Back to Journals » Infection and Drug Resistance » Volume 8
Corynebacterium urealyticum: a comprehensive review of an understated organism
Authors Salem N, Salem L, Saber S, Ismail GA, Bluth MH
Received 23 September 2014
Accepted for publication 6 January 2015
Published 21 May 2015 Volume 2015:8 Pages 129—145
DOI https://doi.org/10.2147/IDR.S74795
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Nagla Salem,1 Lamyaa Salem,2 Sally Saber,2 Ghada Ismail,2 Martin H Bluth1
1Department of Pathology, Wayne State University School of Medicine, Detroit, MI, USA; 2Department of Clinical and Chemical Pathology, Ain Shams University, Cairo, Egypt
Abstract: Corynebacterium urealyticum is a Gram positive, slow-growing, lipophilic, multi-drug resistant, urease positive micro-organism with diphtheroid morphology. It has been reported as an opportunistic nosocomial pathogen and as the cause of a variety of diseases including but not limited to cystitis, pyelonephritis, and bacteremia among others. This review serves to describe C. urealyticum with respect to its history, identification, laboratory investigation, relationship to disease and treatment in order to allow increased familiarity with this organism in clinical disease.
Keywords: Corynebacterium urealyticum, cystitis, pyelonephritis, bacteremia, micro-organisms
Introduction
Corynebacterium urealyticum (formerly known as coryneform CDC 79 group D2) was first recognized to be involved in human infections 80 years ago. It is a Gram positive, slow-growing, asaccharolytic, lipophilic, multi-drug resistant, urease positive micro-organism with diphtheroid morphology. It was named C. urealyticum stressing its strong ability to split urea.1
C. urealyticum is isolated from the groin of elderly patients receiving broad spectrum antibiotics with relative frequency, thus providing an ideal environment that may favor colonization of urinary catheters with subsequent infection of bladder mucosa particularly in the setting of co-morbidities (trauma, tumor, or inflammation).1
C. urealyticum is an opportunistic nosocomial pathogen mainly causing acute cystitis, pyelonephritis, alkaline encrusted cystitis, and encrusted pyelitis and may also cause bacteremia preferential in patients with chronic urological diseases.2
C. urealyticum isolates are usually missed in routine urine cultures since it does not grow well after an overnight incubation.3 However C. urealyticum can be isolated after 48 hours of incubation at 35°C–37°C preferably in 10% CO2 mainly on blood agar. Colonies are characteristically pinpoint, whitish, opaque, smooth, convex, circular, and non-hemolytic.4
The identification of C. urealyticum is one of the biggest challenges facing the laboratory.5 Phenotypic studies using home-made media or commercial systems (API Coryne) can be used for proper identification.6 Identification can also be confirmed by polymerase chain reaction (PCR) amplification.7
The majority of C. urealyticum strains are highly resistant to a large number of antibiotics, including aminoglycosides, B-lactams, and macrolides although teicoplanin and vancomycin remain universally active against these isolates.3
C. urealyticum can be considered an underestimated cause of sepsis among hospitalized patients. However, adequate identification and therapy may be lifesaving.8
The aim of this review is to highlight the historical context of C. urealyticum, its relationship to disease, differentiation from other infectious agents, and detection patterns of antibiotic sensitivity for proper management of patients.
C. urealyticum
Historical overview
C. urealyticum is one of the non diphtherial corynebacteria (coryneforms). Coryneforms were considered to be simple contaminants with little potential pathogenicity. After decades of uncertainty about their clinical significance, coryneforms have emerged as important pathogens causing many serious infections.6,9
In 1972, King described a non-spore-forming, aerobic and Gram-positive bacillus with strong urease activity as coryneform CDC group D2. A case of pneumonia in a debilitated patient from whom such an organism was isolated, was described in 1979 by Jacobs and Perlino. In 1985, Ponte et al published a report where this organism was implicated for the first time in urinary tract infections (UTIs) in four patients with alkaline encrusted cystitis. In the same year, eight cases of UTI caused by this organism were reported with results subsequently confirmed in a larger case series.2
The name C. urealyticum was first proposed in 1986 to describe the formation of bladder stones in vivo in an animal model by urealytic bacteria of the CDC coryneform group D2 (old name of C. urealyticum).3
Taxonomic features
C. urealyticum belongs to the family Corynebacteriaceae and the genus Corynebacterium (Table 1).10 According to Funke et al,11 medically relevant species in the genus Corynebacterium are classified into the following:
  | Table 1 The taxonomic classification of C. urealyticum |
- nonlipophilic, fermentative corynebacteria
- C. diphtheriae group
- C. xerosis and C. striatum
- C. minutissimum
- C. amycolatum/striatum
- C. glucuronolyticum
- C. argentoratense
- C. matruchotii
- nonlipophilic, non-fermentative corynebacteria
- C. afermentans
- C. auris
- C. pseudodiphtheriticum
- C. propinquum
- lipophilic corynebacteria
- C. jeikeium
- C. urealyticum
- C. lipophilum
- C. accolens
- C. macginleyi
- CDC group F-1 and G
- C. bovis.
The taxonomic classification of C. urealyticum was originally characterized based on its biochemical properties, including failure to produce acid from carbohydrates and the ability to hydrolyze urea.12,13 The cell wall peptidoglycan of C. urealyticum contains meso-diaminopimelic acid. Arabinose and galactose are the major cell wall sugars. Tuberculostearic acid and short-chain mycolic acids are present resembling mycobacterium tuberculosis.2 The degree of relatedness of C. urealyticum to other Corynebacterium species can be determined by using DNA–DNA hybridization studies2,14 and 16S ribosomal ribonucleic acid (rRNA) sequence analysis. A 16S rDNA sequence-based phylogenetic tree of the species belonging to the genus is shown in Figure 1.15,16 Studies were done using a small-subunit rRNA sequence data and observed a close phylogenetic relationship between C. urealyticum and C. jeikeium.2 However, the difference between them is that C. jeikeium is non-urealytic and produces acid from glucose among others.17,18
  | Figure 1 Dendrogram representing phylogenetic relationships of Corynebacterium type species and not-well-classified isolates by the neighbor-joining method. |
Genomic structure
In a study done by Tauch et al,3 the genome sequence and lifestyle of C. urealyticum established by pyrosequencing was determined. According to the study, the genome of C. urealyticum DSM7109 consists of a circular chromosome with a size of 2, 369, 219 bp and a mean G + C content of 64.2%.2 Table 2 shows the general features of C. urealyticum DSM7109 genome.
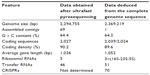  | Table 2 The general features of C. urealyticum DSM7109 genome |
The size of the C. urealyticum DSM7109 chromosome is smaller than that of the genomes from other pathogenic corynebacteria and is about100 kb, suggesting that further gene reduction occurred during the evolution of the C. urealyticum chromosome.19 The C. urealyticum DSM7109 chromosome has three rrn operons on the leading strands, one on the right and two on the left replichore. This is different from C. jeikeium, which contains three rrn operons on the leading strand of the right replichore, indicating the occurrence of structural rearrangements in the chromosome of C. urealyticum DSM7109.20
In addition to that in their study a total of 51 transfer RNA (tRNA) genes were detected in the C. urealyticum DSM7109 genome as well as 2024 protein-coding sequences were identified and classified into the Clusters of Orthologous Groups of proteins.21 Comparison of proteins of the C. urealyticum DSM7109 genome with the proteins encoded by C. jeikeium K411 revealed 1,589 proteins from C. urealyticum DSM7109 (78.5%), which were considered as orthologous with proteins from the C. jeikeium K411 genome. This subset of orthologous proteins indicated the close phylogenetic relationship between the lipid-requiring species C. urealyticum and C. jeikeium.19 Recently a study established the genome sequence of another strain of C. urealyticum; DSM 7111 which contains 79 specific genes.22
Epidemiological aspects of C. urealyticum
Although formerly considered as a contaminant, C. urealyticum is being increasingly reported as the causative organism of many infections, especially in those at risk.12
Incidence
C. urealyticum is the Corynebacterium most frequently isolated from clinical specimens, if the specimens are adequately processed. The incidence varies from 0.016% to 0.32% among all samples sent for routine culture. When specimens with diphtheroid morphology on culture are specifically chosen, the rates of detection are increased by a factor of 3-30.2
The incidence of UTI from C. urealyticum in the general population has been reported to be 1%–2%.23 Rizvi et al24 identified nine C. urealyticum isolates out of 139 Corynebacterium isolates with an incidence of 6.5% which were revealed from three patients with a UTI, two patients with peritonitis, two patients with a wound infection, one with a neonatal umbilical tip infection and one with a soft tissue infection.
Geographic distribution
In five major studies done in France, United Kingdom, South Africa, the United States, and Spain, the incidence of C. urealyticum UTI ranged from 0.5% to 2.5% for UTIs in non-selected populations there. In the United States, infections with C. urealyticum are being reported more frequently, especially those associated with medical devices such as urethral catheters, artificial valves, intravascular catheters, and central nervous system (CNS) drainage devices.23
More infections with C. urealyticum are being reported worldwide in studies done in Saudi Arabia,25 Spain,2 Poland,26 and Russia.27 Although C. Urealyticum infection has been reported in these countries, it may be more widespread yet under-reported.2,25–27 The actual incidence of diseases related to this organism remains unknown.
Age and sex
C. urealyticum causes UTIs including pyelonephritis and encrusted cystitis, which can occur at any age (mean age of 55–65 years). On the other hand, encrusted pyelitis usually occurs at an earlier age (with a mean age of 40–45 years).2
A sex-related affinity of C. urealyticum for skin colonization is supported by the fact that it has been isolated from the skin of 25%–37% of healthy elderly individuals, predominantly females.3
Infection with C. urealyticum is uncommon in children but must be considered. An unusual case of a 19-year-old kidney recipient with necrotizing pyelitis due to C. urealyticum in the absence of mucosal encrustation or calculi was reported. In such cases diagnosis must be rapid, and conservative treatment administered if possible.23
Risk factors
According to Pagnoux et al,12 the risk factors for C. urealyticum infection include the following:
- prolonged use of a urinary catheter.
- Hospitalization for long periods.
- Immunocompromised patients.
- Kidney transplantation.
- Recent urologic procedure.
- Chronic debilitating disease.
- Broad spectrum antibiotic treatment.
- Cytotoxic drug usage.
- History of previous UTIs.
It is noteworthy that patients at highest risk are those with prior urinary tract abnormalities or who have recently undergone urologic procedures as well as elderly patients who have had prolonged surgery.28
Risk factors for developing pyelonephritis include immunosuppression and kidney transplantation. Risk factors for the development of encrusted cystitis include urological diseases and previous UTIs in patients with chronic debilitating conditions as well as bladders damaged by trauma, tumor or intravesical administration of cytotoxic drugs.23
A cohort of 163 renal transplant recipients who were screened for UTIs due to C. urealyticum revealed that infection was closely related to obstructive uropathy. C. urealyticum has been reported in 10% of kidney recepients.29
Nosocomial versus community acquired infections
C. urealyticum has been isolated from the hospital environment which suggests it could be transmitted nosocomially or by air.2,30 Patients who were hospitalized for a prolonged period have shown infection with C. urealyticum. Outbreaks of nosocomial infections have been described. It can be deduced from several studies that nosocomial transmission of C. urealyticum infection is more common than community acquired infection.8
Hospitalized patients, especially ICU patients, tend to have more risk factors than non-hospitalized patients do, and have the highest infection rates. These highly resistant organisms deserve special attention in health care facilities. The availability of vulnerable patients, increased potential for transmission from larger numbers of colonized or infected patients “colonization pressure”, selective pressure exerted by antimicrobial use determine transmission and persistence of the resistant strain.8
Pathogenesis of C. urealyticum
Colonization and adherence to host tissues
C. urealyticum is known to be a natural colonizer of the human skin and urinary tract, especially in patients receiving broad-spectrum antibiotics.12 It is characterized by adherence to uroepithelial cells, which was observed for both urinary isolates and strains from healthy skin.2,23 If a patient is asymptomatic but his/her urine culture was positive for C. urealyticum, this patient is considered to be experiencing urinary colonization with this bacteria.29 Adherence to host tissues may be mediated by adhesive pili. These pili are covalently anchored to the Corynebacterium cell wall by a transpeptidylation mechanism to facilitate contact between the organisms and host tissue. C. urealyticum expresses a proteinaceous pilus with some structural similarity to the pilus of C. diphtheria.31
Adherence may occur in the absence of a pilus shaft. C. urealyticum has a SpaE subunit of its SpaDEF pilus which is covalently anchored to the cell wall. This provides tight contact between the bacterial cell and the host tissue. Similarly, adherence of C. diphtheriae to pharyngeal epithelium is mediated by the minor pilin SpaB of the structurally related SpaABC pilus.31 Previous studies have shown that only few C. urealyticum isolates that strongly adhered to uroepithelium were piliated.3
Moreover, human skin provides a habitat for C. urealyticum growth via provision of appropriate fatty acid composition. The gene repertoire of C. urealyticum DSM7109 reflects its adaptation as a lipophilic species to those habitats. In this context some studies have found that C. urealyticum strains were more frequently isolated from female patients than from males.23 Adherence of C. urealyticum to the skin has also been explained through studying the metabolic analysis of the genome sequence of C. urealyticum, which provided evidence that the lipid-requiring phenotype of C. urealyticum is due to the absence of a microbial type I fatty acid synthase gene provided by human skin that provide an appropriate amount of exogenous fatty acids for growth.3
Furthermore, the presence of C. urealyticum at a specific skin site excludes the colonization of the same body site by C. jeikeium and vice versa. C. jeikeium is also a strictly lipophilic species that is recovered from the axillary, inguinal, and perineal areas of the human body and, unlike C. urealyticum, it is more prevalent on the skin of male patients than on females.12
Consequently, one can assume that C. urealyticum strains commonly colonize the human skin, and after urological instrumentation gain access from the skin to the urinary tract of inpatients.23
Adherence to medical devices
A critical factor in the initiation of nosocomial UTIs by C. urealyticum is its adherence to foreign body materials in the urinary tract and also efficient adherence to intravascular catheters, artificial valves, and CNS drainage devices.12
Adherence of C. urealyticum to catheter materials such as polyvinyl chloride, Teflon and Teflon-coated rubber, may depend on the hydrophobicity of the cell surface of C. urealyticum that promotes the development of biofilms on solid surfaces.32 Analysis of the genome sequence revealed that C. urealyticum DSM7109 contains genes for two putative surface proteins (surA and surB) that display internal repeats in their amino acid sequences and may play important roles in UTIs of catheterized patients. Therefore, the surface proteins SurA and SurB may play important roles in UTIs of catheterized patients.32
Urease activity
C. urealyticum is a micro-organism with strong urease activity, which plays a fundamental role in its pathogenicity.3 The ureABC genes encode the structural subunits of the urease, whereas the ureEFGD genes code for accessory proteins. These proteins are homologous between urease gene clusters and are required for the assembly of the nickel metallocenter within the active site of the urease enzyme.33 When the organism adheres to the urinary tract, it grows under the stimulation of the urea present in the urine. Hydrolysis of urea leads to hyperammonuria and the alkalinization of human urine, which can cause hypersaturation with struvite and calcium phosphate with consequent crystallization of struvite and can also be facilitatory to urological disease.34
Clinical significance of C. urealyticum
C. urealyticum is an opportunistic nosocomial pathogen causing mainly acute cystitis, pyelonephritis, alkaline encrusted cystitis, and encrusted pyelitis and may cause bacteremia mainly in patients with chronic urological diseases.2
UTIs
C. urealyticum was identified as a pathogen causing chronic or recurrent cystitis, bacteriuria, bladder stones, and pyelonephritis.12 UTIs can be acute or chronic.
Acute UTIs
Acute cystitis is the more common clinical picture, although in many reported cases no differentiation between cystitis and pyelonephritis was made.6 Diagnosis of an acute UTI is made if symptoms of a UTI are present with leukocyturia and more than two urine samples positive for C. urealyticum. A UTI is considered acute if symptoms are present for less than 4 weeks. Pyelonephritis is defined as the presence of fever (>38°C), flank pain, with detection of C. urealyticum in urine and/or blood.6 Pyelonephritis due to C. urealyticum has been diagnosed in previous studies, with isolation from urine or from urine and blood or from urine obtained through a perirenal catheter. Patients were usually immunosuppressed.28
Chronic UTIs
Encrusted cystitis
Encrusted cystitis is one of the most frequent chronic infections caused by C. urealyticum, a condition of chronic ulcerative bladder inflammation, with deposits of (magnesium ammonium phosphate) on the surface and on the walls of the ulcer.35 C. urealyticum converts urea into ammonia, making urine alkaline, which precipitates struvite and calcium phosphate crystals, forming stones and encrustations on the infected mucosa. This produces a fibrotic and retractile bladder with reduced capacity, which may produce stenosis of the ureteral meatus, leading to dilatation of the upper urinary tract as shown in Figure 2. Bladder wall histology after resection of calcified encrustations shows three distinct zones: a necrotic layer containing calcified encrustations, an inflammatory layer containing bacterial colonies, lymphocytes and polymorphonuclear cells, and normal tissue (Figure 2).36
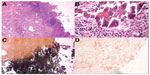  | Figure 2 Bladder wall histology of C. urealyticum. |
Encrusted cystitis is not a life-threatening disease, but is a very painful condition. It causes long-lasting symptoms in the lower urinary tract, with frequent relapses.37,38 It is also possible to demonstrate uretero-hydronephrosis by intravenous pyelography.39
A case of a 61-year-old man who presented with encrusted cystitis associated with bilateral hydronephrosis 4 months after adenomectomy complicated by vesicocutaneous fistula was described.38 Also a case of encrusted cystitis caused by C. urealyticum in a 69-year-old female who was admitted for recurrent cystitis following bladder catheterization after orthopedic surgery was reported, and despite prolonged non-specific and differentiated antibiotic therapy this patient had calcium-oxalate and struvite “stones” expelled daily in her urine with an alkaline (pH 8–9), and severe bladder incontinence.36
Another case of encrusted cystitis due to C. urealyticum was described in a 39-year-old female with ANCA associated vasculitis. She received several immunosuppressant medications since the onset of her vasculitis, with cycles of broad-spectrum antibiotics during the acute initial phase of her disease. Treatment of encrusted cystitis in this patient was successful with Teicoplanin.12
Encrusted pyelitis
Encrusted pyelitis is another serious chronic infection,35 which is characterized by the presence of struvite encrustations on the renal pelvis wall, identified when the pelvis is open during a urological intervention.40 It has been mainly described in renal transplant recipients and has been reported to facilitate graft dectruction.41,42 An unusual case of a 19-year-old kidney recipient with necrotizing pyelitis due to C. urealyticum in the absence of mucosal encrustations or calculi has been reported.23
Encrusted cystitis and encrusted pyelitis are uncommon in children but must be considered. Diagnosis must be rapid, and conservative treatment administered if possible. Nevertheless, graft loss can occur in kidney transplant recipients with encrusted pyelitis.23
Skin and soft tissue infections
C. urealyticum is a colonizer of the skin of hospitalized patients with or without UTIs. There is evidence that C. urealyticum may be transmitted by air so skin colonization in compromised patients could occur by that route. There are also well-documented cases of wound infections caused by C. urealyticum.2,6 A few cases of breast abscesses and other wound and soft tissue infections caused by C. urealyticum have also been reported.8
Bacteremia
C. urealyticum is probably an underestimated cause of sepsis among hospitalized patients mainly due to the difficult microbiologic identification of the organism.8 Prolonged hospitalization, prior antibiotic use, and the presence of intravenous lines are important factors placing patients at risk for C. urealyticum bacteremia. Bacteremia due to C. urealyticum remains with mortality rate up to 20%.8 Previous studies have demonstrated well-documented cases of bacteremia caused by C. urealyticum with mean age 52.9 years, some had urological disorders others had leukemia or AIDS.2
C. urealyticum is the cause of some cases of endocarditis that have been reported. These patients either had endocarditis on native valves or on a prosthetic valve. One of these patients was a drug addict. Another patient had a valvular prosthesis, developed endocarditis, and died.2,6
A case of sepsis by C. urealyticum in an 86-year-old patient acquired in the hospital was described. In this patient, admission for a Pseudomonas aeruginosa infection of the urinary tract was complicated by severe sepsis caused by C. urealyticum. Sepsis occurred through the intravenous line 17 days after P. aeruginosa had been successfully eradicated.8
Other infections caused by C. urealyticum have been documented, for example: osteomyelitis and pneumonia, which were documented in patients with urological disorders, neutropenic patients, patients with breast cancer, and acute leukemia.2,6
Some related species to C. urealyticum may be clinically relevant, and thus must be identified to the species level. For example, C. jeikeium can cause bacteremia; endocarditis; pneumonia; prosthetic joint infection or otitis media. C. pseudodiphtheriticum on the other hand can cause pneumonia (occasionally with pseudomembranous formation), keratitis or conjunctivitis.4
Laboratory diagnosis of C. urealyticum
C. urealyticum is isolated frequently in laboratories but usually not properly identified. Identification is important to differentiate contamination and/or colonization from infection, which influences decisions regarding clinical intervention.17 The correct identification is important because the antimicrobial susceptibilities of different coryneform bacterial isolates are quite variable.44
C. urealyticum should be thoroughly identified when isolated in pure cultures, when isolated from blood or sterile body fluids, when isolated repeatedly from specimens, or when isolated as the predominant organism from a mixed infection.9
The clinician should be notified immediately if C. urealyticum is present in blood culture, and the clinical significance of C. urealyticum must be carefully examined by cooperation between the microbiology laboratory and the clinician. Care must be taken in the interpretation of the results for those patients in whom half or more of the blood specimens taken for culture become positive, because not all blood samples taken from patients with C. urealyticum infection may eventually become positive.2
Sample collection and storage
C. urealyticum can be isolated from different samples including urine, blood, and pus from wounds. Samples are collected under complete aseptic conditions according to standard procedures and transported quickly (within 2 hours) keeping in mind that C. urealyticum is a fastidious organism. No specific transport media are required.4
Urine samples must be collected according to standard procedures. Likewise, culture swabs are carefully taken from the site of infection without touching adjoining areas to avoid commensal contamination from indigenous flora. The correct anatomic site that represents a location of active disease should be selected because even careful collection methods will produce a specimen of low clinical value if it is not obtained from a site where the infection is active.17
Processing, isolation, and identification
Microscopic identification
Gram staining of C. urealyticum shows slightly curved Gram positive rods with sides not parallel to each other and slightly wider ends giving it an atypical club shape. It is non motile, non spore forming and non acid fast.2
Isolation on culture media
According to Pagnoux et al,12 C. urealyticum can be isolated after culture by streak method on selective or non selective media including:
- non selective media:
- sheep blood agar;
- chocolate agar.
- Selective media:
- CBU agar medium: CBU agar is a lipid-enriched peptone-based agar.
Addition of antibiotics (β-lactam antibiotics or clindamycin plus nalidixic acid) to CBU agar medium can be done to inhibit or reduce other organisms present in the sample. - Loeffler’s serum medium. Constituents: three parts of sheep or horse serum plus one part of glucose broth.
Selective media for isolating C. urealyticum have great epidemiological value, but are not useful for management of patients, as most strains isolated only from selective media have no clinical relevance. That is why the use of selective media is not recommended for isolation of C. urealyticum for routine purposes, and sheep blood agar is the recommended medium for isolation.12
Incubation temperature and duration
Most C. urealyticum isolates are missed in routine cultures because the organism does not grow after “overnight” incubation. Therefore, incubation should last longer than 24 hours. C. urealyticum can be easily overlooked because it grows slowly requiring 48 to 72 hours incubation at 35°C–37°C for colonies to grow on agar plates and thus would be missed if the culture plates are examined after overnight incubation and discarded as negative before full 48 to 72 hours incubation. This is why clinical suspicion should be communicated to the microbiologist when samples are sent for culture.34
Identification from culture
Diphtheroid morphology of C. urealyticum could be interpreted falsely as representing contamination from skin or mucous membrane flora.29
- Appearance on sheep blood agar and chocolate agar: the organism grows on blood agar as pinpoint, whitish, opaque, smooth, convex, circular, entire, and non-hemolytic colonies.17
- Appearance on CBU agar: C. urealyticum forms typical colonies surrounded by a red halo on CBU agar. Because of its strong urease activity, when >10 colonies grow on this medium, the whole plate is usually pink and only the colonial morphology of C. urealyticum allows its recognition. Other urease-producing bacteria isolated on the medium grow faster or are glucose fermenting and no clear red halo is usually seen.12
- Appearance on Loeffler’s medium: the color of C. urealyticum isolates tends to be yellowish white on Loeffler’s medium.12
Biochemical identification
Conventional biochemical tests
C. urealyticum is the only known Corynebacterium that is lipophilic, asaccharolytic, and strongly urease positive. Other urease positive Corynebacterium species can be differentiated from C. urealyticum because they produce acid from glucose, except C. pseudodiphtheriticum which is weakly urease positive and asaccharolytic, yet unlike C. urealyticum it is nonlipophilic and nitrate positive,17 Table 3 shows the biochemical characteristics of different Corynebacterium species. A closely related species, C. jeikeium can be easily differentiated from C. urealyticum because it is non-urealytic and produces acid from glucose.17
  | Table 3 Biochemical characteristics of lipophilic Corynebacterium species |
API Coryne (bioMérieux, Marcy l’Etoile, France)
Biochemical identification can be easily performed by phenotypic studies, using home-made media or commercial systems.17 The API Coryne version 2.0 (bioMérieux, Marcy l’Etoile, France) is one of these systems and it is useful in identifying the majority of the non-diphtherial Corynebacterium species with clinical relevance.5
The principal disadvantage of these systems is that large inocula are required, much time as well as expensive strips can be wasted working with insignificant isolates. That is why a minitube was developed to enable a rapid biochemical test for urease production to be carried out expediently and with minimal inocula, providing a presumptive identification of C. urealyticum in much less time.45
The diagnostic performance of the API Coryne system, version 2.0, was evaluated and was able to identify 90.5% of Corynebacterium species.11 In several reports on new species of Corynebacterium, the API Coryne was used to phenotypically characterize the new isolates and biocodes were generated from the patterns of positive and negative reactions.5
A comparative study was done to compare the ability of the API Coryne system to identify Corynebacterium isolates with respect to conventional biochemical tests which were considered as reference methods.
The study showed that the API Coryne was able to identify 91% of Corynebacterium species correctly without need for further testing and it identified all strains of C. urealyticum correctly. It was considered “proper identification” when the system biocodes obtained indicated that it was excellent identification, “low discrimination” when additional tests were needed for proper identification suggested by the manufacturer and “misidentification” when the biocodes corresponded to a species different from that identified by conventional biochemical methods.5 In a study done by Adderson et al,44 comparing the API Coryne system and sequence analysis of 16S rRNA and rpo genes in identification of C. urealyticum isolates, the most sensitive tests for identification were found to be the API Coryne system and amplification and sequencing of the 16S rRNA gene using primers optimized for corynebacteria. Phenotypic combined with genotypic systems correctly identified up to 75.0% of control isolates to the species level.
Previously published studies found that the efficacy of biochemical identification systems is limited by the need to perform additional tests to definitively identify strains to the species level. The accuracies of phenotypic identification systems may be limited by the entries present in their respective databases. Additional difficulties when depending upon biochemical results from organisms that were particularly slow growing and relatively biochemically inert may have arisen. Such characteristics may make it hard to achieve sufficient inocula for testing and may result in weak biochemical reactions that are difficult to discriminate. The latter may affect the reproducibility and accuracy of test results.44
Roux et al46 found that 16S rRNA gene sequencing identified 90.3% of Corynebacterium species correctly whereas the API Coryne system led to the correct identification of only 75.8% of the strains.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing is a phenotypic method where isolates can be differentiated by generating antimicrobial resistance profiles. By understanding antimicrobial use patterns in certain environments, geographic locations, and health care settings, the occurrence of resistance to certain antimicrobials could provide a means to differentiate among isolates of different origins. Various testing methods, such as disc diffusion on Muller Hinton blood agar, broth microdilution, and E-test exist and can be performed to verify the susceptibility of C. urealyticum to antimicrobial agents.34 Table 4 shows possible antibiotic sensitivity of C. urealyticum isolates using microdilution.
  | Table 4 Antibiotic sensitivity of C. urealyticum isolates |
The Clinical and Laboratory Standards Institute guidelines do not provide breakpoints for disc diffusion. CLSI (Clinical and Laboratory Standards Institute)47 recommends penicillin, vancomycin, erythromycin, and gentamicin for primary testing. The European Committee on antimicrobial Susceptibility testing (EUCAST) provides breakpoints for corynebacteria antibiotic susceptibility testing. It recommends the usage of benzylpenicillin, ciprofloxacin, moxifloxacin, gentamicin, vancomycin, clindamycin, tetracycline, linezolid, and rifampicin.47
Molecular identification
Molecular identification systems for Corynebacterium species have been outlined. Progress in molecular taxonomy (DNA–DNA hybridization and 16S rRNA sequencing) and in chemotaxonomy has profoundly modified the classification of Corynebacterium species. The improvements in taxonomy and means of detection, together with an increased interest in Corynebacterium as an opportunistic infectious agent in humans, have resulted in the delineation of a plethora of new Corynebacterium species from human sources. Amplified rDNA analysis, and amplification of the 16S-23S gene spacer regions can differentiate between species that are difficult to be differentiated by biochemical reactions. With the application of molecular methods the number of species in the genus Corynebacterium has expanded.16
PCR
PCR is primarily used for rapid diagnosis of C. urealyticum. PCR is closely patterned after the natural DNA replication process. Each round of PCR synthesis involves three steps: denaturation, annealing, and extension.
This three-step PCR cycle is repeated several times. The number of target DNA copies approximately doubles every cycle (Roche Diagnostics, Germany). C. urealyticum was identified by PCR using mycobacterial primers in a case of suspected tuberculosis. Sequencing was further done for confirmation.7
Types of PCR
Conventional PCR: Molecular detection techniques, like PCR, can be used for identification of C. urealyticum, when other tests remain negative and suspicion is strong.12
Real time PCR (RT-PCR): RT-PCR is also used for detecting C. urealyticum where fluorescent dyes or fluorophore-containing DNA probes are used to measure the amount of the amplified product as the amplification progresses (Roche Diagnostics).
Many studies have been using RT-PCR recently for rapid identification of different Corynebacterium species including C. urealyticum.48
Gene sequencing techniques
16S rRNA gene sequencing: Another molecular technique useful for difficult-to-identify organisms, as Corynebacterium species, is rRNA gene sequencing, because conventional PCR is used for identifying Corynebacterium species. The purpose of conventional PCR is amplification of normal DNA templates using Taq DNA polymerase enzyme (Roche Diagnostics). Of the universal distribution of species-specific variable regions in the genes, the 16S rRNA gene sequence analysis plays an important role in the unambiguous species level identification of corynebacteria in the clinical laboratory. Even more importantly, sequence analysis may provide a degree of resolution that allows discrimination of different Corynebacterium species isolated from the same patient, which may help in determining the clinical significance of these multiple isolates.49
Another genotypic identification system, the MicroSeq 500 16S bacterial sequencing kit, is designed to sequence the first 527 bp of the 16S rRNA gene for bacterial identification. This system is a simplified version of the original MicroSeq system, which uses only two sequencing primers to analyze a single PCR product, thereby significantly reducing the cost and labor required for identification.49
Due to the difficulty in identifying Corynebacterium species with standard phenotypic methods, the MicroSeq system’s ability was compared with that of standard phenotypic identification methods. Corynebacterium identification using the MicroSeq was faster than conventional methods.
Phenotypic identification of Corynebacterium species routinely requires 3 to 4 days while identification based on the MicroSeq can be completed within 48 hours.49
rpoB gene sequencing: Among the universal genes that can be used for taxonomic analysis and gene sequence-based identification, the RNA polymerase beta subunit-encoding gene (rpoB) can also be used for identification of Corynebacterium species.18 rpoB is a single-copy gene, it belongs to the common set of genes and it is long enough to contain phylogenetically useful information. It might be less prone than the 16S rRNA gene to lateral gene transfer due to its housekeeping function and it has already been used for bacterial species delineation in selected cases as well as to estimate the DNA G + C content of whole bacterial genomes. Among some alternative protein-coding genes, the rpoB gene showed the highest correlation to average amino-acid identity, which reflects whole genome-level relatedness.14
Restriction fragment length polymorphism (RFLP)
RFLP is a genotypic method where bacterial isolates are differentiated based on the profile generated following restriction enzyme digestion of bacterial chromosomal DNA or plasmids. This method has the advantage of being rapid and easy to perform.16
RFLP analysis, allowed the identification of Corynebacterium species, three different restriction enzymes are used, it has been demonstrated to be of use for the identification of species within the genus Corynebacterium.18
The abilities of 16S rRNA and rpoB gene sequencing to identify Corynebacterium isolates from clinical specimens were directly compared by Khamis et al,16 who found rpoB sequencing to be the most sensitive assay, positively identifying 91% of isolates, compared to 81% by 16S rRNA gene sequencing.
Matrix-assisted laser desorption/ionization coupled to time of flight (MALDI-TOF) MS
Recently, detection of C. urealyticum has been accomplished by automated methodology including MALDI-TOF, and BD-Phoenix among others. Additional studies have shown that all isolates identified as C. urealyticum by API Coryne were also identified by MALDI-TOF MS.50,51
Antimicrobial resistance to C. urealyticum
The majority of C. urealyticum strains obtained from clinical samples display multiple resistance to antibiotics17 which suggests the organism may be acquired from the hospital environment and, therefore, the use of antibiotics in the hospital setting could favor the appearance of multi-resistant strains.52
A case report of C. urealyticum infection acquired in the hospital demonstrated the potential to achieve a complete recovery even among those patients whose disease is caused by C. urealyticum strains resistant to a wide range of antibiotics (β-lactams and aminoglycosides), if the appropriate treatment with teicoplanin, vancomycin, linezolid or quinupristin-dalfopristin, the only antibiotics to which this organism is universally susceptible, is given and started in time.8,18,30
Antibiotic resistance determinants of the C. urealyticum DSM7109 genome
The presence of an ErmX determinant encoding a 23S rRNA adenine N-6-methyltransferase in the genome of C. urealyticum has been demonstrated. Variants of the erm(X) gene were detected previously in other pathogenic corynebacteria, including C. jeikeium, C. diphtheriae, and C. striatum, as well as in cutaneous propionibacteria and Arcanobacterium.20 The erm(X) gene can be organized as an integral part of transposons that are active in corynebacterial species.53
Antimicrobial groups and mechanism of resistance of each
Macrolides
In a recent study, the frequency of macrolide resistance mechanisms in clinical isolates of Corynebacterium species was described. The study showed high resistance of the different strains to macrolides. Erythromycin, clindamycin as well as other macrolides showed poor activity against C. urealyticum, C. jeikeium, and C. amycolatum. Telithromycin is more active in vitro than erythromycin, but only against erythromycin-susceptible and erythromycin-intermediate isolates, while cethromycin is only poorly active against C. urealyticum.33
In a previous study, antibiotic susceptibility assays with C. urealyticum DSM7109 revealed high minimum inhibitory concentrations (MICs) for the macrolide erythromycin (>128 μg/mL) and the lincosamide lincomycin (>256 μg/mL).3
The erm(X) resistance gene conferred resistance in corynebacteria to telithromycin and to a spectrum of macrolides and lincosamide antibiotics, such as erythromycin, azithromycin, josamycin, midecamycin, roxithromycin, spiramycin, tylosin, clindamycin, and lincomycin. That study concluded that erm(X) confers the high resistance levels for macrolides, lincosamides, and the ketolide telithromycin.54
Quinolones
C. urealyticum isolates were previously found to be susceptible to ofloxacin, norfloxacin, and ciprofloxacin, but more recent studies revealed an increased level of resistance to fluoroquinolones. Another study indicated that only 20.3% of the tested C. urealyticum isolates were susceptible to ciprofloxacin. Newer fluoroquinolones are more effective in vitro than ciprofloxacin and norfloxacin, but may remain ineffective against high-level ciprofloxacin-resistant isolates.55
Aminoglycosides
C. urealyticum is mostly resistant to aminoglycosides. According to a previous study, annotation of the genome sequence of C. urealyticum DSM7109 revealed the presence of the aph(3′)-Ia gene encoding a member of the aminoglycoside 3′-phosphotransferase protein family. (Figure 3). The aph(3′)-Ia gene is part of a larger chromosomal region that is almost identical to a DNA segment of the multi-drug resistance plasmid pTP10 from C. striatum M82B.3 This DNA region also contains the strA-strB tandem pair of resistance genes that encode aminoglycoside-3″-phosphotransferase and aminoglycoside-6-phosphotransferase, which confer high levels of streptomycin resistance. The strA-strB genes are widespread among bacteria and can be associated with transposons, such as Tn5393-related elements.
  | Figure 3 Comparative analysis of antibiotic resistance gene regions detected in corynebacteria. |
The strA-strB coding regions of C. urealyticum DSM7109 are located downstream of the tnpR gene from a Tn5393-like transposon that is disrupted by the Tn5715 element containing the aph(3′)-Ia gene (Figure 3).56
Consequently, the presence of the aph(3’)-Ia gene may be related to the resistance of C. urealyticum strains to a wide range of aminoglycosides, whereas the strA-strB genes may specifically confer the observed resistance to the aminoglycoside streptomycin.55
The MIC of C. urealyticum DSM7109 for the aminoglycosides, kanamycin, and streptomycin are >256 μg/mL and >128 μg/mL, respectively.56
Chloramphenicol
The genome of C. urealyticum DSM7109 contains two identical genes (cmx1 and cmx2) that encode a chloramphenicol exporter of the major facilitator superfamily.3 Like the orthologous counterparts from C. glutamicum and C. striatum, the cmx1 gene of C. urealyticum DSM7109 is part of a structurally unusual transposable element, in conjunction with the transposase gene tnp7109-14. The cmx2 gene is located in the chloramphenicol and aminoglycoside resistance gene region of C. urealyticum DSM7109 (Figure 3).56
The MIC of C. urealyticum DSM7109 for chloramphenicol is >32 μg/mL. Chloramphenicol resistance was detected also in the majority of C. urealyticum isolates during earlier antimicrobial susceptibility assays, revealing MICs in the range from 16 to >128 μg/mL. The mobile cmx gene is thus a likely candidate to mediate chloramphenicol resistance in C. urealyticum strains.3
Tetracyclines
It was found that the genome sequence of C. urealyticum DSM7109 contains a gene region that comprises the tetA-tetB gene pair (Figure 3). The tetAB proteins represent a specific group of tetracycline resistance determinants that use ATP rather than the proton gradient as energy source.57 Antibiotic susceptibility assays reveal that C. urealyticum DSM7109 is resistant to tetracycline (MIC >32 μg/mL) and susceptible to doxycycline (MIC <1 μg/mL). The tetAB genes may contribute to the tetracycline resistance of clinical C. urealyticum isolates, but may not confer resistance against doxycycline that was recognized in some C. urealyticum strains. The tetAB genes are flanked at the 5′ junction by an IS1249, indicating that a Tn3598-related element integrated into the genome of C. urealyticum DSM7109 (Figure 3).58
Multi-drug resistance
As with many organisms, antibiotic use could favor the appearance of multiresistant strains obtained via the transfer of resistance genes,8 which confer multi-drug resistance in corynebacteria.
Treatment, prevention, and control
Treatment
C. urealyticum, being a known multi-drug resistant organism (MDRO), must be treated with the right antibiotics, as failure in therapy leads to persistent infections.59 Vancomycin and teicoplanin have been used with great success in many patients with UTIs due to C. urealyticum. In some acute UTIs, acetohydroxamic acid has been concurrently administered as well as with adjuvant therapy.2,60
A case of C. urealyticum in a 57-year-old man affected by systemic erythematosus lupus with a long history of dysuria and suprapubic pain was reported, he underwent percutaneous nephrostomy drainage with urethral stenting for lupoid obstructive uropathy. Eventually the infection was definitively cured after a course of intramuscular teicoplanin.37
Studies show that bacteremic patients were usually treated with an antibiotic active against C. urealyticum, mainly vancomycin. A case of a neutropenic patient with acute myeloblastic leukemia and catheter related bacteremia was reported. This patient was cured after a course of vancomycin in conjunction with catheter removal.2
Development of resistance during treatment has been observed with beta-lactam antibiotics, fluoroquinolones, macrolides, rifampin, tetracycline, and gentamicin.2 Teicoplanin, vancomycin, and linezolid may be useful alternatives in the treatment of C. urealyticum infections caused by multi-drug resistant strains.52
Prevention and control
According to the CDC61 C. urealyticum is considered an MDRO. Interventions for prevention and control may be grouped into the following categories: education, judicious use of antimicrobials, surveillance (routine and enhanced), standard and contact precautions, environmental measures, education, and decolonization. Other measures that have been associated with a positive impact on prevention efforts are appropriate placement of hand washing sinks and alcohol-containing hand-rub dispensers in the facility and enforcing adherence to recommended infection control practices (eg, hand hygiene, standard and contact precautions) for MDROs.
Education
Educational interventions were included in several successful studies. The focus of the interventions was to encourage a behavior change through improved understanding of the problem (MDRO) that the facility was trying to control through hand hygiene, antimicrobial prescribing patterns, or other outcomes. These factors were viewed as essential to the success of the intervention.61 Educational campaigns to enhance adherence to hand hygiene practices in conjunction with other control measures have been associated temporally with decreases in MDRO transmission in various health care settings.61
Judicious use of antimicrobial agents
Limiting antimicrobial use alone may fail to control resistance due to a combination of factors; including 1) the relative effect of antimicrobials on providing initial selective pressure, compared to perpetuating resistance once it has emerged; 2) inadequate limits on usage; or 3) insufficient time to observe the impact of this intervention. Efforts should be focused on effective antimicrobial treatment of infections, use of narrow spectrum agents, avoiding excessive duration of therapy, and restricting use of broad-spectrum or more potent antimicrobials for treatment of serious infections. Achieving these objectives would likely diminish the selective pressure that favors proliferation of multi-drug resistant strains.34
Surveillance
Surveillance is a critically important component of any control program, allowing detection of newly emerging pathogens, monitoring epidemiologic trends, and measuring the effectiveness of interventions. Multiple surveillance strategies have been employed, such as monitoring of clinical microbiology isolates resulting from tests ordered as part of routine clinical care. This method is particularly useful to detect emergence of new MDROs not previously detected. In addition, this information can be used to prepare facility- or unit-specific summary antimicrobial susceptibility reports that describe pathogen-specific prevalence of resistance among clinical isolates. Such reports may be useful to monitor for changes in known resistance patterns that might signal emergence or transmission of MDROs, and also to provide clinicians with information to guide antimicrobial prescribing practices.27
Standard precautions
Colonization with multi-drug resistant strains is frequently undetected; even surveillance cultures may fail to identify colonized persons due to lack of sensitivity, laboratory deficiencies, or intermittent colonization due to antimicrobial therapy. Therefore, standard precautions must be used in order to prevent transmission from potentially colonized patients. Hand hygiene is an important component of standard precautions.26
Contact precautions
Contact precautions are intended to prevent transmission of C. urealyticum which can be transmitted by direct or indirect contact with the patient or the patient’s environment. A single-patient room is preferred for patients who require contact precautions. For personnel caring for patients on contact precautions, donning gown and gloves upon room entry and discarding before exiting the patient room is done to contain pathogens, especially those that have been implicated in transmission through environmental contamination.26
Conclusion
C. urealyticum belongs to the genus Corynebacterium, it is a Gram positive, lipophilic, asaccharolytic, non-hemolytic, urease positive micro-organism with diphtheroid morphology.
It is an underestimated cause of infections especially: acute cystitis, pyelonephritis, alkaline encrusted cystitis, encrusted pyelitis, and wound infection and it may cause bacteremia.
It is usually acquired nosocomially especially in immunocompromised patients, patients with prolonged hospitalization or catheterization, patients with underlying urological disorders or previous UTIs.
It is usually missed in routine culture because of its diphtheroid morphology (so considered a contaminant) and it requires 48 hours of incubation to grow.
It can be diagnosed using phenotypic (Urease test, API Coryne) and molecular methods (RT-PCR using mycobacterial primers). Recently MALDI-TOF MS has also been used for identification.
It is an MDRO but still sensitive to teicoplanin and vancomycin.
Recommendations
In the case of symptoms of a UTI, high urine pH, bladder stones, recent urologic manipulation, negative routine urine culture or a UTI not responding to treatment; suspect a C. urealyticum infection.
Standard precautions must be used in order to prevent transmission from potentially colonized patients with C. urealyticum. Hand hygiene is an important component. For personnel caring for such patients, gloves upon room entry and discarding before exiting the patient room is important to contain this pathogen.
Avoid unnecessary catheterization of inpatients especially those with an underlying urological disorder.
In the laboratory, do not disregard colonies with a diphtheroid morphology specially if in a pure culture, in the elderly or in immunocompromised patients.
Incubate suspected colonies of C. urealyticum for 48 hours instead of 24 hours. If detected in a patient do not forget to assess the bacteremia status of this patient. Deal with this pathogen as an MDRO yet teicoplanin and vancomycin can be used for treatment.
Further studies on a larger scale are needed to:
- detect the prevalence of C. urealyticum among hospitalized and non-hospitalized patients;
- evaluate the ability of the API Coryne system to correctly identify C. urealyticum isolates compared with molecular methods;
- evaluate the sensitivity of RT-PCR using mycobacterial primers in identification of C. urealyticum compared with the use of other primers.
Disclosure
The authors report no conflicts of interest in this work.
References
Duztas MN, Erdem H, Pahsa A, Emekdas G, Avci A. Epidemiology of Corynebacterial Urinary Infections. Int J Infect Dis. 2006;10(3):264. | |
Soriano F, Tauch A. Microbiological and Clinical Features of Corynebacterium urealyticum: Urinary Tract Stones and Genomics as The Rosetta Stone. Clin Microbiol Infect. 2008;14(7):632–643. | |
Tauch A, Trost E, Tilker A, et al. The lifestyle of Corynebacterium urealyticum Derived from its Complete Genome Sequence Established by Pyrosequencing. J Biotechnol. 2008;136(1–2):11–21. | |
Bernard K. Corynebacterium Species and Coryneforms: An Update on Taxonomy and Diseases Attributed to These Taxa. Clinical Microbiology Newsletter. 2005;27(2):9–18. | |
Almuzara MN, De Mier C, Rodríguez CR, Famiglietti AM, Vay CA. [Evaluation of API Coryne system, version 2.0, for Identification of Gram-positive Diphtheroid of Clinical Relevance]. Rev Argent Microbiol. 2006;38(4):197–201. Spanish. | |
Soriano F. [Corynebacterium urealyticum: From Bedside to Complete Genome Sequence]. Enferm Infecc Microbiol Clin. 2009;27(1):5–6. Spanish. | |
Simoons-Smit AM, Savelkoul PH, Newling DW, Vandenbroucke-Grauls CM. Chronic Cystitis Caused by Corynebacterium urealyticum Detected by Polymerase Chain Reaction. Eur J Clin Microbiol Infect Dis. 19:949–952. | |
Famularo G, Minisola G, Nicotra GC, Parisi G, De Simone C. A Case Report and Literature Review of Corynebacterium urealyticum Infection Acquired in the Hospital. Intern Emerg Med. 2008;3(3):293–295. | |
Balci I, Eksi F, Bayram A. Coryneform Bacteria isolated from blood cultures and their Antibiotic susceptibility. J Int Med Res. 2002;30(4):422–427. | |
Pitcher D, Soto A, Soriano F, Valero-Guillen P. Classification of Coryneform Bacteria Associated With Human Urinary Tract Infection (group D2) As Corynebacterium urealyticum sp. nov. Int J Syst Bacteriol. 1992;42(1):178–181. | |
Funke G, von Graevenitz A, Clarridge JE 3rd, Bernard KA. Clinical Microbiology of Coryneform Bacteria. Clin Microbiol Rev. 1997;10(1):125–159. | |
Pagnoux C, Bérezné A, Damade R, et al. Encrusting Cystitis Due to Corynebacterium urealyticum in A Patient With ANCA-associated Vasculitis: Case Report and Review of The Literature. Semin Arthritis Rheum. 2011;41(2):297–300. | |
Chung CS, Liao CH, Cheng SL, Lin TH, Hsueh PR. Percutaneous Nephrostomy Tube-associated Bacteremia Caused By Corynebacterium urealyticum. J Microbiol Immunol Infect. 2008;41(6):525–527. | |
Adékambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB Gene Sequencing As a Suitable Supplement to DNA-DNA Hybridization For Bacterial Species and Genus Delineation. Int J Syst Evol Microbiol. 2008;58(Pt 8):1807–1814. | |
Van den Velde S, Lagrou K, Desmet K, Wauters G, Verhaegen J. Species Identification of Corynebacteria by Cellular Fatty Acid Analysis. Diagn Microbiol Infect Dis. 2006;54(2):99–104. | |
Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA Gene Sequencing for Molecular Identification of 168 Clinical Isolates of Corynebacterium. J Clin Microbiol. 2005;43(4):1934–1936. | |
Johnson MH, Strope SA. Encrusted Cystitis. Urology. 2012;79(3):e31–e32. | |
Funke G, Bernard KA. Coryneform Gram-positive Rods. In: Murray PR, Baron EJ, Jorgensen JG, et al, editors. Manual of Clinical Microbiology, 9th ed. Washington, DC: ASM Press; 2007:485–514. | |
Yukawa H, Omumasaba CA, Nonaka H, et al. Comparative Analysis of the Corynebacterium glutamicum group and Complete Genome Sequence of Strain R. Microbiology. 2007;153(Pt 4):1042–1058. | |
Tauch A, Kaiser O, Hain T, et al. Complete Genome Sequence and Analysis of the Multiresistant Nosocomial Pathogen Corynebacterium jeikeium K411, a Lipid Requiring Bacterium of the Human Skin Flora. J Bacteriol. 2005;187(13):4671–4682. | |
Barzantny H, Brune I, Tauch A. Molecular Basis of Human Body Odour Formation: Insights Deduced from Corynebacterial Genome Sequences. Int J Cosmet Sci. 2012;34(1):2–11. | |
Guimarães LC, Soares SC, Albersmeier A, et al. Complete Genome Sequence of Corynebacterium urealyticum Strain DSM 7111, Isolated from a 9-Year-Old Patient with Alkaline-Encrusted Cystitis. Genome Announc. 2013;1(3):e00264-13. | |
Fontana I, Bertocchi M, Rossi AM, et al. Corynebacterium urealyticum Infection in a Pediatric Kidney Transplant Recipient: Case Report. Transplant Proc. 2010;42(4):1367–1368. | |
Rizvi M, Khan F, Raza A, Shukla I, et al. Coryneforms the Opportunistic Pathogens-An Emerging Challenge for Immunocompetent Individuals. J Scien Res. 2011;6:165–171. | |
Babay AH, Kambal M. Isolation of Coryneform Bacteria from Blood Cultures of Patients at a University Hospital in Saudi Arabia. Saudi Med J. 2004;25(8):1073–1079. | |
Olender A, Łetowska I. [Wound Infections Due To opportunistic Corynebacterium Species]. Med Dosw Mikrobiol. 2010;62(2):135–140. Polish. | |
Cherkasov SV, Gladysheva IV. [Antibiotic Resistance of Coryneform Bacteria Isolated From The Reproductive Tract of Women]. Antibiot Khimioter. 2010;55(9–10):45–49. Russian. | |
Remacha Esteras MA, Guerra Laso JM, Esteban Martin A, Moran Blanco A. [Urinary Tract Infection Caused By Corynebacterium urealyticum]. Actas urologicas españolas. 2004;28(1):75–76. Spanish. | |
López-Medrano F, García-Bravo M, Morales JM, et al. Urinary Tract Infection due to Corynebacterium urealyticum in Kidney Transplant Recipients: An Underdiagnosed Etiology for Obstructive Uropathy and Graft Dysfunction-Results of a Prospective Cohort Study. Clin Infect Dis. 2008;46(6):825–830. | |
Nieto E, Vindel A, Valero-Guillén PL, Saez-Nieto JA, Soriano F. Biochemical, Antimicrobial Susceptibility and Genotyping Studies on Corynebacterium urealyticum Isolates From Diverse Sources. J Med Microbiol. 2000;49(8):759–763. | |
Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae Employs Specific Minor Pilins to Target Human Pharyngeal Epithelial Cells. Mol Microbiol. 2007;64(1):111–124. | |
Kwaszewska AK, Brewczyńska A, Szewczyk EM. Hydrophobicity and Biofilm Formation of Lipophilic Skin Corynebacteria. Pol J Microbiol. 2006;55(3):189–193. | |
Walter B, Hänssler E, Kalinowski J, Burkovski A. Nitrogen metabolism and Nitrogen Control in Corynebacteria: Variations of a Common Theme. J Mol Microbiol Biotechnol. 2007;12(1–2):131–138. | |
Ortiz-Pérez A, Martín-de-Hijas NZ, Esteban J, Fernández-Natal MI, García-Cía JI, Fernández-Roblas R. High Frequency of Macrolide Resistance Mechanisms in Clinical Isolates of Corynebacterium Species. Microb Drug Resist. 2010;16(4):273–277. | |
Lieten S, Schelfaut D, Wissing KM, Geers C, Tielemans C. Alkaline-encrusted Pyelitis and Cystitis: An Easily Missed and Life-threatening Urinary Infection. BMJ Case Rep. 2011;2011:bcr1220103613. | |
Del Prete D, Polverino B, Ceol M, et al. Encrusted cystitis by Corynebacterium urealyticum: A Case Report With Novel Insights Into Bladder Lesions. Nephrol Dial Transplant. 2008;23(8):2685–2687. | |
Perciaccante A, Pompeo ME, Fabi F, Venditti M. Corynebacterium urealyticum Encrusted Cystitis: A Case Report and Literature Review. Infez Med. 2007;15(1):56–58. | |
El Sayegh H, Elouardani M, Iken A, et al. [Encrusted Cystitis By Corynebacterium urealyticum]. Rev Med Interne. 2008;29(4):328–330. French. | |
Khallouk A, Wallerand H, Kleinclauss F, Bittard H, Bernardini S. [Conservative management of Corynebacterium urealyticum encrusted cystitis]. Prog Urol. 2006;16(4):496–498. French. | |
Meria P, Margaryan M, Haddad E, Dore B, Lottmann HB. Encrusted Cystitis and Pyelitis in Children: An Unusual Condition with Potentially Severe Consequences. Urology. 2004;64(3):569–573. | |
Vázquez V, Morales MD, Serrano C, Reus M, Llorente S, García J. [Corynebacterium urealyticum in renal trasplantation. CT and sonography imaging characteristics of encrusted cistitis and pielitis]. Nefrologia. 2004;24(3):288–293. Spanish. | |
Audard V, Garrouste-Orgeas M, Misset B, et al. Fatal septic shock caused by Corynebacterium D2. Intensive Care Med. 2003;29(8):1376–1379. | |
Adderson EE, Boudreaux JW, Cummings JR, et al. Identification of Clinical Coryneform Bacterial Isolates: Comparison of Biochemical Methods and Sequence Analysis of 16S rRNA and rpoB Genes. J Clin Microbiol. 2008;46(3):921–927. | |
Otsuka Y, Kawamura Y, Koyama T, Iihara H, Ohkusu K, Ezaki T. Corynebacterium resistens sp. Nov., a New Multidrug-resistant Coryneform Bacterium Isolated From Human Infections. J Clin Microbiol. 2005;43(8):3713–3717. | |
Roux V, Drancourt M, Stein A, Riegel P, Raoult D, La Scola B. Corynebacterium species isolated from bone and joint infections identified by 16S rRNA gene sequence analysis. J Clin Microbiol. 2004;42(5):2231–2233. | |
Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated Fastidious Bacteria; Approved Guideline-Second Edition. M45-A2. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. | |
European Committee on Antimicrobial Susceptibility Testing [homepage on the Internet]. EUCAST breakpoints tables. Available from: http://www.eucast.org/clinical-breakpoints/-EUCAST:clinicalbreakpoints. Accessed January 11, 2015. | |
Sing A, Berger A, Schneider-Brachert W, Holzmann T, Reischl U. Rapid Detection and Molecular Differentiation of Toxigenic Corynebacterium diphtheriae and Corynebacterium ulcerans Strains by LightCycler PCR. J Clin Microbiol. 2011;49(7):2485–2489. | |
Tang YW, Von Graevenitz A, Waddington MG, et al. Identification of Coryneform Bacterial Isolates by Ribosomal DNA Sequence Analysis. J Clin Microbiol. 2000;38(4):1676–1678. | |
Konrad R, Berger A, Huber I, et al. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria- associated bacteria. Euro Surveill. 2010;15(43):19699. | |
Vila J, Juiz P, Salas C, et al. Identification of clinically relevant Corynebacterium spp., Arcanobacterium haemolyticum and Rhodococcus equi by MALDI-TOF MS. J Clin Microbiol. 2012;50(5):1745–1747. | |
Gomez-Garces JL, Alos JI, Tamayo J. In vitro Activity of Linezolid and 12 other Antimicrobials Against Coryneform Bacteria. Int J Antimicrob Agents. 2007;29(6):688–692. | |
Rosato AE, Lee BS, Nash KA. Inducible Macrolide Resistance in Corynebacterium jeikeium. Antimicrob Agents Chemother. 2001;45(7):1982–1989. | |
Tauch A, Krieft S, Kalinowski J, Pühler A. The 51,409-bp R-plasmid pTP10 From the Multiresistant Clinical Isolate Corynebacterium striatum M82B is Composed of DNA segments Initially Identified in Soil Bacteria and in Plant, Animal, and Human Pathogens. Mol Gen Genet. 2000;263(1):1–11. | |
Sánchez Hernández J, Mora Peris B, Yagüe Guirao G, et al. In vitro Activity of Newer Antibiotics Against Corynebacterium jeikeium, amycolatum and urealyticum. Int J Antimicrob Agents. 2003;22(5):492–496. | |
McManus PS, Stockwell VO, Sundin GW, Jones AL. Antibiotic Use in Plant Agriculture. Annu Rev Phytopathol. 2002;40:443–465. | |
Ventura M, Canchaya C, Tauch A, et al. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol Mol Biol Rev. 2007;71(3):495–548. | |
Martínez-Martínez L, Joyanes P, Suárez AI, Perea EJ. Activities of Gemifloxacin and Five Other Antimicrobial Agents Against Listeria monocytogenes and Coryneform Bacteria Isolated From Clinical Samples. Antimicrob Agents Chemother. 2001;45(8):2390–2392. | |
Krishna S, Bairy I. Urinary Tract Infection by Corynebacterium Urealyticum – Look Twice. J Clin Diagn Res. 2011;5:1646–1647. | |
Thoumas D, Darmallaicq C, Pfister C, et al. Imaging Characteristics of Alkaline-Encrusted Cystitis and Pyelitis. AJR Am J Roentgenol. 2002; 178(2):389–392. | |
Siegel JD, Rhinehart E, Jackson M, Chiarello L; Healthcare Infection Control Practices Advisory Committee. Management of Multidrug Resistant Organisms in Healthcare Settings. Am J Infec Control. 2007;35(10 Suppl 2):165–193. | |
Lehmann KB and Neumann RO, Bacteriological Diagnosis, I, ed. Munich: J.F. Lishmann. 1896;390(2). |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
