. Alcohol Clin Exp Res. 2005;29(11 Suppl):121s–131s. doi:10.1097/01.alc.0000189284.98684.22
25. Leask A, Abraham DJ. TGF-β signaling and the fibrotic response. FASEB J. 2004;18(7):816–827. doi:10.1096/fj.03-1273rev
26. Hannon GJ. RNA interference. Nature. 2002;418(6894):244. doi:10.1038/418244a
27. Tijsterman M, Ketting RF, Plasterk RH. The genetics of RNA silencing. Annu Rev Genet. 2002;36(1):489–519. doi:10.1146/annurev.genet.36.043002.091619
28. Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu Rev Med. 2005;56:401–423. doi:10.1146/annurev.med.56.082103.104606
29. Sledz CA, Williams BR. RNA interference in biology and disease. Blood. 2005;106(3):787–794. doi:10.1182/blood-2004-12-4643
30. Leung RK, Whittaker PA. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Ther. 2005;107(2):222–239. doi:10.1016/j.pharmthera.2005.03.004
31. McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3(10):737–747. doi:10.1038/nrg908
32. Shiver JW, Fu T-M, Chen L, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415(6869):331–335. doi:10.1038/415331a
33. Song E, Lee S-K, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9(3):347–351. doi:10.1038/nm828
34. Fu R, Wu J, Ding J, et al. Targeting transforming growth factor betaRII expression inhibits the activation of hepatic stellate cells and reduces collagen synthesis. Exp Biol Med. 2011;236(3):291–297. doi:10.1258/ebm.2010.010231
35. Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4(6):457–467. doi:10.1038/nrm1129
36. Xiong X-B, Uludağ H, Lavasanifar A. Biodegradable amphiphilic poly (ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nanocarriers for efficient siRNA delivery. Biomaterials. 2009;30(2):242–253. doi:10.1016/j.biomaterials.2008.09.025
37. Lächelt U, Wagner E. Nucleic acid therapeutics using polyplexes: a journey of 50 years (and beyond). Chem Rev. 2015;115(19):11043–11078.
38. Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi:10.1038/nrg1066
39. Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi:10.1038/nrd1775
40. Jiang B, He H, Yao L, et al. Harmonizing the intracellular kinetics toward effective gene delivery using cancer cell-targeted and light-degradable polyplexes. Biomacromolecules. 2017;18(3):877–885. doi:10.1021/acs.biomac.6b01774
41. Merkel OM, Kissel T. Quo vadis polyplex? J Control Release. 2014;190:415–423. doi:10.1016/j.jconrel.2014.06.009
42. Shim MS, Kwon YJ. Acid-responsive linear polyethylenimine for efficient, specific, and biocompatible siRNA delivery. Bioconjug Chem. 2009;20(3):488–499. doi:10.1021/bc800436v
43. Bonnet ME, Erbacher P, Bolcato-Bellemin AL. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm Res. 2008;25(12):2972–2982. doi:10.1007/s11095-008-9693-1
44. Dehshahri A, Oskuee RK, Shier WT, Hatefi A, Ramezani M. Gene transfer efficiency of high primary amine content, hydrophobic, alkyl-oligoamine derivatives of polyethylenimine. Biomaterials. 2009;30(25):4187–4194. doi:10.1016/j.biomaterials.2009.04.036
45. Masotti A, Moretti F, Mancini F, et al. Physicochemical and biological study of selected hydrophobic polyethylenimine-based polycationic liposomes and their complexes with DNA. Bioorg Med Chem. 2007;15(3):1504–1515. doi:10.1016/j.bmc.2006.10.066
46. Yamazaki Y, Nango M, Matsuura M, Hasegawa Y, Hasegawa M, Oku N. Polycation liposomes, a novel nonviral gene transfer system, constructed from cetylated polyethylenimine. Gene Ther. 2000;7(13):1148–1155. doi:10.1038/sj.gt.3301217
47. Liang Z, Wu T, Lou H, et al. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004;64(12):4302–4308. doi:10.1158/0008-5472.CAN-03-3958
48. Huang EH, Singh B, Cristofanilli M, et al. A CXCR4 antagonist CTCE-9908 inhibits primary tumor growth and metastasis of breast cancer. J Surg Res. 2009;155(2):231–236. doi:10.1016/j.jss.2008.06.044
49. Gerlach LO, Skerlj RT, Bridger GJ, Schwartz TW. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J Biol Chem. 2001;276(17):14153–14160. doi:10.1074/jbc.M010429200
50. Li J, Zhu Y, Hazeldine ST, Li C, Oupicky D. Dual-function CXCR4 antagonist polyplexes to deliver gene therapy and inhibit cancer cell invasion. Angew Chem Int Ed Engl. 2012;51(35):8740–8743. doi:10.1002/anie.201203463
51. Wang Y, Li J, Oupický D. Polymeric plerixafor: effect of PEGylation on CXCR4 antagonism, cancer cell invasion, and DNA transfection. Pharm Res. 2014;31(12):3538–3548. doi:10.1007/s11095-014-1440-1
52. Tsolaki E, Athanasiou E, Gounari E, et al. Hematopoietic stem cells and liver regeneration: differentially acting hematopoietic stem cell mobilization agents reverse induced chronic liver injury. Blood Cells Mol Dis. 2014;53(3):124–132. doi:10.1016/j.bcmd.2014.05.003
53. Zhou Y, Yu F, Zhang F, et al. Cyclam-modified PEI for combined VEGF siRNA silencing and CXCR4 inhibition to treat metastatic breast cancer. Biomacromolecules. 2018;19(2):392–401. doi:10.1021/acs.biomac.7b01487
54. Sung DK, Kong WH, Park K, et al. Noncovalenly PEGylated CTGF siRNA/PDMAEMA complex for pulmonary treatment of bleomycin-induced lung fibrosis. Biomaterials. 2013;34(4):1261–1269. doi:10.1016/j.biomaterials.2012.09.061
55. Ding L, Zhu C, Yu F, et al. Pulmonary delivery of polyplexes for combined PAI-1 gene silencing and CXCR4 inhibition to treat lung fibrosis. Nanomedicine. 2018;14(6):1765–1776. doi:10.1016/j.nano.2018.05.005
56. Yang C, Wang Y, Liu H, et al. Ghrelin protects H9c2 cardiomyocytes from angiotensin II-induced apoptosis through the endoplasmic reticulum stress pathway. J Cardiovasc Pharmacol. 2012;59(5):465–471. doi:10.1097/FJC.0b013e31824a7b60
57. Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi:10.1016/S0142-9612(02)00445-3
58. Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum Gene Ther. 1996;7(16):1947–1954. doi:10.1089/hum.1996.7.16-1947
59. Fischer D, Bieber T, Li Y, Elsässer H-P, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16(8):1273–1279. doi:10.1023/A:1014861900478
60. Godbey W, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly (ethyleneimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45(3):268–275. doi:10.1002/(SICI)1097-4636(19990605)45:3<268::AID-JBM15>3.0.CO;2-Q
61. Thomas M, Lu JJ, Chen J, Klibanov AM. Non-viral siRNA delivery to the lung. Adv Drug Deliv Rev. 2007;59(2–3):124–133. doi:10.1016/j.addr.2007.03.003
62. Lin C, Zhong Z, Lok MC, et al. Linear poly (amido amine) s with secondary and tertiary amino groups and variable amounts of disulfide linkages: synthesis and in vitro gene transfer properties. J Control Release. 2006;116(2):130–137. doi:10.1016/j.jconrel.2006.09.009
63. Connolly KM, Bogdanffy MS. Evaluation of proliferating cell nuclear antigen (PCNA) as an endogenous marker of cell proliferation in rat liver: a dual-stain comparison with 5-bromo-2ʹ-deoxyuridine. J Histochem Cytochem. 1993;41(1):1–6. doi:10.1177/41.1.7678022
64. Wang Y, Hazeldine ST, Li J, Oupicky D. Development of functional poly(amido amine) CXCR4 antagonists with the ability to mobilize leukocytes and deliver nucleic acids. Adv Healthc Mater. 2015;4(5):729–738. doi:10.1002/adhm.201400608
65. Wang Y, Kumar S, Rachagani S, et al. Polyplex-mediated inhibition of chemokine receptor CXCR4 and chromatin-remodeling enzyme NCOA3 impedes pancreatic cancer progression and metastasis. Biomaterials. 2016;101:108–120. doi:10.1016/j.biomaterials.2016.05.042
66. Blasi P, Giovagnoli S, Schoubben A, Ricci M, Rossi C. Solid lipid nanoparticles for targeted brain drug delivery. Adv Drug Deliv Rev. 2007;59(6):454–477. doi:10.1016/j.addr.2007.04.011
67. Bertrand N, Leroux JC. The journey of a drug-carrier in the body: an anatomo-physiological perspective. J Control Release. 2012;161(2):152–163. doi:10.1016/j.jconrel.2011.09.098
68. Clayton KN, Salameh JW, Wereley ST, Kinzer-Ursem TL. Physical characterization of nanoparticle size and surface modification using particle scattering diffusometry. Biomicrofluidics. 2016;10(5):054107. doi:10.1063/1.4962992
69. Colmenares Roldán GJ, Agudelo Gomez LM, Carlos Cornelio JA, Rodriguez LF, Pinal R, Hoyos Palacio LM. Production of polycaprolactone nanoparticles with low polydispersity index in a tubular recirculating system by using a multifactorial design of experiments. J Nanopart Res. 2018;20(3):68. doi:10.1007/s11051-018-4168-8
70. Kim J, Lee SH, Choe J, Park TG. Intracellular small interfering RNA delivery using genetically engineered double-stranded RNA binding protein domain. J Gene Med. 2009;11(9):804–812. doi:10.1002/jgm.1365
71. Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104(39):15549–15554. doi:10.1073/pnas.0707461104
72. Hong S, Leroueil PR, Janus EK, et al. Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjug Chem. 2006;17(3):728–734. doi:10.1021/bc060077y
73. Chen J, Hessler JA, Putchakayala K, et al. Cationic nanoparticles induce nanoscale disruption in living cell plasma membranes. J Phys Chem B. 2009;113(32):11179–11185. doi:10.1021/jp9033936
74. Zhang C, Wu F-G, Hu P, Chen Z. Interaction of polyethylenimine with model cell membranes studied by linear and nonlinear spectroscopic techniques. J Phys Chem C. 2014;118(23):12195–12205. doi:10.1021/jp502383u
75. Hancock RD, Motekaitis RJ, Mashishi J, Cukrowski I, Reibenspies JH, Martell AE. The unusual protonation constants of cyclam. A potentiometric, crystallographic and molecular mechanics study. J Chem Soc Perkin Trans. 1996;2(9):1925–1929. doi:10.1039/p29960001925
76. Chen G, Wang K, Hu Q, et al. Combining fluorination and bioreducibility for improved siRNA polyplex delivery. ACS Appl Mater Interfaces. 2017;9(5):4457–4466. doi:10.1021/acsami.6b14184
77. Li J, Oupicky D. Effect of biodegradability on CXCR4 antagonism, transfection efficacy and antimetastatic activity of polymeric Plerixafor. Biomaterials. 2014;35(21):5572–5579. doi:10.1016/j.biomaterials.2014.03.047
78. Duncan R, Richardson SC. Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: opportunities and challenges. Mol Pharm. 2012;9(9):2380–2402. doi:10.1021/mp300293n
79. Chen L, Yao X, Yao H, Ji Q, Ding G, Liu X. Exosomal miR-103-3p from LPS-activated THP-1 macrophage contributes to the activation of hepatic stellate cells. FASEB J. 2020;34(4):5178–5192. doi:10.1096/fj.201902307RRR
80. Ramesh S, Qi XJ, Wildey GM, et al. TGF β-mediated BIM expression and apoptosis are regulated through SMAD3-dependent expression of the MAPK phosphatase MKP2. EMBO Rep. 2008;9(10):990–997. doi:10.1038/embor.2008.158
81. Schwall RH, Robbins K, Jardieu P, Chang L, Lai C, Terrell TG. Activin induces cell death in hepatocytes in vivo and in vitro. Hepatology. 1993;18(2):347–356. doi:10.1016/0270-9139(93)90018-i
82. Fabregat I, Moreno-Caceres J, Sanchez A, et al. TGF β signalling and liver disease. FEBS J. 2016;283(12):2219–2232. doi:10.1111/febs.13665
83. Chen -C-C, Lau LF. Deadly liaisons: fatal attraction between CCN matricellular proteins and the tumor necrosis factor family of cytokines. Cell Commun Signal. 2010;4(1):63–69. doi:10.1007/s12079-009-0080-4
84. Johnson A, DiPietro LA. Apoptosis and angiogenesis: an evolving mechanism for fibrosis. FASEB J. 2013;27(10):3893–3901. doi:10.1096/fj.12-214189
85. Calon A, Tauriello DV, Batlle E. TGF-beta in CAF-mediated tumor growth and metastasis. Semin Cancer Biol. 2014;25:15–22. doi:10.1016/j.semcancer.2013.12.008
86. Delhaye M, Louis H, Degraef C, et al. Relationship between hepatocyte proliferative activity and liver functional reserve in human cirrhosis. Hepatology. 1996;23(5):1003–1011. doi:10.1002/hep.510230510
87. Fernandez IE, Eickelberg O. The impact of TGF-β on lung fibrosis: from targeting to biomarkers. Proc Am Thorac Soc. 2012;9(3):111–116. doi:10.1513/pats.201203-023AW
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.