Back to Journals » International Journal of Women's Health » Volume 14
Current Perspectives of Prenatal Cell-free DNA Screening in Clinical Management of First-Trimester Septated Cystic Hygroma
Authors Sherer DM , Hsieh V, Hall A, Gerren A, Walters E, Dalloul M
Received 28 April 2022
Accepted for publication 8 August 2022
Published 25 October 2022 Volume 2022:14 Pages 1499—1518
DOI https://doi.org/10.2147/IJWH.S328201
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Marleen van Gelder
David M Sherer, Vicky Hsieh, Anika Hall, Allison Gerren, Erin Walters, Mudar Dalloul
The Division of Maternal Fetal Medicine, the Department of Obstetrics and Gynecology, State University of New York (SUNY), Downstate Health Sciences University, Brooklyn, New York, USA
Correspondence: David M Sherer, Division of Maternal-Fetal Medicine, Department of Obstetrics and Gynecology, State University of New York (SUNY), Downstate Health Sciences University, 450 Clarkson Avenue, Box 24, Brooklyn, NY, 11203, USA, Tel +001-718-270-2081, Fax +001-718-270-4122, Email [email protected]
Abstract: First-trimester septated cystic hygroma occurs in approximately 1 in 268 pregnancies and has long been associated with a markedly increased risk of fetal aneuploidy and, among euploid fetuses, an increased risk of structural anomalies primarily affecting the cardiac and skeletal systems. Invasive prenatal diagnosis – chorionic villus sampling and/or amniocentesis – encompasses the time-honored clinical tools for the next step in management following prenatal sonographic diagnosis of first-trimester septated cystic hygroma. Currently, prenatal cell-free DNA (cfDNA) screening for fetal aneuploidy with select microdeletions is gradually replacing the considerably less sensitive, and labor-intensive combined first-trimester screening. These new technologies have opened potential new venues in the clinical management of this ominous late first-trimester sonographic diagnosis. Advances in cfDNA technologies are now permitting detection of chromosomal copy number variants (CNV) larger than 7Mb across genome and select serious single-gene disorders (mainly impacting skeletal and neurological development), affecting quality of life and may benefit from medical and/or surgical management. This commentary will address the available non-invasive prenatal screening technologies, which clearly enhance immediate genetic analysis modalities applicable in the presence of the complex sonographic finding of first-trimester septated cystic hygroma.
Keywords: prenatal ultrasound, first-trimester septated cystic hygroma, cell free DNA, cfDNA, noninvasive prenatal screening (NIPS), transvaginal ultrasound, fetal aneuploidy
Prenatal Screening for Fetal Aneuploidy
First-trimester screening for aneuploidy described by Nicolaides et al in 1992 utilized first-trimester nuchal translucency.1 Prospective assessment of 827 women referred for first-trimester fetal karyotyping (due to advanced maternal age, parental anxiety, or familial history of a chromosomal abnormality in the absence of balanced parental translocation), reported a 3% incidence of chromosomal defects (28/827).1 In 51 fetuses with increased nuchal translucency thickness (3–8 mm), the incidence of chromosomal defects was 35% (18/51). In contrast, only 10 of the remaining 776 (1%) fetuses manifested chromosomal abnormalities. Thus, fetal nuchal translucency ≥3 mm was found a useful marker for fetal chromosomal abnormalities.1 This original observation was subsequently confirmed by others.2,3
Following, Snijders et al in 1998 incorporated maternal age and assessment of nuchal-translucency thickness in calculating risk for fetal trisomy 21.4 Risk of fetal trisomy 21 was originally estimated among 96,127 women (median age of 31 years) with singleton pregnancies. Risk of trisomy 21 was calculated by maternal age and gestational age prevalence, multiplied by a likelihood ratio depending on the deviation from normal of nuchal-translucency thickness for crown-rump length (CRL).4 The estimated risk of trisomy 21 from maternal age and fetal nuchal-translucency, was calculated at 1 in 300 or higher in 7907 (8.3%) of 95,476 normal pregnancies, 268 (82.2%) of 326 pregnancies with trisomy 21, and 253 (77.9%) of 325 fetuses with other chromosomal abnormalities. Of note, 5% of the population with the highest calculated risk included 77% of the trisomy 21 fetuses.4 In 2004, Nicolaides et al reviewed prospective Down syndrome screening studies, which utilized first-trimester NT in the previous 10 years. Above 200,000 first-trimester patients were evaluated, including 871 with Down syndrome. First-trimester NT was noted to have a detection rate of 76.8% (with a false-positive rate of 4.2%).5
Fetal nuchal-translucency thickness (a gestational age-dependent parameter hence reported in MoM) was subsequently incorporated with first-trimester maternal serum analytes including Pregnancy Associated Plasma Protein-A (PAPP-A), and free beta human chorionic gonadotropin (hCG). The latter screening modality resulted in an increased sensitivity for detecting fetal trisomy 21.6–15
Canick et al demonstrated that although levels of free beta-hCG in affected pregnancies were higher earlier than the levels of either total hCG or inhibin A, there was no significant difference in screening performance when either of the three markers was utilized with nuchal translucency and PAPP-A between 11 and 13 weeks of pregnancy.16
Huang et al in 2013, reported that pregnancies with unbalanced translocations exhibited significantly higher free beta-hCG (MoM) levels and larger nuchal translucency thickness than pregnancies with normal karyotype or those with balanced translocations. These authors concluded that first-trimester combined screening is not only effective in the screening of trisomy 21 and other trisomies, but exhibits a high sensitivity in the detection of fetal unbalanced translocations.17
A recent Cochrane database systematic review (of 126 studies including 1,604,040 fetuses with 8454 Down’s syndrome), compared the ten most frequently evaluated test modalities. The study demonstrated that a combined nuchal translucency thickness, serum PAPP-A, free-beta hCG and maternal age test modality was significantly superior to ultrasound markers alone (with or without maternal age) except nasal bone, detecting approximately nine out of every 10 Down’s syndrome fetuses with a 5% false positive rate (FPR).18 Although the sonographic finding of an absent nasal bone appeared to have high diagnostic accuracy, only five out of ten affected Down’s pregnancies were detected with a 1% false positive rate.18 In both direct and indirect assessments, the combined nuchal translucency (NT) thickness, PAPP-A, free beta hCG and maternal age test strategy exhibited superior diagnostic accuracy to nuchal translucency thickness and maternal age test strategy (P < 0.0001). Based on the indirect evaluations of all available studies for the two tests, the sensitivity (95% confidence interval) estimated with a 5% FPR for the combined nuchal translucency (NT) thickness, PAPP-A, free-beta hCG and maternal age test strategy (69 studies; including 1,173,853 fetuses, 6010 of these with Down’s syndrome) was 87% (86 to 89). In comparison, the sensitivity for nuchal translucency thickness and maternal age test strategy (50 studies; 530,874 fetuses including 2701 Down’s syndrome pregnancies) was 71% (66 to 75). Combinations of nuchal translucency thickness with other ultrasound markers, PAPP-A, and free beta hCG were evaluated in one or two studies and exhibited sensitivities of >90% and specificities of >95%.18
Notwithstanding, it is worthy of consideration that inexperienced sonographers are noted to under-measure nuchal translucency. Experience, training and continued monitoring of data are essential in order to assure that measurements are precise. Evans et al calculated the impact of mathematically modifying nuchal translucency measurements to either a 25% or a 0.5 mm decrease in measurement.19 These authors calculated that such minor inaccuracies in nuchal translucency measurements of Trisomy 21 and other abnormalities will have a very significant impact upon abnormality detection lowering the sensitivity from 81.7% and 70.5%, respectively, to 67.1% and 62.3% (P < 0.01) while reducing detection rates by up to 18%.19
Despite the above objective concerns regarding continued quality assurance of performance of the accuracy of the nuchal translucency measurement, combined first-trimester screening for fetal aneuploidy gained relatively rapid utilization world-wide and rapidly became the accepted standard of care in many nations.
The American College of Obstetricians and Gynecologists (ACOG) and the Society for Maternal-Fetal Medicine (SMFM) recommend genetic counseling and diagnostic testing for nuchal translucency at ≥3.0 mm (or above the 99th centile for crown rump length).20 Hui et al recently published a retrospective study of women residents of Victoria, Australia, who underwent combined first-trimester screening during the two year period between January 2015 and December 2016.21 Linkages between statewide results for combined first-trimester screening, prenatal diagnostic procedures, and postnatal cytogenetic results from products of conception and infants up to 12 months of age were applied to confirm frequency and type of chromosome abnormality by gestation and nuchal translucency (NT) measurement. Atypical chromosome abnormality was defined as any major chromosome abnormality other than whole chromosome aneuploidy of chromosomes 21, 18, 13, X, or Y.
Of 81,244 singleton pregnancies who underwent combined first-trimester screening, 491 (0.60%) exhibited a nuchal translucency (NT) of ≥3.5 mm, 534 (0.66%) nuchal translucency (NT) of 3.0 to 3.4 mm, and 80,219 (98.74%) a nuchal translucency (NT) of <3.0 mm. Grouped by nuchal translucency (NT) multiples of the median (MoM), 192 (0.24%) had a nuchal translucency (NT) of ≥3.0 MoM, 513 (0.63%) had a nuchal translucency of 1.9 to 2.9 MoM, and 80,539 (99.13%) had a nuchal translucency (NT) <1.9 MoM. A total of 1779 pregnancies underwent prenatal or postnatal diagnostic testing, of which 89.60% were performed by whole-genome single-nucleotide polymorphism (SNP) chromosomal microarray. The frequency of total major chromosome abnormalities was significantly higher in the group with nuchal translucency (NT) of ≥3.5 mm (147 of 491, 29.94%) in comparison the group with a nuchal translucency of 3.0 to 3.4 mm (21 of 534, 3.93%) or a nuchal translucency of <3.0 mm (71 of 80,219, 0.09%) (P < 0.001). A total of 93 pregnancies with atypical chromosome abnormalities were noted in the total screened group of patients. The frequency of an atypical chromosome abnormality was 4.07% (95% confidence interval, 2.51–6.22), 0.37% (95% confidence interval, 0.05–1.35), and 0.09% (95% confidence interval, 0.07–0.11) in the groups with a nuchal translucency (NT) measurements of ≥3.5 mm, 3.0 to 3.4 mm, and <3.0 mm, respectively. In contrast, frequency of atypical chromosome abnormalities was 4.69% (95% confidence interval, 2.17–8.71), 2.53% (95% confidence interval, 1.36–4.29), and 0.09% (95% confidence interval, 0.07–0.11) in groups with nuchal translucencies of ≥3.0 MoM, 1.9 to 2.9 MoM and <1.9 MoM, respectively. When thresholds were defined for offering diagnosis with chromosomal microarray at 11 to 13 weeks, both a nuchal translucency threshold of 1.9 MoM and a fixed threshold of 3.0 mm identified 22 of 93 fetuses (23.7%) with an atypical chromosome abnormality. Of these, 50.0% had a coexisting fetal abnormality on ultrasound. However, the gestation-specific threshold of 1.9 MoM had a better specificity than 3.0 mm. The positive predictive value of an enlarged nuchal translucency (NT) for any atypical chromosome abnormality was 1 in 47 for nuchal translucency (NT) of >3.0 mm and 1 in 32 for nuchal translucency of >1.9 MoM. The nuchal translucency (NT) threshold of 1.9 MoM identified 0.87% of fetuses, approximating the 99th centile. These authors concluded that a gestational age-adjusted nuchal translucency (NT) threshold of 1.9 MoM or 99th centile is superior to the fixed cutoff of 3.0 mm for the identification of atypical chromosome abnormalities. Risk of an atypical chromosome abnormality in a fetus with an enlarged nuchal translucency (NT) is more than triple in the presence of an additional sonographic abnormality.20
Currently, universal cell free DNA (cfDNA) screening (commonly known as non-invasive prenatal screening, NIPS or NIPT) for fetal aneuploidy and select microdeletions is gradually replacing the considerably less specific, sensitive, and labor intensive combined first-trimester screening modalities. cfDNA screening has clearly been demonstrated as the most sensitive and specific test for detection of common fetal aneuploidies.20 The fetal fraction of cfDNA is derived from placental trophoblasts (and syncytiotrophoblast) which are released into the maternal circulation through apoptosis. The fetal fraction typically constitutes between 3% and 13% of the total cfDNA in maternal blood.22,23 The amount of fetal cfDNA increases with advancing gestational age. In contrast to timing sensitive maternal serum analytes, this screening modality may be utilized starting at 9 weeks’ gestation with detection rates increasing throughout gestation.
In a 2015 prospective, international blinded study conducted at 35 centers, patients presenting for fetal aneuploidy screening between 10 and 14 weeks’ gestation undergoing both standard screening (with nuchal translucency measurement and serum analytes) and cfDNA screening were compared.24 Primary outcome assessed was detection of trisomy 21 with cfDNA versus standard screening. Also assessed, were cfDNA and standard screening to assess the risk of trisomies 18 and 13. Of 15,841 patients for whom cfDNA screening for Down syndrome was compared with first-trimester screening (NT and serum analytes) in the general population (mean maternal age = 30.7), Down syndrome was detected in 38/38 patients [100%; 95% confidence interval (CI), 90.7–00] in the cfDNA group compared with 30 of 38 patients (78.9%; 95% CI, 62.7–90.4) in the standard screening group. cfDNA exhibited a lower false-positive rate 0.06% (95% CI, 0.03–0.11) versus 5.4% (95% CI 5.1–5.8) in the standard screening group. Positive predictive value for cfDNA screening = 80.9% (95% CI, 66.7–90.9) versus 3.4% (95% CI, 2.3–4.8) in the standard screening group. The authors concluded that cfDNA screening has higher sensitivity, lower false positive rate and higher positive predictive value than standard screening (NT and serum analytes).24
In the primary analysis population there were ten cases of trisomy 18. Of these, cfDNA testing identified nine while standard testing identified eight; cfDNA testing had one false positive result for a false positive rate of 0.01% (95% CI, 0–0.04) and a positive predictive value of 90.0% (95% CI, 55.5–99.7) in contrast to 49 false positive results on standard screening, for false positive rate of 0.311% (95% CI, 0.23–0.41) and a positive predictive values of 14.0% (95% CI, 6.3–25.8) (P < 0.001 for both comparisons). Of the 11,185 patients who underwent cfDNA testing and standard screening for trisomy 13, there were 2 confirmed cases. Of these, cfDNA identified two while standard screening identified one. One false positive result was noted on cfDNA screening and 28 false positive results on standard screening, for false positive rates of 0.02% (95% CI, 0–0.06) and 0.25% (95% CI, 0.17–0.36), respectively (P < 0.001).24 It is important to note that 3% of cfDNA screening did not yield results due to assay variation or low fetal fraction, as previously reported.25,26
Kagan et al in 2018 performed a controlled trial in which patients with normal first-trimester ultrasound assessment between 11 and 13 weeks’ gestation (nuchal translucency ≤ 3.5 mm) were randomized into two groups.27 In the first group, risk of fetal aneuploidy was evaluated according to maternal age, gestational age, nuchal translucency thickness (NT), maternal serum screen levels of PAPP-A and free beta hCG. In the second group, risk assessment was based upon ultrasound findings and cfDNA analysis. Overall, 1518 patients with singleton pregnancies underwent first-trimester assessment. Thirty-one (2.0%) pregnancies were not eligible for inclusion in the study due to increased NT (>3.5 mm) and/or fetal defect. Following exclusion of patients who declined randomization (n = 87) and cases of fetal demise and absent follow-up (n = 24), 688 patients were randomized to standard screening and 688 patients to ultrasound and with cfDNA screening. No differences were noted in maternal and gestational age, maternal weight and BMI, ethnicity, use of assisted reproductive technologies (ART) and smoking between the two groups. In the ultrasound combined with cfDNA screening group, median risk for trisomy 21 was 1 in 10,000. None of the patients had a risk above 1:100 (95% CI, 0.0–0.5%). In the standard screening group, the median risk for trisomy 21 was 1 in 3787. In 17 patients, the risk was higher than 1:100, corresponding to 2.5% (95% CI, 1.5–3.9%) of the standard screening group.27 These results were considered to confirm that first-trimester risk screening for trisomy 21, which includes a detailed ultrasound examination and nuchal translucency measurement followed by cfDNA testing, is associated with a significant decrease in the false-positive rate in comparison with standard screening technology. Thus, the screening modality combining first-trimester ultrasound with cfDNA analysis negates the need for maternal serum free beta hCG and PAPP-A in screening for fetal aneuploidy.
Carrara et al assessed the benefit and reliability of cfDNA in screening for trisomies 21, 18 and 13 for patients with free hCG < 0.25 MoM, hCG > 5.0 MoM, and/or PAPP-A < 0.5 MoM, PAPP-A > 2.5 MoM. This group of investigators assessed 798 patients who were assessed with cfDNA assay in this context.28 Twenty-one patients (4.2%) were unavailable for follow up. Of 477 cases remaining, cfDNA screening failed in two patients. cfDNA was positive for Trisomy 21 (n = 19), Trisomy 18 (n = 6) and Trisomy 13 (n = 1) and negative in 449 patients. The sensitivity of cfDNA screening for Trisomy 21 screening was 100% (19/19) (95% CI: 82.4–100), and specificity 100% (458/458) (95% CI: 99.2–100). Interestingly, fetal growth restriction was significantly associated with a low fetal fraction (OR = 0.87, 95% CI: 0.79–0.96), P = 0.006). These authors concluded that cfDNA is a reliable and effective clinical tool for women with abnormal first-trimester serum biomarker profiles.28
Migliorini et al in a randomized clinical trial of patient’s experience comparing pre- and post-test survey questionnaires addressing measure reassurance, satisfaction, and anxiety between patients undergoing standard first-trimester screening (nuchal translucency and maternal serum analytes) versus ultrasound findings followed by cfDNA analysis found that mean score for reassurance was significantly higher in the cfDNA group in comparison with standard screening in both pre- and post-test questionnaires.29 Forty women with singleton gestations were included in the study. Women were randomized at the time of their first prenatal visit to either first-trimester risk assessment based on first-trimester combined screening or first-trimester risk assessment based on sonographic findings and cfDNA. First-trimester combined screening included ultrasound evaluation with crown-rump length (CRL), nuchal translucency (NT) measurement, and a detailed sonographic assessment, in addition to maternal serum analyte (PAPP-A and free beta hCG). In this group, invasive diagnostic testing was offered to patients with risk >1 in 100, or NT >3.5 mm, or alternatively any sonographic fetal abnormalities. Patients were randomized in the intervention group were evaluated with first-trimester risk assessment based on sonographic findings and cfDNA. Cell-free DNA screening consisted of simultaneous microarray-based assay of non-polymorphic (chromosomes 13, 18, 21, X and Y) and polymorphic loci evaluating chromosome proportion and fetal fraction. In the intervention group, invasive diagnostic testing was offered to patients with abnormal cfDNA screening, or increased nuchal translucency thickness (NT) >3.5 mm, or any sonographic fetal abnormalities.
Mean score for reassurance was significantly higher in the cfDNA group in comparison to the first-trimester combined screening group in the pre-test survey (MD 0.80 points, 95% CI 0.27–1.33) and in the post-test survey (MD 16.50 points, 95% CI 2.18–0.82).29 Patients randomized to cfDNA screening exhibited higher satisfaction and lower mean anxiety scores as assessed in the STAI (State-Trait Anxiety Inventory) pre-test questionnaire. These authors thus concluded that first-trimester screening for fetal aneuploidy combined with detailed sonographic examination and cfDNA is associated with improved maternal satisfaction and maternal reassurance compared to the standard first-trimester combined screening with nuchal translucency (NT), and maternal serum analytes.29
Iwarsson and Conner from the Karolinska Institute, studied detection rates and residual risk for postnatal diagnosis of an atypical chromosomal aberration after combined first-trimester screening.30 Among 129,493 patients, 852 (0.7%) clinically significant chromosome aberrations, including those detected after birth were noted. Of these, a total of 12% were atypical chromosome aberrations. Following consideration that 40% were found subsequent to early pregnancy loss, fetal demise or sonographic detection of a structural anomaly, there was a 0.05% (1:2000) background risk of a postnatal diagnosis of a live-born infant with an atypical chromosomal aberration if no further test is performed throughout pregnancy. If all patients with an increased risk (above or equal to 1:200) had an invasive test and cfDNA screening was performed up to a risk of 1:1000, 95% of common trisomies/sex chromosome aberrations and 55% of atypical aberrations would have been detected.30 These authors concluded that if only cfDNA screening was offered to all women with a combined first-trimester screen without diagnostic testing, three-fold as many children would be born with an atypical chromosome aberration, not identifiable on cfDNA screening.30
Bjerrgaard et al reported, that following cfDNA screening implementation, invasive testing rates decreased considerably from 70% to 48% among women with high-risk pregnancies (P < 0.01). In addition, high-risk women declining further testing decreased from 26% to 3% (P < 0.01).31
More recently, cfDNA technology has continued to expand far beyond its initial capability of detecting common chromosome aneuploidies, and has extended into genome-wide copy number evaluations, detection of microdeletion and microduplication syndromes, and even single gene disorders.32–41 These recent updates in technology have enabled increased and superior detection of underlying etiologies in the presence of first-trimester sonographic finding of cystic hygroma.41
It is important here to note that maternal serum screening has the capability of detecting pregnancies at increased risk for overall adverse perinatal outcome. These include (but are not limited to): pregnancy loss/stillbirth, fetal growth restriction (FGR), premature rupture of membranes, preterm delivery, placental abruption, and preeclampsia.42–44 Evaluation of such risks using cfDNA fetal fraction is being evaluated but not currently well-defined.
First-Trimester Septated Cystic Hygroma
This striking and clear sonographic entity consists of the finding of sacculated, fluid-filled cystic structures distending/projecting from the posterior-lateral aspect of the fetal neck (Figures 1–5). First-trimester septated cystic hygroma is most often noted during the following indications for sonographic evaluation:
- During the complete first-trimester screening protocol for aneuploidy.
- During independent fetal nuchal translucency thickness assessment.
- In centers practicing detailed late first-trimester sonographic evaluation of fetal anatomy.
- Not uncommonly, first-trimester septated cystic hygroma is an anecdotal sonographic finding during the assessment of first-trimester uterine hemorrhage, pain or threatened miscarriage.
 |
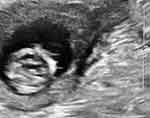
Figure 2 First-trimester septated cystic hygroma in a 29 year-old, P2 at 11 and 3/7 weeks’ gestation. Non-invasive prenatal screening was low risk for fetal aneuploidy, yet maternal Noonan’s syndrome was suspected and later confirmed by conventional testing. Fetal Noonan’s syndrome was confirmed at amniocentesis. The patient elected to continue her pregnancy and delivered at 39 weeks’ gestation. The male neonate exhibited facial dysmorphic features consistent with established Noonan’s syndrome (see original report, Sherer DM, Hsieh V, Kheyman M, Makanjuola O, Dalloul M. Cell-free DNA screening following first-trimester septated cystic hygroma leading to diagnosis of previously unknown familial Noonan Syndrome. Eur J Obstet Gynecol Reprod Biol. 2021 Sep;264:389. doi: 10.1016/j.ejogrb. 2021.07.0566).100 |
Morphogenesis
Cystic hygromas likely result from a defect in the development of lymphatic vessels. The fetal lymphatic vessels drain into two large sacs located lateral to the jugular veins. These jugular lymph sacs eventually will communicate directly with the venous system becoming the terminal portions of the right lymphatic duct and thoracic duct, respectfully.45 In the event that lymphatic and venous structures do not connect, the jugular lymph sacs enlarge and lymph accumulates in tissues, resulting in cystic hygromas of the posterior triangles of the neck.46,47 Thus, van der Putte suggested, following study of seven spontaneously aborted fetuses characterized by large cystic hygromas, that this disorder essentially represents generalized hypoplasia with partial agenesis of the lymphatic system, which failed to extend peripherally during early embryonic development.46,47 Cystic hygroma thus represents a congenital anomaly of the fetal lymphatic system in which obstruction between the lymphatic and venous vessels of the fetal neck results in accumulation of lymphatic fluid in the jugular lymphatic sacs. Large and extensive cystic hygromas may extend anatomically and include axillary lymphatic collections.
It appears that if the jugular lymph sacs and the jugular veins subsequently connect or an alternative route of lymphatic drainage develops prior to fetal demise, the cystic hygromas may regress and peripheral edema may spontaneously resolve. Notwithstanding, it is important to recognize that resolution of first-trimester septated cystic hygroma does not infer euploid fetal status nor normal structural fetal anatomy as resolution of this late first-trimester sonographic finding has been reported despite the presence of significant underlying aneuploidy and/or structural fetal anomalies.
Fetal/Neonatal Outcome in Pregnancies Complicated by the Presence of First-Trimester Septated Cystic Hygroma
The late first-trimester sonographic finding of septated cystic hygroma has long been recognized as a marker for fetal aneuploidy.48,49 Initially, prior to the advent (and later widespread utilization) of transvaginal sonography, this ominous finding was noted at transabdominal mid-trimester scanning and was found strongly associated with monosomy X (45,X, Turner syndrome) and other aneuploidies.48–63 A study of 15 fetuses with cystic hygroma appearing as single or multiloculated fluid-filled cavities noted that none of the fetuses survived. Eleven of the fetuses (73%) had karyotypes consistent with Turner’s syndrome.48 Application of transvaginal sonography has greatly enhanced earlier, first-trimester ultrasonographic diagnosis of septated cystic hygroma.63–81 Utilization of this sonographic modality enabled depiction of subtle septations, which became widely recognized as independent risk factors for chromosomal anomalies,77,78 consistent with earlier second-trimester observations of septations.53,54 Application of first-trimester screening for fetal aneuploidy and the National Institute for Child Health and Human Development (NICHD) multi-center First And Second Trimester Evaluation of Risk (FASTER) trial in the United States validated the European experience with first-trimester screening for aneuploidy prior to recommendation by governing bodies of this diagnostic tool in the USA. These data have led to increased knowledge regarding the ominous first-trimester sonographic finding of first-trimester septated cystic hygroma.82 In addition, recently published data regarding 410 cases of first-trimester septated cystic hygroma from among 110,000 pregnancies in Ireland, have confirmed associated abnormal chromosomal findings among such fetuses and an increased likelihood of structural anomalies among euploid fetuses with first-trimester septated cystic hygroma.83
In a study of 134 cases of first-trimester septated cystic hygroma (generated from the FASTER Trial Research Consortium data (among 38,167 screened patients)) Malone et al in 2005, reported 132 patients with complete follow up.82 Chromosomal abnormalities were confirmed in 67 (51%) patients. These included 25 cases of trisomy 21, 19 cases of monosomy X, 13 cases of trisomy 18, and ten others.82 Major structural fetal malformations (predominantly cardiac and skeletal systems) were documented in 22 of the remaining 65 patients (34%). There were 5 cases (8%) of fetal demise and 15 cases of voluntary pregnancy termination, without evidence of abnormality. One of 23 (4%) normal surviving infants was later diagnosed with developmental delay and cerebral palsy. In all, survival with normal neonatal outcome was documented in 17% of cases (22 of 132).82 In comparison with simple increased nuchal translucency, cystic hygroma is associated with 5, 12, and 6-fold increased risk of aneuploidy, cardiac malformation, and perinatal death, respectively. These authors concluded that first-trimester cystic hygroma is a frequent finding in a general obstetric screening program.82 This sonographic finding exhibited the strongest prenatal association with aneuploidy hitherto, with significantly worse outcome in comparison with simple increased nuchal translucency thickness (NT). The majority of pregnancies with normal evaluation at the completion of the second trimester resulted in a healthy infant with a normal neonatal outcome.82
Visualization of nuchal septations at first-trimester genetic screening as a powerful risk factor for chromosomal anomalies, independent of increased nuchal translucency has been confirmed in subsequent studies. Mack et al in a retrospective cohort study of all patients who underwent first-trimester genetic screening during 25 months (between 2011 and 2014), attempted to determine whether nuchal septations represent a risk factor for chromosomal abnormalities independent of nuchal translucency (NT).78 The 95th percentile for the NT measurements were calculated for each gestational week. Multivariable logistic regression analysis determined whether depiction of nuchal septations constitute an independent risk factor for chromosomal analysis while controlling for existing confounding variables. Chromosomal abnormalities were present in 1.0% of the study group (33/3275). The prevalence of chromosomal abnormalities was significantly higher among fetuses with nuchal septations in comparison with fetuses with normal nuchal translucency without septations (P < 0.001) and those with NT above the 95th percentile without septations (P < 0 0.001). The sonographic evidence of septations was associated with an increased risk of chromosomal abnormalities (odds ratio, 40.0; 95% confidence interval, 9.1–174.0) after controlling for NT measurements and other confounding variables. These authors concluded that sonographic depiction of nuchal septations during first-trimester genetic screening is a powerful risk factor for chromosomal anomalies, independent of increased NT.78
Recently, Malone et al performed a large retrospective cohort study between 2007 and 2017 of data generated at a single tertiary referral prenatal diagnosis center with the aim of establishing contemporary outcome data, especially in the setting of normal karyotype.83 Data were analyzed from a prospective fetal anomaly database. All cases were confirmed to have nuchal translucency (NT) >3 mm, with septations. Cases of simple increased nuchal translucency thickness without septations were excluded from the analysis. Throughout the study, in excess of 110,000 pregnancies were delivered at this center, including 410 cases of first-trimester septated cystic hygroma (diagnosed prior to 14 weeks’ gestation) (1:268) an incidence striking parallel to previously established FASTER trial data (1:285).83 Pregnancy outcome was documented in 99% (405/410) of patients, with detailed pathology outcome available in 92% (378/410) of cases. A total of 87% of patients (351/405) underwent invasive prenatal testing, and postnatal chromosome status was documented in another 27 cases. A total of 61% of patients (230/378) had confirmed abnormal chromosomal abnormalities. Of the 39% of patients (148/378) with normal chromosome analysis, only 13% (19/148) had a significant structural fetal abnormality (including 7 cardiac and 12 non-cardiac abnormalities). Overall, perinatal loss was 62% (253/405). Interestingly, the total survival rate of euploid fetuses/neonates manifesting first-trimester septated cystic hygroma without structural abnormalities was 84% (108/129).83
These data clearly substantiate the established strong likelihood of an abnormal karyotype, which occurs in 61% of patients. Notwithstanding, once fetal chromosomal abnormality is excluded, these results demonstrated a 13% incidence of major structural fetal abnormality, a considerably lower incidence than previously reported. Euploid fetuses have a 77% survival rate. This study represents the largest single-center study of first-trimester septated cystic hygroma with complete outcome data to date, and clearly should be incorporated in current patient counseling, with an emphasis on the favorable outcome of euploid fetuses without structural fetal anomalies. Thus, these data have been incorporated in our current counseling of these not infrequent patients.83
Role of cfDNA Screening in the Assessment of First-Trimester Septated Cystic Hygroma
First-trimester septated cystic hygroma is typically detected between 11 weeks to 14 weeks gestation. Due to the timing of such sonogram finding, diagnostic testing offerings may be limited. Chorionic villus sampling (CVS) is typically offered between 10 and 12 weeks’ gestation and amniocentesis is typically performed between 15 and 22 weeks’ gestation. Early amniocentesis (11–14 weeks’ gestation) is not offered at all medical centers and the availability of CVS varies in different parts of the United States. For this reason, this potentially ominous sonogram finding uniquely qualifies for immediate cfDNA screening (also known as NIPS/NIPT). Currently, universal cfDNA screening for fetal aneuploidy and select microdeletions are gradually replacing the labor intensive combined first-trimester screening.20,84 Since ACOG opinion of 2020 stating that all women, irrespective of age, should be offered non-invasive cfDNA screening, the uptake of NIPS has increased substantially.20 As a result, a considerable decrease in the number of diagnostic (invasive) testing has been reported.31,85,86
In recent years, cell-free DNA technology has improved to include the option of genome-wide analysis of copy number variants larger than 7Mb.87 Such testing has the capability of detecting rare chromosome aneuploidy and copy number variants with resolution similar to a traditional karyotype, which detects CMV between 5 and 10 Mb.87 In the 3 years of clinical experience of genome-wide NIPS, 25% of positive results can only be detected through this technology and would otherwise be missed by traditional NIPS. In the presence of first-trimester septated cystic hygroma, cfDNA screening with the highest diagnostic yield should be considered especially in the absence of diagnostic testing such as amniocentesis or chorionic villus sampling, or according to patient preference following counseling.
In addition to genome-wide analysis, cfDNA screening has expanded further into single gene disorders. Currently, there is widely available cfDNA screening, which is capable of detecting single gene pathogenic mutations across 30 genes. Conditions screened are autosomal or X-linked dominant in inheritance with reported cases of de novo mutations.88 These conditions affect quality of life and affected individuals may benefit from medical and/or surgical intervention (mainly impacting skeletal and neurological development). This unique cell-free DNA screening option is able to detect conditions such as Noonan syndrome, achondroplasia, osteogenesis imperfecta, and Rett syndrome.88,89 First-trimester septated cystic hygroma has been associated with an increased risk of Noonan syndrome in pregnancy. Traditionally, single gene analysis with diagnostic testing has been restricted by insurance coverage and laboratory selection. Moreover, turnaround time is typically between 4 and 6 weeks due requirement of cell cultures by some laboratories. Cell-free DNA analysis of these conditions improves diagnostic yield and can be performed between the timing of detection of cystic hygroma on sonogram and the time in which amniocentesis can be offered.
Cell-free DNA screening technologies continue to improve with additional sensitivity for copy number variants and have expanded into single gene disorders. In the presence of first-trimester finding of septated cystic hygroma, diagnostic testing typically includes karyotype, microarray, and Noonan syndrome panel. However, due to the timing of such sonogram findings, patients are often in limbo between the timing of CVS and amniocentesis, without an immediate diagnostic testing option. Cell-free DNA screening can be implemented in this time-frame to provide additional information for the patient and inform providers of the probable outcome. The typical turnaround time for genome-wide cell-free DNA screening is 1–2 weeks, and the typical turnaround time for single gene cell-free DNA screening is 2–3 weeks. Results from cfDNA screening can also guide prioritization of different diagnostic tests for confirmation.
Both of the cfDNA screening technologies discussed are available for patients starting at 9 weeks gestation. Even in the event of pregnancy loss, maternal samples collected at the time of the identification of cystic hygroma would likely provide some analytic yield and may be offered to the patient who has elected for expectant management in cases of first-trimester non-immune fetal hydrops when invasive diagnostic testing following counseling is declined (Figures 6 and 7).
The difference between screening and diagnostic testing modalities cannot be oversimplified. Although cfDNA analysis provides a relatively robust and accurate means of prenatal genetic evaluation, it remains a screening tool. It is widely understood cfDNA in maternal circulation are placental in origin and a well-known limitation of the test is its inability to evaluate placental mosaicism.31 The fetus may have different expressions of mosaicism than the placenta. Although placental mosaicism has been well documented as a limitation for atypical chromosome aneuploidies and copy number variants, it is not currently well understood for single gene disorders. The current working understanding is single gene disorders are less likely to be impacted by placental mosaicism, unless they are due to larger chromosomal CMV.32 It is also important to state cfDNA analysis is cannot be performed for a subset of patients due to a variety of factors. Patients with increased BMI or on certain medications may have repeated low fetal fraction in which cfDNA analysis is not possible. Patients with maternal blood malignancies, uterine fibroids, or other malignancies contributing cfDNA in circulation are also not candidates for cfDNA analysis.90 Currently, genome-wide cfDNA screening and single gene cfDNA screening are not available for twin pregnancies or other higher order multiple pregnancies.91 Although cfDNA analysis has created an exciting new frontier of prenatal testing, it may not be available for all patients under all circumstances.
Esoteric Chromosomal Findings Associated with Septated Cystic Hygroma
In addition to the relatively common aneuploidies noted in association with septated cystic hygroma, unusual, esoteric chromosomal abnormalities have been reported.92–99 Chen et al reported that amniocentesis performed at 16 weeks’ gestation in a fetus with a large septated cystic hygroma revealed a terminal deletion in the long arm of chromosome 10.89 The paternal karyotype was subsequently confirmed 46, XY, t(10;18)(q25.3;q23), confirming an inherited unbalanced translocation. The maternal karyotype was 46XX. The fetal karyotype was 46, XY, der(10)t(10;18)(q25.3;q23)pat. The patient underwent pregnancy termination.
Vivic et al reported prenatal diagnosis of 18p deletion and isochromosome 8q mosaicism in a fetus with a cystic hygroma.92 Cytogenetic analysis depicted a mosaic 46, XX, del(18)(p11.2)/46, XX, i(18)(q10). The patient underwent pregnancy termination.
Stipoljev et al in 2021 reported prenatal diagnosis of partial trisomy 2p and partial monosomy 3p (unbalanced translocation (2;3)(p25.1;p25.3) of paternal origin), in association with first-trimester septated cystic hygroma.93 The duplicated region at 2p25.1p25.3 contains 45 different genes, among which 12 are reported as OMIM morbid genes with various phenotypical implications. The deleted region at 3p26.3-p25.3 contains 65 genes, of which 27 (41.5%) are OMIM genes.
Ples et al reported prenatal diagnosis of Niemann-Pick disease type C2 following first-trimester sonographic depiction of a septated cystic hygroma.94 Following normal karyotype at chorionic villus sampling, additional evaluation of 1,024 genes underlying structural anomalies was performed. Homozygous mutation of the NPC2 gene (OMIM 601015), was found, consistent with Niemann-Pick disease type C2. The patient underwent pregnancy termination. In a subsequent pregnancy, interestingly, despite normal sonographic findings, molecular testing for the previous familial mutation identified the fetus as homozygous for this mutation. The patient underwent termination of pregnancy.94
Tica et al described a unique case of recurrent first-trimester cystic hygroma occurring in subsequent pregnancies of a 27 year-old P1. In each of these cases, unusual chromosomal abnormalities were noted.95 In the first case, a fetal inherited heteromorphism of chromosome 1 (1qh+) was noted and found to be inherited from the mother (46, XX1qh+,14ps, 21ps). The patient underwent termination of pregnancy. The karyotype of the fetus in the recurrent first-trimester septated cystic hygroma was triploidy 69, XXX.95 In 2009 Teague et al reported a patient with recurrent fetal septated cystic hygroma (at 11 weeks’ gestation and mid-trimester in a subsequent pregnancy).96 The patient also had two previous pregnancies complicated by cystic hygroma and nonimmune hydrops fetalis (NIHF) with 46, XX karyotypes.96 Following these events, the authors suggested that cystic hygroma associated with a normal karyotype may be inherited as an autosomal dominant trait. Recurrent first-trimester septated cystic hygroma with hydrops fetalis was also previously reported in 2009 by Baxi et al five fetuses (four pregnancies) of a 39 year-old P8 of Ashkenazi Jewish descent.97 In three separate fetuses (two pregnancies), the authors excluded aneuploidy, Noonan syndrome, Fryns syndrome and Gunther’s disease and suggested an undetermined autosomal recessive disorder.
First-trimester septated cystic hygroma (increased nuchal translucency and nonimmune hydrops) has been associated with Noonan syndrome (an autosomal dominant disorder characterized by short stature, congenital heart defects and distinctive facies).98–102 This disorder is genetically heterogeneous with approximately 50% of patients demonstrating PTPN11 mutations. Studies of fetuses with cystic hygroma suggest a prevalence of 1–3% of Noonan syndrome. In a retrospective review, Lee et al assessed the utility of PTPN11 testing based on prenatal sonographic findings (n = 134).98 Most commonly reported indications for testing were increased nuchal translucency (NT) and cystic hygroma. Analysis demonstrated that 12 fetuses had heterozygous missense mutations, corresponding to a positive test rate of 9%. PTPN11 mutations were identified in 16% and 2% of fetuses with cystic hygroma and increased nuchal translucency, respectively. Among fetuses with isolated cystic hygroma, PTPN11 mutation prevalence was 11%.98 Mutations observed in the three fetuses with hydrops fetalis have been reported as somatic cancer mutations. Prenatal PTPN11 testing has diagnostic and potential prognostic properties that may assist in risk assessment and genetic counseling. Negative PTPN11 testing cannot exclude the diagnosis in that Noonan syndrome is genetically heterogeneous, requiring further studies regarding the other Noonan syndrome genes.
Sherer et al reported a case of first-trimester septated cystic hygroma in which cfDNA screening for serious single-gene disorders identifying a maternal pathogenic variant c.188A > G in PTPN11 (Figure 2).100 Due to the nature of cfDNA screening and the maternal contribution of cfDNA, this test was unable to confirm whether the fetus had inherited the maternal Noonan syndrome mutation (a 50% risk of direct inheritance). Confirmatory testing was performed for both the patient and cultured amniocytes, concluding a direct inheritance of Noonan syndrome. The patient herself was later recognized with subtle facial structures consistent with Noonan syndrome, yet reported no personal or familial history of developmental delays, intellectual disabilities, congenital heart defects or short stature, and hence elected to continue her pregnancy. This case was the first report of cfDNA screening after a finding of first-trimester septated cystic hygroma leading to the diagnosis of previously unestablished familial Noonan syndrome.100
Pan et at al in 2020 described two cases of first-trimester septated cystic hygroma with normal karyotype and microarray workup in whom no additional structural anomalies were noted.101 Subsequently the two infants manifested early developmental delays within two years of age. Exome sequencing of the two children confirmed one child with mental retardation, autosomal dominant 23 (MRD23) with a c.646delC variant in SETD5 gene, and the other child had Smith-Magenis syndrome, c.3103dupC variant in RA1 gene.101
In addition to its association with chromosome aneuploidy and microarray copy number variants, prenatal diagnosis of septated cystic hygroma conveys increased risk for single gene disorders. Until fetal whole exome/genome sequencing (WES/WGS) becomes available clinically, evaluation of single gene disorders in the presence of first-trimester septated cystic hygroma, relies upon clinical judgment of the provider patient insurance, and patient socioeconomic status. A guideline-based gene panel for septated cystic hygroma would be helpful in the patient management of this sonographic finding. Long-term follow up studies following euploid fetuses with septated cystic hygroma without structural anomalies, have not been published and hence neurodevelopmental deviations in this clinical scenario have not been assessed.
First-Trimester Septated Cystic Hygroma Associated with Generalized Fetal Subcutaneous Edema [Nonimmune Hydrops Fetalis (NIHF)]
In contrast, the combination of first-trimester septated cystic hygroma in the presence of additional signs of edema, often total body subcutaneous edema, and other abnormal collections of fluid [pleural effusion, pericardial fluid and ascites – all consistent with nonimmune fetal hydrops (NIHF)] are considered ominous findings often preceding or associated with existing fetal demise (Figures 6 and 7). We believe that despite the ominous prognosis, these cases, which carry a considerably increased risk of fetal aneuploidy and hence, following patient consent, merit a detailed genetic workup.
In a retrospective cohort study of all nonimmune hydrops fetalis (NIHF) cases diagnosed prenatally (excluding NIHF due to twin–twin transfusion syndrome) at the University of California, San Francisco between 2008 and 2018, Mardy et al described the overall yield of chromosomal microarray (CMA) in the diagnostic evaluation of NIHF, comparing isolated cases to those with concurrent structural anomalies.102 Prenatally diagnosed NIHF was reported in 131 cases. In 43/44 cases with a CMA data, results were grouped as normal, or likely benign. One case was found on CMA analysis to have a large pathogenic duplication of 21p11.2q22.3, which could have been detected by karyotype and was consistent with Trisomy 21. There was no incremental yield demonstrated for detailed CMA analysis over karyotype. Thus, these authors concluded that, in their study of prenatally diagnosed NIHF cases, CMA analysis did not identify any copy number variants beyond those detectable by karyotype, and the clear majority of CMAs were normal. These results suggest that CMA analysis has low diagnostic utility for NIHF.102
In contrast, Sparks et al reported a series of 127 consecutive unexplained cases of NIHF following the presence of fetal ascites, pleural or pericardial effusions, skin edema, increased nuchal translucency or cystic hygroma, or any combination of these findings.103 Primary outcome was the diagnostic yield of exome-sequencing for the detection of genetic variants that were classified as either pathogenic or likely pathogenic according to criteria of the American College of Medical Genetics and Genomics. Secondary outcomes consisted of the percentage of cases associated with specific genetic disorders and the proportion of variants that were inherited. In 37 of the 127 cases (29%), diagnostic genetic variants were identified, including those for disorders affecting the RAS-MAPK cell-signaling pathway (otherwise termed RASopathies) (30% of genetic diagnoses); inborn errors of metabolism and musculoskeletal disorders (11% each); lymphatic, neurodevelopmental, cardiovascular, and hematologic disorders (8% each); and others. Prognoses varied and included a range from relatively mild outcomes to perinatal death. Overall, 68% of the cases (25 of 37) with diagnostic variants were autosomal dominant (12% were inherited and 88% de novo), 27% (10 of 37) were autosomal recessive (95% were inherited and 5% de novo), one case was X-linked recessive inherited, and one was of undetermined inheritance. Potentially diagnostic variants were identified in an additional 12 cases.103
Mone et al in a prospective study determined the incremental yield of whole exome-sequencing (WES) over chromosomal microarray analysis (CMA) or karyotyping in prenatally diagnosed non-immune hydrops fetalis (NIHF).104 The study included 28 patients with prenatally diagnosed NIHF who underwent trio exomic sequencing following negative CMA or karyotyping. These cases were combined with data from a systematic literature review (including a meta-analysis). Additional diagnostic yield of WES over CMA or karyotyping was 25.0% (7/28) in all patient with NIHF, 21.4% (3/14) in patients with isolated NIHF and 28.6% (4/14) in patients with non-isolated NIHF. In the meta-analysis, pooled incremental yield based on 21 studies (306 cases) was 29% (95% CI, 24–34%; P < 0.00001; I2 = 0%) in all NIHF, 21% (95% CI, 13–30%; P < 0.00001; P = 0%) among patients with isolated NIHF and 39% (95% CI, 30–49%; P < 0.00001; I2 = 1%) among patients with NIHF associated with an additional fetal structural malformation. In the latter group, congenital limb contractures were the most prevalent additional structural anomaly associated with a causal pathogenic variant, occurring in 17.3% (19/110) of cases. Incremental yield did not differ according to hydrops severity. Most common genetic disorders identified were RASopathies, which occurred in 30.3% (27/89) of patients with a causal pathogenic variant, most commonly due to a PTPN11 variant (44.4%; 12/27). The predominant inheritance pattern in causal pathogenic variants was autosomal dominant in monoallelic disease genes (57.3%; 51/89), with most representing de novo events (86.3%; 44/51).104 These authors concluded that prenatal next-generation sequencing in patients with either isolated or non-isolated NIHF should be considered in the development of clinical pathways. Following the wide range of potential syndromic conditions and heterogeneity of the prenatal phenotype of NIHF, exome or whole-genome sequencing may represent a more appropriate testing modality than targeted gene panel testing.104
Norton et al, recently performed a secondary analysis of a study of exome-sequencing for NIHF, which included exome sequencing and phenotype-driven variant analyses that were completed in 127 patients with such fetuses. Exome-sequencing identified a pathogenic or likely variant in 37/127 (29%) of patients, with a total of 29 genes.105 A variant of undetermined significance strongly suspected associated with the phenotype was identified in an additional 12 patients (9%). These authors noted that when a NIHF targeted gene panel is utilized in lieu of exome-sequencing, 13 to 15 of the 29 genes (45–52%) identified in the NIHF cohort would have been sequenced, and 19 to 24 of the pathogenic variants (51–62%) would have been detected. The yield was predicted to be the lowest with the metabolic panel (11%) and the highest with the largest NIHF panel (62%). The largest NIHF gene panel would have a diagnostic yield of 18% compared with 29% with exome-sequencing. These authors concluded that the broader coverage of exome-sequencing for genetically heterogeneous conditions such as NIHF, was a superior alternative to targeted gene specific panels.105 Notwithstanding, the commercial availability and insurance coverage for WES in the perinatal setting has been limited, causing disparities in patient access to such testing.
In contrast, Mellis et al evaluated the utilization of prenatal exome-sequencing in the assessment of isolated nuchal translucency (NT) among 213 patients with fetuses with increased nuchal translucency (NT) ≥3.5 mm between 11 and 14 weeks’ gestation.106 Cases were grouped based on additional structural anomalies in the first-trimester including later in pregnancy or nuchal translucency (NT) at presentation. Diagnostic variants were detected in 12/54 (22.2%) fetuses presenting with non-isolated increased nuchal translucency, 12/37 (32.4%) fetuses with isolated increased nuchal translucency and additional anomalies later in gestation, and 2/111 (1.8%) fetuses with isolated increased nuchal translucency in the first-trimester and no other anomalies later in pregnancy. These authors concluded that diagnostic yield of prenatal exome-sequencing is low in the assessment of fetuses with truly isolated increased nuchal translucency (NT) yet is considerably higher in the presence of additional fetal malformation noted later in pregnancy. Continued sonographic follow up of fetuses with increased nuchal translucency (NT) is recommended, in that in the event a fetal anomaly is noted later in gestation, prenatal exome sequencing may be warranted.106
First-Trimester Screening for Structural Anomalies
Karim et al utilizing a systematic review and meta-analysis of all relevant publications determined sensitivity and specificity rates of first-trimester ultrasound for the detection of fetal anomalies and established factors which might impact screening.107 Thirty studies published between 1991 and 2014 were included. The pooled estimate for the detection of major anomalies in low-risk or unselected populations (19 studies, 115,731 fetuses) was 46.1% (95% CI: 36.88–55.46%). The detection rate for all anomalies in low-risk or unselected populations (14 studies, 97,976 fetuses) was 32.35% (95% CI: 22.45–43.12%). In contrast, in high-risk populations (six studies, 2841 fetuses) the pooled estimate was 61.18% (95% CI: 37.71–82.19%). Thus, these authors found that detection rates of first-trimester fetal structural malformations range between 32% in low-risk patients to above 60% in high-risk patients.107 A similar detection rate of 50% with first-trimester sonographic evidence of structural anomalies was reported recently by Achiron et al.108
In contrast, a retrospective study of patients at a tertiary referral center determined that the introduction of cfDNA screening as an alternative to first-trimester screening has resulted in fewer patients receiving first-trimester ultrasound.109 It is worth emphasizing the importance of first-trimester nuchal translucency sonogram as part of routine prenatal follow up. In the age of new genome testing such as cfDNA analysis, it is important to emphasize the benefits of traditional tried and tested sonographic evaluations.
Products of Conception Testing
Due to the considerably elevated risk of fetal demise/spontaneous pregnancy loss, product of conception testing should also be considered when first-trimester septated cystic hygroma is observed on sonogram. Currently the standard of testing is chromosome karyotype with reflex to chromosome microarray if the patient miscarries in a medical center. Additional options for home collection kits are also on the market where patients save a sample of the product of conception in a take-home kit and mail the sample in for analysis. In the presence of such ominous sonogram finding, a take-home pregnancy loss analysis kit may be considered for patients interested in further workup.
In a study of 340 patients who underwent spontaneous miscarriages, Zhang et al noted 165 patients in whom conception products were diagnosed with chromosomal anomalies, which included: 135 aneuploidies, 11 triploidies, 10 complex abnormalities and 9 segmental aneuploidies.110 In a recent study, Xu et al demonstrated that next-generation sequencing (NGS) in combination with multiplex PCR, is an effective modality to test trisomies in products of conception following spontaneous miscarriage.111
Fan et al similarly evaluated the clinical utility of NGS technology in the identification of chromosomal abnormalities associated with first-trimester pregnancy loss.112 A total of 1010 miscarriage specimens were assessed. Total DNA and NGS analyses were performed. A total of 634 patients with chromosomal variants were detected. Of these, 462 (72.9%) exhibited numerical variants, which included 383 (60.4%) aneuploidies, 44 (6.9%) polyploidies, and 34 (5.5%) mosaicisms.112 The remaining 127 (27.1%) cases demonstrated structural variants including 19 (3.0%) benign copy number variations (CNVs), 52 (8.2%) pathogenic CNVs and 101 (16%) variants of undetermined significance. These data were considered to support that NGS may be utilized for the successful determination of genetic anomalies in pregnancy loss, and the authors recommend that fetal chromosomal analysis be offered routinely for all patients with pregnancy losses, irrespective of their frequency.112
Summary
Late first-trimester sonographic assessment of fetal anatomy, or alternatively, targeted nuchal translucency thickness assessment leading to prenatal diagnosis of septated cystic hygromas requiring genetic assessment of the fetus by invasive testing (chorionic villus sampling or amniocentesis), has been augmented considerably by recent rapidly advancing non-invasive screening technologies. Early data suggest that expanding cfDNA screening including aneuploidy screening for Trisomies 21, 18 and 13, supplemented by select microdeletion technology, genome-wide cfDNA screening and single gene cfDNA screening focusing on a number of autosomal or X-linked dominant conditions, are gaining their respective place in the genetic assessment of first-trimester septated cystic hygroma. It is important to recognize that previously, single gene analysis for prenatal diagnosis, which depended upon availability of prenatal single gene disorder panels and could only be offered as part of invasive prenatal testing, is now becoming increasingly available with non-invasive technologies and better insurance coverage. Although the differences between screening and diagnostic testing cannot be oversimplified, physicians (and patients) should be increasingly aware of and comfortable with the potential benefits (and limitations) of these novel capabilities. cfDNA screening does not replace diagnostic testing, but offers a time-sensitive method of providing patients with more information without delays in waiting for invasive diagnostic procedures. This non-invasive technology represents a truly unique capability to provide highly accurate and timely analysis following notation of early first-trimester sonographic abnormalities, which overall will clearly empower patients in their decision-making.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nicolaides KH, Azar G, Byrne D, Mansur C, Marks K. Fetal nuchal translucency: ultrasound screening for chromosomal defects in first trimester of pregnancy.. BMJ. 1992;304(6831):867–869. doi:10.1136/bmj.304.6831.867
2. van Vugt JM, van Zalen-Sprock RM, Kostense PJ. First-trimester nuchal translucency: a risk analysis on fetal chromosome abnormality. Radiology. 1996;200(2):537–540. doi:10.1148/radiology.200.2.8685353
3. Taipale P, Hiilesmaa V, Salonen R, Ylöstalo P. Increased nuchal translucency as a marker for fetal chromosomal defects. N Eng J Med. 1997;337(23):1654–1658. doi:10.1056/NEJM199712043372303
4. Snijders RJ, Noble P, Sebire N, Souka A, Nicolaides KH. UK multicentre project on assessment of risk by maternal age and fetal nuchal-translucency thickness at 10-14 weeks gestation. Fetal Medicine Foundation First Trimester Screening Group. Lancet. 1998;352(9125):343–346. doi:10.1016/s0140-6736(97)11280-6
5. Nicolaides KH. Nuchal translucency and other first-trimester sonographic markers of chromosomal anomalies. Am J Obstet Gynecol. 2004;191(1):45–67. doi:10.1016/j.acog.2004.03.090
6. Crossley JA, Aitken DA, Cameron AD, McBride E, Connor JM. Combined ultrasound and biochemical screening for Down’s syndrome in the first trimester: a Scottish multicentre study. BJOG. 2002;109(6):667–676. doi:10.1111/j.1471-0528.2002.01394.x
7. Muller F, Benattar C, Audibert F, Roussel N, Dreux S, Cuckle H. First-trimester screening for Down syndrome in France combining fetal nuchal translucency measurement and biochemical markers. Prenat Diagn. 2003;23(10):833–836. doi:10.1002/pd.77
8. Chasen ST, Sharma G, Kalish RB, Chervenak FA. First-trimester screening for aneuploidy with fetal nuchal translucency in the United States. Ultrasound Obstet Gynecol. 2003;22(2):149–151. doi:10.1002/uog.174
9. Muller F, Benattar C, Audibert F, Roussel N, Dreux S, Cuckle H. First-trimester screening for Down syndrome in France combining fetal nuchal translucency measurement and biochemical markers. Prenat Diag. 2003;23(10):833–836. doi:10.1002/pd.700
10. Scott F, Peters H, Bonifacio M, et al. Prospective evaluation of a first trimester screening program for Down syndrome and other chromosomal abnormalities using maternal age, nuchal translucency, and biochemistry in an Australian population. Aust NZ J Obstet Gynaecol. 2004;44(3):205–209. doi:10.1111/j.1479-828X.2004.00205.x
11. Malone FD, Canick JA, Ball RH, et al. First- and Second-Trimester Evaluation of Risk (FASTER) Research Consortium. First-trimester or second-trimester screening or both, for Down’s syndrome. N Engl J Med. 2015;353(19):2001–2011. doi:10.1056/NEJMMoa043693
12. Kublickas M, Crossley J, Aitken D. Screening for Down’s syndrome in the first trimester: combined risk calculation, methodology, and validation of a web-based system. Acta Obstet Gynecol Scand. 2009;88(6):635–638. doi:10.1080/00016340902859299
13. Norton ME. First-trimester screening for chromosomal abnormalities: advantages of an instant results approach. Clin Lab Med. 2010;30(3):565–571. doi:10.1016/j.cli.2010.04015
14. Torring N, Petersen OB, Uldbjerg N. Ten years of experience with first-trimester screening for fetal aneuploidy employing biochemistry from gestational weeks 6+0 to 13+6. Fetal Diagn Ther. 2015;37(1):51–57. doi:10.1159/000362665
15. Lindquist A, Poulton A, Halliday J, Hui L. Prenatal diagnostic testing and atypical chromosome abnormalities following combined first-trimester screening: implications for contingent models of non-invasive prenatal testing. Ultrasound Obstet Gynecol. 2018;51(4):487–492. doi:10.1002/uog.18979
16. Canick JA, Lambert-Messerlian GM, Palomaki GE, et al. Comparison of serum markers in first-trimester down syndrome screening. Obstet Gynecol. 2006;108(5):1192–1199. doi:10.1097/01AOG.0000241095.19638.f2
17. Huang S, Chang C, Cheng P, Hsiao C, Soong Y, Duan T. First-trimester combined screening is effective for the detection of unbalanced chromosomal translocations at 11 to 12 weeks of gestation. Reprod Sci. 2017;21(5):594–600. doi:10.1177/1933719113508818
18. Alldred SK, Takwoingi Y, Guo B, et al. First trimester ultrasound test alone or in combination with first trimester serum test for Downs’ syndrome screening. Cochrane Database Syst Rev. 2017;3(3):CD012600. doi:10.1002/14651858.CD012600
19. Evans MI, Van Decruyes H, Nicolaides KH. Nuchal translucency measurements for first-trimester screening: the ‘price’of inaccuracy. Fetal Diagn Ther. 2007;22(6):400–404. doi:10.1159/000106342
20. Dugoff L. ACOG Practice Bulletin Number 226: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2020;136:e48–e69.
21. Hui L, Pynaker C, Bonacquisto L, et al. Reexamining the optimal nuchal translucency cutoff for diagnostic testing in the cell-free DNA and microarray era: results from the Victorian Perinatal Record Linkage study. Am J Obstet Gynecol. 2021;225(5):
22. Ashoor G, Syngelaki A, Poon LC, Rezende JC, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks’ gestation relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. 2013;41:26–32.
23. Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effect on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013;33:662–666.
24. Norton ME, Jacobsson N, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589–1597. doi:10.1056/NEJMpa1407349
25. Rava RP, Srinivasan A, Sehnert AJ, Bianci DW. Circulating fetal cell-free DNA fractions differ in autosomal aneuploidies and monosomy X. Clin Chem. 2014;1:855.
26. Pergament E, Cukle H, Zimmerman B, et al. Single nucleotide polymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet Gynecol. 2014;124(2 Pt 1):210–218. doi:10.1097/AOG.0000000000000363
27. Kagan KO, Sroka F, Sonek J, et al. First-trimester risk assessment based on ultrasound and cell-free DNA vs. combined screening a randomized controlled trial. Ultrasound Obstet Gynecol. 2018;51(4):437–444. doi:10.1002/uog.18905
28. Carrara J, Vivanti A, Jani JC, Demain A, Costa JM, Benachi A. Usefulness and reliability of cell-free DNA screening for main trisomies in case of atypical profile on first-trimester maternal serum screening. J Transl Med. 2019;17(1):398. doi:10.1186/s12967-019-02152-7
29. Migliorini S, Saccone G, Silvestro F, et al. First-trimester screening based upon cell-free DNA vs combined screening: a randomized clinical trial on women’s experience. Prenat Diagn. 2020;40(11):1482–1488. doi:10.1002/pd.5800
30. Iwarsson E, Conner P. Detection rates and residual risk for a postnatal diagnosis of an atypical chromosome aberration following combined first-trimester screening. Prenat Diagn. 2020;40(7):852–859. doi:10.1002/pd.5698
31. Shi P, Wang Y, Liang H, et al. The potential of expanded noninvasive prenatal screening for detection of microdeletion and microduplication syndromes. Prenat Diagn. 2021;41(10):1332–1342. doi:10.1002/pd6002
32. Mohan P, Lemoine J, Trotter C, et al. Clinical experience with non-invasive prenatal screening for single-gene disorders (NIPT-SGD). Ultrasound Obstet Gynecol. 2021.
33. Vogel I, Petersen OB, Christensen R, Hyett J, Louu S, Vestergaard EM. Chromosomal microarray as primary diagnostic genomic tool for pregnancies at increased risk with a population-based combined first-trimester screening program. Ultrasound Obstet Gynecol. 2018;51(4):480–486. doi:10.1002/uog.17548
34. Lefkowitz RB, Tynan JA, Liu T, et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am J Obstet Gynecol. 2016;215(2):
35. Zhang J, Li J, Feng Y, et al. Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat Med. 2019;25(3):439–447. doi:10.1038/s41591-018-0334x
36. Peters D, Chu T, Yatsenko SA, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011;365(19):1847–1848. doi:10.1056/NEJMc1106975
37. Chu T, Yeniterzi S, Rajovic WA, et al. High resolution non-invasive detection of a fetal microdeletion using the GCREM algorithm. Prenat Diagn. 2014;34(5):469–477. doi:10.1002/pd.4331
38. Che H, Villela D, Dimitriadou D, et al. Noninvasive prenatal diagnosis by genome-wide haplotyping of cell-free DNA. Genet Med. 2020;22(5):962–973. doi:10.1038/s41436-0190748-y
39. Vermeulen C, Geeven G, de Wit E, et al. Sensitive monogenic noninvasive prenatal diagnosis by targeted haplotyping. Am J Med Genet. 2017;101(3):326–339. doi:10.1016/j.ajhg2017.07.012
40. Scotchman E, Shaw B, Chandler N, Chitty LS. Non-invasive prenatal diagnosis and screening for monogenic disorders. Eur J Obstet Gynecol Reprod Biol. 2020;253:320–327. doi:10.1016/j.ejogrb.2020.08.001
41. Dugoff L, Hobbins JC, Malone FD, et al. FASTER Consortium. Obstet Gynecol. 2005;106(2):260–267. doi:10.1097/10.AOG.0000172419.374.eb
42. Gagnon A, Douglas-Wilson R. Obstetrical complications associated with abnormal maternal serum markers analytes. J Obstet Gynaecol Can. 2008;30(10):918–932. doi:10.1016/S1701-2163(16)32973-5
43. Baer RJ, Currier RJ, Norton ME, Flessel MC. Obstetric, perinatal, and fetal outcomes in pregnancies screening results. Obstet Gynecol. 2014;123(3):603–609. doi:10.1097/AOG.0000000000000145
44. Van der Putte SCJ. The development of the lymphatic system in man. Adv Anat Embryol Cell Biol. 1975;5:3–60.
45. Van der Putte SC. Lymphatic malformation in human fetuses. A study of fetuses with Turner’s syndrome or status Bonnevie-Ullrich. Virchows Arch a Pathol Anat Histol. 1977;376(3):233–246. doi:10.1007/BF00432399
46. Wiegand S, Eivazi B, Barth PJ, et al. Pathogenesis of lymphangiomas. Virchow Arch. 2008;453(1):1–8. doi:10.1007/s00428-008-0611-z
47. Chervenak FA, Issacson G, Blakemore KJ, et al. Fetal cystic hygroma. Cause and natural history. N Engl J Med. 1983;309(14):822–825. doi:10.1056/NEJM198310063091403
48. Pijpers L, Reuss A, Stewart PA, Wladimiroff JW, Sachs ES. Fetal cystic hygroma: prenatal diagnosis and management. Obstet Gynecol. 1988;72(2):223–224.
49. Bernstein HS, Filly RA, Goldberg JD, Golbus MS. Prognosis of fetuses with a cystic hygroma. Prenat Diagn. 1991;11(6):349–355. doi:10.1002/pd.1970110603
50. Azar GB, Snijders RJ, Gosden C, Nicolaides KH. Fetal nuchal cystic hygromata: associated malformations and chromosomal defects. Fetal Diagn Ther. 1991;6(1–2):46–57. doi:10.1159/000263624
51. Nadel A, Bromley B, Benacerraf BR. Nuchal thickening or cystic hygromas in the first- and early second-trimester fetuses: prognosis and outcome. Obstet Gynecol. 1993;82(1):43–48.
52. Brumfield CG, Wenstrom KD, David RO, Owen J, Cosper P. Second-trimester cystic hygroma: prognosis of septated and nonseptated lesions. Obstet Gynecol. 1996;88(6):979–982. doi:10.1016/s0029-7844(96)00358-4
53. Bronshtein M, Bar-Hava I, Blumenfeld I, Bejar J, Toder V, Blumenfeld Z. The difference between septated and non-septated nuchal cystic hygroma in the early second-trimester. Obstet Gynecol. 1993;81:863–867.
54. Suchet IB, Van der Westhuizen NG, Labatte MF. Fetal cystic hygromas: further insights into their natural history. Can Assoc Radiol J. 1992;43(6):420–424.
55. Bronshtein M, Rottem S, Yoffe N, Blumenfeld Z. First-trimester and early second-trimester diagnosis of nuchal cystic hygroma by transvaginal sonography: diverse prognosis of the septated and the nonseptated lesion. Am J Obstet Gynecol. 1989;161(1):78–82.
56. Ganapathy R, Guven M, Sethna F, Vivekanada U, Thialagnathan N. Natural history and outcome of prenatally diagnosed cystic hygroma. Prenat Diagn. 2004;24:965–968. doi:10.1002/pd.991
57. Basgul A, Güdücü N, Kavak ZN, Gökaslan H, Uyar E. Three fetuses karyotyped as Turner syndrome with cystic hygroma developing hydrops: prognosis and outcome. Clin Exp Obstet Gynecol. 2007;34(3):182–184.
58. Gedikbasi A, Gul A, Sargin A, Ceylan Y. Cystic hygroma and lymphangioma: associated findings, prenatal outcome, and prognostic factor in live-born infants. Arch Gynecol Obstet. 2007;276(5):491. doi:10.1007/s00404-007-0364-y
59. Gedlikbasi A, Oztarhan K, Aslan G, Demirali O, Akyol A. Multidisciplinary approach in cystic hygroma: prenatal diagnosis, outcome and postnatal follow up. Pediatr Int. 2009;51(5):670–677. doi:10.1111/j.1442-200X.2009.02846.x
60. Beke A, Joó JG, Lázár L, Bán Z, Tóth-Pál E, Papp Z. Incidence of chromosomal abnormalities in the presence of fetal subcutaneous oedema, such as nuchal oedema, cystic hygroma and non-immune hydrops. Fetal Diagn Ther. 2009;25(1):83–92. doi:10.1159/000201946
61. Levy AT, Berghella V, Al-Kouatly HB. Outcome of 45, X fetuses with cystic hygroma: a systematic review. Am J Med Genet A. 2016;185(1):26–32. doi:10.1002/ajmg.a.61902
62. Chen CP, Chern SR, Chang CL, et al. Prenatal diagnosis and genetic analysis of X chromosome polysomy 49, XXXXXY. Prenat Diagn. 2000;20(9):754–757. doi:10.1002/1097-0223(200009)20:9<754::aid-pd896>3.0.co:2-n
63. Rottem S, Bronshtein M, Thaler I, Brandes J. First trimester transvaginal sonographic diagnosis of fetal anomalies. Lancet. 1989;1(8635):55–445. doi:10.1016/0002-9378(89)90237-8
64. Podobnik M, Singer Z, Podobnik-Sarkanji S, Bulic M. First-trimester diagnosis of cystic hygroma by transvaginal sonography. Ultrasound Obstet Gynecol. 1992;2(2):124–125. doi:10.1046/j.1469-0705.1992.02020124.x
65. Souka AP, Von Kaisenberg CS, Hyett JA, Sonek JD, Nicolaides KH. Increased nuchal translucency with normal karyotype. American J Obstet Gynecol. 2005;192(4):1005–1021. doi:10.1016/j.ajog.2004.12.903
66. Bronshtein M, Zimmer EZ, Blazer S. A characteristics cluster of fetal sonographic markers that are predictive of fetal Turner syndrome in early pregnancy. Am J Obstet Gynecol. 2003;188(4):45.
67. Shulman LP, Emerson DS, Felker RE, Phillips OP, Simpson JL, Elias S. High frequency of cytogenetic abnormalities in fetuses with cystic hygroma diagnosed in the first trimester. Obstet Gynecol. 1992;80(1):80–82.
68. Sanhal CY, Mendilcioglu I, Ozekinci M, et al. Prenatal management, pregnancy and pediatric outcomes in fetuses with septated cystic hygroma. Braz J Med Biol Res. 2014;47(9):799–803. doi:10.1590/1414x20143895
69. Graesslin O, Derniaux E, Alanio E, et al. Characteristics and outcome of fetal cystic hygroma in the first-trimester. Acta Obstet Gynecol Scand. 2007;86:1442–1446.
70. Rosati P, Guariglia L. Prognostic value of ultrasound findings of fetal cystic hygroma detected in early pregnancy by transvaginal sonography. Obstet Gynecol. 2000;16:245–250. doi:10.1159/000264452
71. Scholl J, Durfee SM, Russell MA, et al. First-trimester cystic hygroma: relationship of nuchal translucency thickness and outcomes. Obstet Gynecol. 2012;120:551–559. doi:10.1097/AOG.Ob013e318264f829
72. Molina F, Avigdou K, Kagan KO, Poggi S, Nicolaides KH. Cystic hygromas, nuchal edema, and nuchal translucency at 11-14 weeks of gestation. Obstet Gynecol. 2006;107(3):678–683. doi:10.1097/01/AOG.0000201979.23031.32
73. Paoloni-Giacobino A, Extermann D, Extermann P, Dahoun SY. Pregnancy outcome of 30 fetuses with cystic hygroma diagnosed during the first 15 weeks of gestation. Genet Couns. 2003;14(4):413–418.
74. Lajeunesse C, Stadler A, Trombert B, et al. First-trimester cystic hygroma: prenatal diagnosis and fetal outcome. J Gynecol Obstet Biol Reprod. 2014;43(6):455–462. doi:10.1016/j.jgyn.2013.04005
75. Santanes N, Guigue V, Kohler M, et al. Nuchal translucency and cystic hygroma colli in screening for fetal major congenital effects in a series of 12910 euploid pregnancies. Ultrasound Obstet Gynecol. 2010;35:273–279. doi:10.1002/uog.7534
76. Schreurs L, Lanoo L, De Catte L, Van Schoubroeck D, Devriendt K, Richter J. First trimester cystic hygroma colli: retrospective analysis in a tertiary center. Eur J Obstet Gynecol Reprod Biol. 2018;231:60–64. doi:10.1016/j.ejogrb.2018.10.019
77. Shulman LP, Raafat NA, Mace PC, et al. Significance of septations in isolated fetal cystic hygroma detected in the first trimester. Prenat Diagn. 1994;14(3):223–226. doi:10.1002/pd1970140315
78. Mack LM, Lee W, Mastrobattista JM, et al. Are first-trimester septations independent risk factors for chromosomal anomalies. J Ultrasound Med. 2017;36(1):155–161. doi:10.7863/ultra.16.01066
79. Sherer DM, Dalloul M, Miller MJ, Kheyman M, Zinn H, Abulafia O. First-trimester septated cystic hygroma and cavum velum interpositum cyst. J Clin Ultrasound. 2011;39(6):356–358. doi:10.1002/jcu.20834
80. Phupong V, Sittisomwong T. Spontaneous resolution of cystic hygroma in 47, XYY fetus. Arch Gynecol Obstet. 2007;276(1):77–80. doi:10.1007/s00404-006-0295-z
81. Malone FD, Ball RH, Nyberd DA, et al. FASTER Trial Research Consortium. Obstet Gynecol. 2005;106(2):288–294. doi:10.1097/01.AOG.0000173318.54978.1f
82. Malone CM, Mullers S, Kelliher N, et al. Euploid first-trimester cystic hygroma: a more benign entity than previously thought? Fetal Diagn Ther. 2021;1–5. doi:10.1159/000519056
83. Wu Y, Zhang L, Ly H, Li Y, Tian W, Zhao L. Applying high-throughput sequencing to identify and evaluate foetal chromosomal deletion and duplication. J Cell Mol Med. 2020;24:9936–9944.
84. Pettit KE, Hull AD, Korty L, Jones MC, Pretorius DH. The utilization of circulating cell-free fetal DNA testing and decrease in invasive diagnostic procedures: an institutional experience. J Perinatol. 2014;34(10):750–753. doi:10.1038/jp.2014.102
85. Lostchuck E, Poulton A, Halliday J, Hui L. Population-based trends in invasive prenatal diagnosis for ultrasound-based indications: two decades of change from 1994 to 2016. Ultrasound Obstet Gynecol. 2019;53(4):503–511. doi:10.1002/uog19107
86. Soster E, Boomer T, Hicks S, et al. Three years of clinical experience with a genome-wide cfDNA screening test for aneuploidies and copy-number variants. Genet Med. 2021;23(7):1378. doi:10.1038/s41436-021-01190-1
87. Zhang J, Saucier JB, Feng Y, et al. Non-invise prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free DNA. Nature Med. 2019;25(3):439–447. doi:10.1038/s41591-018-0334-x
88. Mohan P, Lemoine J, Trotter C, et al. Clinical experience with non-invasive prenatal screening for single-gene disorders. Ultrasound Obstet Gynecol. 2022;59(1):33–39. doi:10.1002/uog.2356
89. Chen CP, Chern SR, Wang TH, et al. Prenatal diagnosis and molecular cytogenetic analysis of partial monosomy 10q(10q25.3 – >qter) and partial trisomy 18q23 qter) in a fetus associated with cystic hygroma and ambiguous genitalia. Prenat Diagn. 2005;25(6):492–496. doi:10.1002/pd.1179
90. Thorpe J, Osei-Owusu IA, Avigdor BE, Tupler R, Pevsner J. Mosaicism in Human Health and Disease. Annu Rev Genet. 2020;54:487–510. doi:10.1146/annurev-genet-041720-093403
91. Badeau M, Lindsay C, Blais J, et al. Genomics-based non-invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women. Cochrane Database Syst Rev. 2017;11(11):CD011767. doi:10.1002/14651858.CD011767.pub2
92. Vicic A, Hafner T, Wagner J, Stipoljev F. Prenatal diagnosis of 18p deletion and isochromosome 18q mosaicism in fetus with a cystic hygroma. Coll Antropol. 2014;38(3):1059–1062.
93. Stipoljev F, Barbalic M, Logara M, et al. Fetal cystic hygroma associated with terminal 2p25.1 duplication and terminal 3p25.3 deletion: cytogenetic, fluorescent in situ hybridization and microarray familial characterization of two different chromosomal structural rearrangements. Balkan J Med Genet. 2021;(2):79–86. doi:10.2478/bmjg-2020-0023
94. Ples L, Sima RM, Nedlelea F, Moga M. First prenatal diagnosis of a Niemann-Pick Disease Type C2 revealed by a cystic hygroma: a case report and review of the literature. Front Endocrinol (Lausanne). 2018;292. doi:10.3389/fendo.2018.00292
95. Tica OS, Gug C, Tica AA, et al. A unique case of recurrent fetal cystic hygroma: first fetus with an inherited heteromorphism of chromosome 1 (1qh+) and the second fetus with 69XXX triploidy. Rom J Morphol Embryol. 2020;61(3):935–940.
96. Teague KE, Eggleston MK, Muffley PE, Gherman RB. Recurrent fetal cystic hygroma with normal chromosomes: case report and review of the literature. J Matern Fetal Med. 2000;9(6):366–369. doi:10.1002/1520-6661(200011/12)9:6<366::AID-MFM1010>3.0.CO;2-E
97. Baxi L, Brwon S, Desai K, Thaker H. Recurrent cystic hygroma with hydrops. Fetal Diagn Ther. 2009;25(1):127–129. doi:10.1159/000207553
98. Lee KA, Williams B, Roza K, et al. PTPN11 analysis for the prenatal diagnosis of Noonan syndrome in fetuses with abnormal ultrasound. Clin Genet. 2009;75(2):190–194.
99. Gezdirici A, Ekiz EY, Kaya B, Sezer S, Aydin AA. How necessary is to analyze PTPN11 gene on fetuses with first trimester cystic hygroma and normal karyotype? J Matern Fetal Neonatal Med. 2017;30(8):938–941. doi:10.1080/14767058.2016.1191463
100. Sherer DM, Hsieh V, Kheyman M, Makanjuola O, Dalloul M. Cell-free DNS screening following first-trimester septated cystic hygroma leading to diagnosis of previously unknown familial Noonan syndrome. Eur J Obstet Gynecol Reprod Biol. 2021;264:389. doi:10.1016/j.erogrb.2021.07.056
101. Pan M, Xu Y, Li DZ. First-trimester cystic hygroma and neurodevelopmental disorders: the association to remember. Taiwan J Obstet Gynecol. 2020;59(6):960–962.
102. Mardy AH, Rangwala N, Hernandez-Cruz Y, et al. Utility of chromosomal microarray for diagnosis in cases of nonimmune hydrops fetalis. Prenat Diagn. 2020;40(4):492–496. doi:10.1002/pd.5617
103. Sparks TN, Lianoglu BR, Adami RR, et al. University of California Fetal-Maternal Consortium and the University of California, San Francisco Center for Maternal-Fetal Precision Medicine. Exome sequencing for prenatal diagnosis in nonimmune hydrops fetalis. N Engl J Med. 2020;383(18):1746–1756. doi:10.1056/NEJMoa2023643
104. Mone F, Eberhardt RY, Hurles ME, et al. Fetal hydrops and the incremental yield of Next generation sequencing over standard prenatal Diagnostic testing (FIND) study: prospective cohort study and meta-analysis. Ultrasound Obstet Gynecol. 2021;58(4):509–518. doi:10.1002/uog.23652
105. Norton ME, Van Ziffle J, Lianoglu BR, Hodoglugil U, Devine WP, Sparks TN. Exome sequencing vs targeted gene panels for the evaluation of nonimmune hydrops fetalis. Am J Obstet Gynecol. 2022;226(1):
106. Mellis R, Eberhart RY, Mailtion SJ, et al. Fetal exome sequencing for isolated increased nuchal translucency: should we be doing it? BJOG. 2022;129(1):52–61. doi:10.1111/1471-0528.16869
107. Karim JN, Roberts NW, Salomon LJ, Papageorghiou AT. Systematic review of first-trimester ultrasound screening for detection of fetal structural anomalies and factors that affect screening performance. Ultrasound Obstet Gynecol. 2017;50(4):429–441. doi:10.1002/uog/17246
108. Achiron R, Adamo L, Kassif E. From screening chromosomal anomalies to early diagnosis of fetal malformations. Curr Opin Obstet Gynecol. 2020;32(2):128–133.
109. Doulaveris G, Igel CM, Trejo FE. Impact of introducing cell-free DNA screening into clinical care on first trimester ultrasound. Prenat Diagn. 2022. doi:10.1002/pd.6086
110. Zhang X, Fan J, Chen Y, et al. Cytogenetic analysis of the products of conception after spontaneous abortion in the first-trimester. Cytogenet Genome Res. 2021;161(3–4):120–131. doi:10.1159/000514088
111. Xu Chen M, Liu QY, Li LR, Li J, Ma RM. Detecting trisomy 21 in products of conception from first-trimester spontaneous miscarriages by next-generation sequencing (NGS). Medicine. 2020;99(5):e18731. doi:10.1097/MD0000000000018731
112. Fan L, Wu J, Wu Y, et al. Analysis of chromosomal copy number in first-trimester pregnancy loss using next-generation sequencing. Front Genet. 2020;11:545856. doi:10.3389/fgene.2020.545856
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.