Back to Journals » International Journal of Nanomedicine » Volume 19
Controlled siRNA Release of Nanopolyplex for Effective Targeted Anticancer Therapy in Animal Model
Authors Jia J, Yang J, Qian L, Zhou B, Tang X, Liu S, Wu L, Chen J , Kuang Y
Received 8 October 2023
Accepted for publication 16 January 2024
Published 7 February 2024 Volume 2024:19 Pages 1145—1161
DOI https://doi.org/10.2147/IJN.S443636
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor R.D.K. Misra
Jingchao Jia,1,2,* Jing Yang,3,* Leimin Qian,2 Biao Zhou,2 Xiaodong Tang,2 Shuanghai Liu,2 Li Wu,4 Jifeng Chen,5 Yuting Kuang1
1Department of General Surgery, The First Affiliated Hospital of Soochow University, Suzhou, People’s Republic of China; 2Department of General Surgery, Jiangyin Hospital Affiliated to Nantong University, Wuxi, People’s Republic of China; 3Jiangnan University Medical Center, Wuxi, People’s Republic of China; 4Department of Pharmaceutics, People’s Hospital of Shanggao, Yichun, People’s Republic of China; 5The Guangzhou Key Laboratory of Molecular and Functional Imaging for Clinical Translation, Jinan University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuting Kuang, Department of General Surgery, The First Affiliated Hospital of Soochow University, Suzhou, People’s Republic of China, Email [email protected] Jifeng Chen, The Guangzhou Key Laboratory of Molecular and Functional Imaging for Clinical Translation, Jinan University, Guangzhou, People’s Republic of China, Email [email protected]
Introduction: Spatiotemporally controlled release of siRNA for anti-tumor therapy poses significant challenges. Near-infrared (NIR) light, known for its exceptional tissue penetration and minimal tissue invasiveness, holds promise as a viable exogenous stimulus for inducing controlled siRNA release in vivo. However, the majority of light-responsive chemical bonds exhibit absorption wavelengths in the ultraviolet (UV) or short-wavelength visible light range.
Methods: To achieve NIR-controlled siRNA release, the study synthesized a UV-sensitive triblock copolymer cRGD-poly(ethylene glycol)-b-poly(aspartic acid ester-5-(2’-(dimethylamino)ethoxy)-2-nitrobenzyl alcohol)-b-polyphenylalanine, abbreviated as cRGD-PEG-PAsp(EDONB)-PPHE. This copolymer is composed of a cRGD-capped PEG block (cRGD-PEG), a poly(aspartate) block modified with cationic moieties through UV-cleavable 2-nitrobenzyl ester bonds [PAsp(EDONB)], and a hydrophobic polyphenylalanine block (PPHE). The cationic amphiphilic polymer cRGD-PEG-PAsp(EDONB)-PPHE can assemble with hydrophobic upconversion nanoparticles (UCNPs) to form a cationic micelle designated as T-UCNP, which subsequently complexes with siRNA to create the final nanopolyplex T-si/UCNP. siRNA-PLK1 was employed to prepare T-PLK1/UCNP nanopolyplex for anti-tumor therapy.
Results: T-PLK1/UCNP not only exhibited outstanding tumor cell targeting through cRGD modification but also achieved 980 nm NIR-controlled PLK1 gene silencing. This was achieved by utilizing the encapsulated UCNPs to convert NIR into UV light, facilitating the cleavage of 2-nitrobenzyl ester bonds. As a result, there was a significant suppression of tumor growth.
Conclusion: The UCNPs-encapsulated nanopolyplex T-si/UCNP, capable of co-delivering siRNA and UCNPs, enables precise NIR-controlled release of siRNA at the tumor site for cancer RNAi therapy. This nanopolyplex can enhance the controllability and safety of RNAi therapy for tumors, and it also holds the potential to serve as a platform for achieving controlled release and activation of other drugs, such as mRNA and DNA.
Keywords: NIR-controlled siRNA release, nanopolyplex, UCNPs, anti-tumor therapy
Introduction
Small interfering RNA (siRNA)-based therapeutics represent a highly promising approach for cancer therapy.1–3 However, the lack of safe and controllable siRNA delivery vehicles has significantly impeded its application and advancement.4,5 Currently, only a limited number of siRNA drugs have obtained clinical approval from the FDA, and they are exclusively indicated for the treatment of hepatic disorders.6,7 This is due to the challenge of achieving specific siRNA delivery exclusively to non-hepatic tissues.8 Even siRNA drugs with excellent targeting properties are predominantly metabolized in the liver, leading to adverse effects.9 Therefore, a controllable delivery carrier is crucial for triggering siRNA function at the pathological site, thereby expanding the effective application of siRNA therapy to a broader range of diseases, including cancer.
Until now, various controlled-release strategies have been employed in the development of siRNA delivery carriers, including light irradiation,10 pH response,11 enzymatic hydrolysis,12 and redox reactions.13 Among these strategies, light irradiation, as an external stimulus, has proven to be an effective approach for achieving precise spatiotemporal control over siRNA release.14,15 The delivery of siRNA is typically achieved using cationic carriers, allowing for the construction of light-controllable siRNA release vehicles by linking sensitive bonds to cationic moieties.16,17 There are primarily two approaches: one involves the direct cleavage of chemical bonds induced by light, thereby releasing siRNA. This can be achieved using light-sensitive moieties such as o-nitrobenzyl ester bond.18 The other approach involves the indirect cleavage of chemical bonds induced by light, leading to siRNA release. For example, red light-induced photosensitizers at 660 nm can generate ROS-induced cleavage of TK bonds.19,20 However, the light sources required for cleaving these chemical bonds are typically of short wavelengths, including UV or visible light. These light sources exhibit poor tissue penetration and are susceptible to absorption by tissues, resulting in undesired side effects and thereby impeding further in vivo.21 In contrast, NIR light, especially as the wavelength increases, exhibits superior tissue penetration capabilities compared to visible or ultraviolet light, and also reduces damage to biological tissues, thereby offering enhanced safety.22,23 To effectively bridge this disparity, UCNP is a promising choice as they can efficiently absorb longer-wavelength NIR light and subsequently convert it into shorter-wavelength emission light, such as ultraviolet or visible light.24,25
Herein, we demonstrated a light-responsive polymeric nanocarrier capable of on-demand co-delivery of siRNA and UCNPs in various ratios, enabling precise NIR-controlled release of siRNA at the tumor site. In order to prepare the light-responsive polymeric nanocarrier, we synthesized a UV-sensitive triblock copolymer cRGD-PEG-PAsp(EDONB)-PPHE consisting of a cRGD-capped PEG block (cRGD-PEG), a poly(aspartate) block grafted with cationic moieties via the UV-cleavable 2-nitrobenzyl ester bonds [PAsp(EDONB)], and a hydrophobic polyphenylalanine block (PPHE). Subsequently, the cationic amphiphilic cRGD-PEG-PAsp(EDONB)-PPHE was employed to assemble with hydrophobic UCNPs, creating a cationic micelle termed T-UNCP. Finally, siRNAs were complexed within the cationic interlayer of T-UNCP to generate the nanopolyplex designated as T-si/UCNP (Scheme 1). Owing to the presence of the cRGD peptide, the nanopolyplex can be more effectively and specifically internalized by tumor cells through the specific binding between the cRGD peptide and integrin αvβ3, which is highly expressed in tumor tissue.26 Subsequently, upon irradiation with 980 nm NIR light at the tumor site, the encapsulated UCNPs underwent energy conversion and emitted 365 nm UV light, inducing the cleavage of 2-nitrobenzyl ester bonds. This photolytic process results in the detachment of cationic moieties and the complete release of siRNA. In this study, we selected the HCT116 colorectal cancer model for therapeutic experiments. In previous research, colorectal cancer has commonly been treated with chemotherapy, often resulting in the development of drug resistance and subsequent therapeutic failure.27 However, RNA interference offers an effective means to silence genes associated with tumor proliferation, promising a more efficient approach to tumor treatment. Polo-like kinase 1 (PLK1) is associated with tumor proliferation and is overexpressed in various cancer types, including colorectal cancer, making it considered an effective therapeutic target for cancer.27,28 Therefore, we utilized the nanopolyplex to load siRNA-PLK1, preparing T-PLK1/UCNP for the treatment of HCT116 colorectal cancer. T-PLK1/UCNP demonstrated NIR-controlled siRNA release in HCT116 xenograft tumor-bearing mice, resulting in a notable suppression of tumor growth. This nanocarrier not only can enhance the controllability and safety of RNAi therapy for tumors, but also have the potential to serve as a platform for achieving controlled release and activation of other drugs, such as mRNA and DNA.
Materials and Methods
Materials
Allyl-PEG-NH2 (APEG-NH2), with a molecular weight of 2.4 kDa, was sourced from 3A Chemicals Company (Shanghai, China). Chemicals such as azobisisobutyronitrile (AIBN), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), 2-(dimethylamino)ethyl chloride hydrochloride, and 5-hydroxy-2-nitrobenzyl alcohol were obtained from J&K Scientific Company (Beijing, China). Phenylalanine-N-carboxyanhydride (Phe-NCA) and β-benzyl-L-aspartate-N-carboxyanhydride (BLA-NCA) were procured from Bide Pharmatech Company (Shanghai, China). The cyclic RGD peptides with the sequence cyclo(-Arg-Gly-Asp-D-Phe-Cys) were provided by Haode Peptide Company (Wuhan, China). Upconversion nanoparticles (UCNPs, approximately 20 nm) which can convert the 980 nm NIR into 365nm emission were purchased from Xi’an ruixi Biological Technology Co., Ltd (Xi’an, China). Dimethyl ether, methanol, DCM, DMSO, DMF, and other standard chemical reagents were acquired from Guangzhou Chemical Reagent Company (Guangzhou, China). HCT116 cells were sourced from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). siRNAs targeting PLK1 (siRNA-PLK1), the luciferase gene (siRNA-Luc), and fluorescence-labeled scramble siRNA were purchased from Guangzhou RiboBio Co. (Guangzhou, China). Phosphate-buffered saline (PBS), Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), and 0.25% trypsin were procured from Gibco BRL (Carlsbad, CA, USA). Additional detection reagents were obtained from Thermo Fisher (NJ, USA). The siRNA-PLK1 was synthesized according to following sequence: 5′-UAUUUAAGGAGGGUGAUCUdTdT-3′ (antisense), 5′-AGAUCACCCUCCUUAAAUAdTdT-3′ (sense) (GenePharma, China).
Synthesis of Allyl-Poly(Ethylene Glycol)-b-Poly(β-Benzyl-Aspartic Acid)-b-Polyphenylalanine
The triblock copolymer Allyl-poly(ethylene glycol)-b-poly(β-benzyl-aspartic acid)-b-polyphenylalanine (APEG-PBLA-PPHE) was synthesized by a ring-opening polymerization of BLA-NCA and PHE-NCA using APEG-NH2 as a macroinitiator. Briefly, 1.0 g of APEG2.4k-NH2 (0.4 mmol) was vacuum-dried at 75 °C for 1.5 h in a 100 mL Schlenk flask. After the temperature dropped to 35 °C, 20 mL anhydrous dichloromethane and 3 g of BLA-NCA (12 mmol) dissolved in 3 mL of anhydrous DMF were added into the reaction system under an argon atmosphere. The polymerization lasted for 2 d. Then, 2.3 g of PHE-NCA (12 mmol) dissolved in 3 mL of anhydrous DMF were added into the reaction system under an argon atmosphere. The polymerization lasted for another 2 d and the final mixture was precipitated in diethyl ether, filtered, washed with diethyl ether for three times, and dried in vacuum. The results of 1H-NMR (Figure 1B) showed that the polymer APEG-PBLA-PPHE was successfully synthesized.
Synthesis of Allyl-Poly(Ethylene Glycol)-b-Poly(Aspartic Acid)-b-Polyphenylalanine
2 g of APEG-PBLA-PPHE was dissolved in 10 mL of trifluoroacetic acid and cooled in an ice-water bath. 5 mL of 33 wt.% HBr-containing acetic acid solution was added and stirred for 2 h at room temperature. The product was precipitated into excess ether, filtered, washed with ether, and dried under vacuum. The product was redissolved in 5 mL of DMSO, dialyzed against water for 24 h, and lyophilized. The results of 1H-NMR (Figure 1B) showed that the polymer Allyl-poly(ethylene glycol)-b-poly(aspartic acid)-b-polyphenylalanine (APEG-PAsp-PPHE) was successfully synthesized.
Synthesis of cRGD-Poly(Ethylene Glycol)-b-Poly(Aspartic Acid)-b-Polyphenylalanine
cRGD-poly(ethylene glycol)-b-poly(aspartic acid)-b-polyphenylalanine (cRGD-PEG-PAsp-PPHE) was synthesized by addition reaction of allyl group with thiol group of cRGD-SH using a little AIBN as initiator. In brief, 1.5 g of APEG-PAsp-PPHE, 0.5 g of cRGD-SH and 15 mg of AIBN were mixed in 5 mL of DMF. After being deoxygenated by N2 bubble for 15 min, reaction mixture was stirred at 65 °C for 12 h under N2 atmosphere, dialyzed (MWCO: 3.5 kDa) against methanol for 1 d, rotary-evaporated, and vacuum-dried. The results of 1H-NMR (Figure 1B) showed that the polymer cRGD-PEG-PAsp-PPHE was successfully synthesized.
Synthesis of 5-(2’-(Dimethylamino)Ethoxy)-2-Nitrobenzyl Alcohol
A 500 mL round-bottom flask was charged with 5-hydroxy-2-nitrobenzyl alcohol (10.0 g, 59.13 mmol), 2-(dimethylamino)ethyl chloride hydrochloride (7.63 g, 60.95 mmol), sodium hydroxide (5.2 g, 130 mmol), toluene (300 mL), and ethanol (60 mL). The reaction mixture was heated to reflux and stirred for 48 h. After cooling to room temperature, the dark solution was poured into 500 mL of water. The organic layer was isolated and the aqueous phase was further extracted with toluene. The combined organic phase was washed successively with saturated aqueous solution of sodium bicarbonate and water, dried over anhydrous sodium sulfate, and then evaporated to dryness. The crude product was purified by silica gel column chromatography using methanol as the eluent to obtain 5-(2’-(dimethylamino)ethoxy)-2-nitrobenzyl alcohol (DMAENA) as a yellowish solid.
Synthesis of cRGD-PEG-PAsp(DMAENA)-PPHE
1 g of cRGD-PEG-PAsp-PPHE and 1 g DMAENA were dissolved in 10 mL of anhydrous DMSO. 1.2 g of 1-ethyl-3(3-dimethylpropylamine) carbodiimide hydrochloride (EDCI) and 0.5 g of DMAP were added and stirred for 24 h. The above solution was dialyzed against methanol (MWCO: 3.5 kDa) for 2 d, rotary-evaporated, and vacuum-dried. The results of 1H-NMR (Figure 1B) showed that the polymer cRGD-PEG-PAsp(DMAENA)-PPHE was successfully synthesized.
Preparation of Nanoparticles
The fabrication of UCNP-encapsulated micelles (referred to as T-UCNP) was achieved using a self-assembly approach. In a concise methodology, 25 mg of cRGD-PEG-PAsp(DMAENA)-PPHE and 3 mg of UCNPs were solubilized in 2 mL of tetrahydrofuran (THF). This resulting solution was subsequently added dropwise into 20 mL of PBS employing sonication, and subjected to ultrafiltration with a molecular weight cutoff of 100 kDa for concentration. The obtained solution was then passed through a syringe filter featuring a pore size of 220 nm. The resultant solution of UCNP-encapsulated micelles (2 μg/μL) was thus prepared. Subsequently, a siRNA solution was mixed with the T-UCNP solution at varying N/P ratios. This mixture was allowed to stand undisturbed for 0.5 hours, ultimately yielding the nanopolyplex designated as T-si/UCNP.
Characterization of Copolymers and Nanoparticles
Proton nuclear magnetic resonance (1H-NMR) spectroscopy was performed utilizing a nuclear magnetic resonance spectrometer (Bruker, 400 MHz) operating at room temperature. Fourier transform infrared (FT-IR) spectrums were acquired employing FT-IR (Frontier, PerkinElmer, USA) within the spectral range spanning from 4000 to 400 cm−1. The hydrodynamic sizes and zeta potentials of nanopolyplexes with varying N/P ratios were assessed using a 90 Plus/BI-MAS instrument (Brookhaven Instruments Corporation, USA). For transmission electron microscopy (TEM) analysis, 3 µL of nanoparticle solution (1 μg/μL) was deposited onto a copper grid coated with amorphous carbon and subsequently dried in drier. After staining, The grids were air-dried for a day and finally observed using TEM (JEM-ARM200P, JEOL, Japan) operating at 200 kV for visualization of nanoparticle morphology and diameter distribution. Fluorescence spectra were captured using a fluorescence spectrophotometer (Shimadzu, RF5301). The upconversion luminescence spectra of nanoparticles were recorded utilizing a fluorescence spectrophotometer (Edinburgh, FLS980) equipped with a 980 nm laser as the excitation source.
Agarose Gel Electrophoresis
The solutions of T-si/UCNP with different N/P ratios (ie, 1, 2, 4, and 6) were subjected to electrophoresis for a duration of 15 minutes on a 1% agarose gel containing GoldenView at an applied voltage of 120 V. Following the electrophoresis, the resultant gel image was visualized under UV light employing a DNR bio-imaging systems.
siRNA Release in vitro
Upon NIR irradiation, the UCNPs encapsulated into the core of T-si/UCNP generate UV emissions that facilitate the cleavage of 2-nitrobenzyl linkers, subsequently resulting in the release of siRNA. To validate the siRNA release from T-si/UCNP nanoparticles, an agarose gel electrophoresis was conducted. Specifically, T-si/UCNP nanoparticles, both with and without NIR irradiation, were subjected to electrophoresis on a 1% (w/v) agarose gel to analyze the migration of siRNA. Additionally, the siRNA release from T-si/UCNP nanoparticles was assessed using siRNA-FITC as a substitute for native siRNA. T-FITC/UCNP solutions were introduced into dialysis bags with a molecular weight cut-off of 100 kDa and subjected to dialysis against 8 mL buffer solutions (pH 7.4), both with and without NIR irradiation. In the event of NIR irradiation (980 nm, 2 W cm−2) for a duration of 10 minutes, the irradiation procedure was repeated twice with a 3-hour interval. The solutions were maintained at 37 °C under agitation. The concentrations of released siRNA-FITC at various time intervals were quantified using a spectrofluorometer (PerkinElmer Ltd., UK).
Cell Culture
HCT116 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) complete culture medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, and cultured at 37°C with a 5% CO2 atmosphere. Upon reaching approximately 80% confluence in the culture dish, the cells were detached using a 0.25% trypsin solution for a duration of 1 minute, subsequently being either subcultured or cryopreserved for further experiments.
Cellular Uptake and Lysosomal Escape
A total of 1×105 HCT116 cells were plated in each well of 12-well plates and incubated overnight. Subsequently, the cells were exposed to the targeting T-FITC/UCNP and the non-targeting NT-FITC/UCNP separately for a duration of 4 hours. Following a wash with PBS, trypsin digestion was utilized to detach the cells, and centrifugation was employed to collect the HCT116 cells. The suspended HCT116 cells were then analyzed using a Gallios flow cytometer (Beckman, Coulter) to assess cellular uptake. To investigate of the lysosomal escape of nanopolyplexes, HCT116 cells were seeded in 35 mm confocal dishes at a density of 1×104 cells per well and transfected with various nanopolyplexes (T-FITC/UCNP and NT-FITC/UCNP) for 4 hours. Following the treatment, the culture medium was substituted with the fresh. Nuclei and endosomes were respectively stained with DAPI and Lyso-Tracker Red (Invitrogen). The intracellular distribution of nanoparticles was examined using a confocal laser scanning microscope system (Carl Zeiss LSM 710, Germany).
NIR-Controlled siRNA Release
To investigate NIR-controlled siRNA release, two distinct siRNA samples were individually labeled with a fluorescence dye (Cy3) and a quencher molecule (BHQ2). The mixture of siRNA-Cy3 and siRNA-BHQ2 at a 1:1 molar ratio was then complexed with cationic micelle T-UCNP, yielding nanoparticle denoted as T-Cy3&BHQ2/UCNP. Additionally, for control purposes, siRNA-Cy3 and siRNA-SCR were mixed at a 1:1 molar ratio and complexed with cationic micelle T-UCNP, forming nanoparticle T-Cy3&Blank/UCNP. Subsequently, HCT116 cells were separately incubated with these nanoparticle, T-Cy3&Blank/UCNP and T-Cy3&BHQ2/UCNP, for 4 hours. After being washed with fresh medium, HCT116 cells were exposed to NIR irradiation (980 nm, 2 W cm−2) for 10 minutes if required. Finally, the NIR-controlled release of siRNA was analyzed using confocal laser scanning microscopy (CLSM) and flow cytometry analysis.
qRT-PCR Test and Western Blotting Assay
HCT116 cells were plated in six-well dishes with a cell density of 5×104 cells per well and incubated for 12h. Following that, the cells were subjected to various treatments [PBS, T-SCR/UCNP (+L), T-PLK1/UCNP (-L), NT-PLK1/UCNP (+L), and T-SCR/UCNP (+L)] containing 1 µg/mL of siRNA-PLK1. For quantitative real-time PCR (RT-qPCR) analysis, total RNA was isolated from the treated cells using TRIzolTM reagent (Invitrogen, USA). The extracted RNA was then reverse transcribed into cDNA using a PrimeScriptTM RT reagent Kit (Takara, Japan) and subsequently subjected to RT-qPCR amplification. Relative quantification of PLK1 mRNA levels was determined employing the ΔΔCt method, with β-actin serving as an endogenous control. The primer sequences for RT-qPCR were as follows: 5′-TCAACACGCCTCATCCTCTA-3′ (forward) and 5′-GTGCTCGCTCATGTAATTGC-3′ (reverse) for the PLK1 gene, and 5′-GGACTTCGAGCAAGAGATGG-3′ (forward) and 5′-GAAGGAAGGCTGGAAGAGTG-3′ (reverse) for the β-actin gene.
For Western blotting assay, total cellular protein of HCT116 cells subjected to various treatments was extracted and quantified using CytoBusterTM protein extraction reagent and bicinchoninic acid protein assay kit respectively. Following that, these total proteins underwent separation through a 12% SDS-polyacrylamide gel electrophoresis and were subsequently transferred via electrotransfer onto a polyvinylidene fluoride (PVDF) membrane. The resultant membranes were immersed in a solution containing 5% nonfat milk. Afterward, following rinsing with tris-buffered saline Tween 20 (TBST) solution, the membranes were subjected to an overnight incubation at 4°C with the primary antibody. Following this incubation period, the membranes were again washed with TBST solution and subsequently exposed to an HRP-conjugated secondary antibody for signal amplification. Lastly, the protein bands were visualized using Alpha Imager 3400 Transilluminator Gel Imaging Detection System (Alpha Innotech, USA).
The Anti-Tumor Effect in vitro
The anti-tumor effect in vitro of the nanopolyplexes was evaluated using the MTT and apoptosis analysis. HCT116 cells were seeded in 96-well plates at a density of 8×104 cells per well and were allowed to culture overnight. Subsequently, the cells were subjected to treatment with various nanopolyplexes at different siRNA-PLK1 concentrations of 0.1, 0.25, 0.5, 0.75, and 1 µg/mL. After 4 h incubation, the culture medium with nanopolyplexes was exchanged for fresh medium. When necessary, cells were exposed to NIR light (980 nm, 2 W/cm2) for 20 minutes. Following an additional 12 h incubation, the cells received a medium containing MTT for 3 h. After the removal of the medium, DMSO was introduced to dissolve the resultant substrate, and this mixture was allowed to incubate for 15 minutes. Finally, the cell viability was quantified using an Infinite F200 Multimode plate reader (Tecan, Switzerland).
For the apoptosis assay, HCT116 cells were cultured in 6-well plates at a density of 3×105 cells per well and treated with PBS, T-SCR/UCNP (+L), T-PLK1/UCNP (-L), NT-PLK1/UCNP (+L), and T-SCR/UCNP (+L) at 1 µg/mL of siRNA-PLK1. Subsequently, the cells were transfected for 12 h. Following this incubation period, the cells were harvested and resuspended in a PBS solution. Annexin V-FITC and PI were used to stain the cells for 30 minutes in the dark. After three PBS washes, the cell apoptosis analysis was performed utilizing a Gallios flow cytometer (Beckman, Coulter).
Animal Model
Male nude mice, aged 5–6 weeks, were procured from the Laboratory Animal Center of Soochow University All procedures involving mice were conducted in compliance with the guidelines of the National Institutes of Health of China for the care and use of laboratory animals. Furthermore, all animal-based investigations received approval from the Laboratory Animal Center of Soochow University. To establish the HCT116 tumor xenograft model, a total of 1×106 HCT116 cells were injected into the foreleg of the mice.
Biodistribution and Tumor Accumulation of Nanopolyplexes
To assess tumor accumulation and biodistribution, we utilized siRNA-Cy7 to fabricate two types of nanoparticles: the targeted nanopolyplexe T-Cy7/UCNP and the non-targeted nanopolyplexe NT-Cy7/UCNP. Following intravenous injection of 80 μL of these nanopolyplexes, each containing siRNA-Cy7 at a dosage of 2 mg per kg body weight, into HCT116 xenograft-bearing mice, the fluorescence of Cy7 was monitored at distinct time intervals (1, 3, 6, 12, and 24 hours) employing an in vivo imaging system (Berthold LB983, Germany). Subsequently, mice were euthanized at 24 hours after nanopolyplexe injection, and both tumor tissues and major organs were collected for ex vivo imaging.
NIR-Controlled Luciferase Gene Silencing Assay in vivo
An implanted tumor model was established via subcutaneous injection of 1×105 recombinant HCT116 cells expressing luciferase (HCT116-Luc) into both thighs of mice. Subsequently, mice harboring HCT116-Luc tumors were randomly allocated into two groups (n = 3), and they received intravenous injections of either 80 µL of PBS solution or 80 µL of T-Luc/UCNP solution, with a siRNA-Luc dosage of 2 mg per kg body weight. After six hours post-injection, the tumor situated on the right thigh was subjected to NIR irradiation (980 nm, 2 W/cm2) for a duration of 30 minutes. Ten minutes prior to image capture, mice were administered with 80 µL of D-luciferin (20 µg/µL). The bioluminescent signals were monitored at various time intervals (0, 12, 24, and 36 hours) following the administration of nanoparticles or PBS, utilizing an in vivo imaging system (Berthold LB983, Germany).
Anti-Tumor Outcome in vivo
Upon reaching a tumor volume of approximately 100 mm3, mice bearing HCT116 xenografts were randomly allocated into 5 groups (n = 10) and treated with various treatments. Every three days, a 100 μL injection of either PBS or a nanoparticle solution was administered to each mouse through the tail vein. Notably, the siRNA-PLK1 was administered at a dosage of 2 mg per kg body weight, totaling five injections over the experimental period. After six hours post-injection, if required, the tumors of some groups were subjected to NIR irradiation (980 nm, 2 W/cm2) for a duration of 30 minutes. Tumor volume was assessed using caliper measurements and calculated using the formula: (length × width^2)/2. After 21 days from the initiation of treatment, some mice were humanely euthanized, while others were continued to be monitored for survival assessment. Tumors from euthanized mice were photographed, weighed, and subsequently subjected to paraffin embedding and preservation in OCT reagent for subsequent investigations.
Statistical Analysis
The results are presented as mean ± SD. Group comparisons were assessed employing the two-tailed Student’s t-test. Significance levels were denoted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Results and Discussion
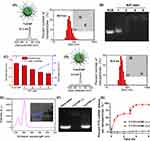
In order to prepare the nanopolyplex T-si/UCNP, a triblock amphiphilic polymer cRGD-PEG-PAsp(DMAENA)-PPHE was prepared following the schematic depicted in Figure 1A. Initially, allyl-polyethylene-NH2 (APEG-NH2, Mn = 2.4 kDa) served as the initiator for sequential ring-opening polymerizations of BLA-NCA and PHE-NCA, resulting in the formation of the allyl-capped triblock copolymer APEG-PBLA-PPHE. The degrees of polymerization for the PBLA block and PPHE block were both 30, based on integral calculations of the 1H-NMR peak areas (Figure 1B). Subsequently, the protective group of the PBLA block was removed, converting it into the PAsp block within APEG-PAsp-PPHE. Moving forward, the cRGD-capped copolymer cRGD-PEG-PAsp-PPHE was prepared through an addition reaction involving alkene and thiol groups. Lastly, the UV-sensitive cationic molecule DMAENA was grafted onto the PAsp block of cRGD-PEG-PAsp-PPHE through esterification, yielding the final copolymer cRGD-PEG-PAsp(DMAENA)-PPHE. The successful synthesis of cRGD-PEG-PAsp(DMAENA)-PPHE was confirmed through proton nuclear magnetic resonance (1H-NMR), Fourier-transform infrared (FTIR) spectroscopy, and mass spectrometry (MS) analyses (Figure 1B–D).
Next, the amphiphilic copolymer was self-assembled with hydrophobic UCNPs (~20 nm) to form micellar nanoparticle T-UCNP which featured a hydrophilic shell of cRGD-PEG, a cationic interlayer, and a hydrophobic core encapsulating the UCNPs. Dynamic light scattering and zeta potential analysis revealed that T-UCNP had a hydrodynamic size of 65.9 nm and a zeta potential of +27.2 mV (Figure 2A). As shown in TEM (Figure 2A), due to the low electron density of the copolymer, only the aggregated UCNPs can be observed, indicating that approximately three UCNPs are encapsulated within a single T-UCNP. In theory, the cationic nanoparticle T-UCNP has the capacity to complex with siRNA. As demonstrated by the gel electrophoresis result (Figure 2B), the mobility of siRNA in the agarose gel was completely retarded at N/P ratios of ≥ 4. This observation indicates that the negative charges of the siRNAs were effectively neutralized by the cationic nanoparticle T-UCNP. Upon complexation with siRNA, the T-si/UCNP nanopolyplex exhibited a decreasing particle size and an increasing Zeta potential with the rising N/P ratio, maintaining a stable particle size at N/P ratios of ≥ 4 (Figure 2C). At an N/P ratio of 4, the T-si/UCNP nanopolyplex exhibited a hydrodynamic size of 55.6 nm and a zeta potential of +4.3 mV (Figure 2D), which is beneficial for facilitating cellular uptake and mitigating cytotoxicity. The TEM image showed that siRNA was loaded in the interlayer of T-si/UCNP. In order to confirm the upconverted UV light property of T-si/UCNP, we conducted measurements of the upconversion luminescence spectra. As depicted in Figure 2E, under 980 nm excitation, T-si/UCNP exhibited a UV emission peak at 365 nm which could cleave the 2-nitrobenzyl ester linkers.
Upon 980 nm excitation, the upconverted UV light cleaves the 2-nitrobenzyl ester linker, leading to the detachment of cationic groups (EDONB) from T-si/UCNP. This process induced the siRNA release from T-si/UCNP. Subsequently, the gel electrophoresis analysis was performed to assess whether NIR irradiation could induce the siRNA release (Figure 2F). When exposed to NIR light, the migratory behavior of siRNA within T-si/UCNP closely resembled that of naked siRNA, but was completely hindered when 980 nm NIR exposure was absent. These results strongly suggested 980 nm NIR light can efficiently trigger the siRNA release from T-si/UCNP. The quantification of siRNA release was carried out using a fluorescence spectrophotometry (Figure 2G). To measure the quantity of released siRNA, FITC-labeled siRNA molecules (siRNA-FITC) were employed. Few siRNAs-FITC were observed to be released from T-FITC/UCNP without NIR irradiation. In contrast, approximately 63% of siRNA-FITC was rapidly released from T-FITC/UCNP with NIR irradiation during the first input cycle, and more than 95% of siRNA-FITC was released after two cycles, demonstrating the highly efficient siRNA release triggered by 980 nm NIR light from T-si/UCNP.
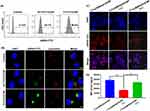
To investigate the cellular uptake and endosomal escape of T-UCNP, those copolymers both with and without cRGD modification were employed to encapsulate hydrophobic UCNPs and then complex with siRNA-FITC, resulting in the preparation of two types of nanoparticles: the targeting nanopolyplex T-FITC/UCNP and the non-targeting nanopolyplex NT-FITC/UCNP, respectively. The cellular internalization efficiency of T-FITC/UCNP and NT-FITC/UCNP was assessed using flow cytometry (Figure 3A). HCT116 cells transfected with T-FITC/UCNP exhibited a significantly higher proportion of FITC-positive cells (96.8% vs 40.1%) compared to cells treated with NT-FITC/UCNP, highlighting the substantial enhancement in siRNA transfection efficiency achieved by the cRGD peptide. Subsequently, the intracellular trafficking of siRNA-FITC was observed via CLSM after HCT116 cells were transfected with various nanopolyplexed for 4 h. The endosomal and nuclei were stained with LysoTracker Red and DAPI, respectively. As illustrated in Figure 3B, the fluorescence signal in cells transfected with T-FITC/UCNP was noticeably greater than in those transfected with NT-FITC/UCNP, aligning with the findings of the flow cytometry analysis. Moreover, after 6 h incubation, HCT116 cells treated with T-FITC/UCNP exhibited a distinct separation of green fluorescence (siRNA-FITC) and red fluorescence (lysosomes). This separation suggested the effective endosomal escape of T-FITC/UCNP, likely facilitated by the proton sponge effect attributed to the tertiary amines in the polymer cRGD-PEG-PAsp(DMAENA)-PPHE.
To investigate NIR-controlled siRNA release in cytoplasm, two distinct siRNA samples were individually labeled with a fluorescence dye (Cy3) and a quencher molecule (BHQ2). The mixture of siRNA-Cy3 and siRNA-BHQ2 at a 1:1 molar ratio was then complexed with cationic micelle T-UCNP, yielding nanoparticle denoted as T-Cy3&BHQ2/UCNP. Additionally, for control purposes, siRNA-Cy3 and scramble siRNA (siRNA-SCR) were mixed at a 1:1 molar ratio and complexed with cationic micelle T-UCNP, forming nanoparticle T-Cy3&Blank/UCNP. The Cy3 fluorescence signal of cells with various treatments were recorded through CLSM. As shown in Figure 3C, HCT116 cells receiving T-Cy3&BHQ2/UCNP exhibited a significantly reduced fluorescence signal of siRNA-Cy3 compared to those receiving T-Cy3&Blank/UCNP (control group). This decrease in fluorescence intensity can be attributed to the fluorescence quenching caused by the aggregation of siRNA-Cy3 and siRNA-BHQ2 within T-Cy3&BHQ2/UCNP. However, after NIR irradiation, the Cy3 fluorescence signal of the cells treated with T-Cy3&BHQ2/UCNP was recovered to a level close to that of the control group. Furthermore, the Cy3 mean fluorescence intensities (MFI) of cells subjected to various treatments were quantitatively assessed using flow cytometry. As shown in Figure 3D, the results of flow cytometry were in accordance with the results obtained from CLSM analysis. These findings validated that 980 nm NIR light can trigger the release of siRNA within the cytoplasm, leading to the dequenching effect between Cy3 and BHQ2.
In order to explore the NIR-controlled gene silencing and anti-tumor potential of T-si/UCNP, we prepared the nanoparticle encapsulating siRNA targeting PLK1 (siRNA-PLK1), denoted as T-PLK1/UCNP. Polo-like kinase 1 (PLK1) is a protein that belongs to the family of serine/threonine kinases and often found to be overexpressed in many types of cancer. Due to its significance in cell cycle regulation and its association with cancer, PLK1 has been a target for cancer research and drug development.29,30 Inhibiting PLK1 activity is being explored as a potential therapeutic strategy for anti-tumor therapy.31
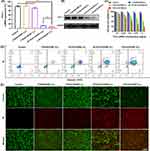
The gene silencing impact of T-PLK1/UCNP was assessed at both the mRNA and protein levels (Figure 4A and B). In contrast to the PBS group, it was observed that the PLK1 mRNA levels in HCT116 cells treated with T-SCR/UCNP (+L) did not decrease, suggesting that neither the carrier nor NIR light had a regulatory effect. Furthermore, in the absence of NIR irradiation, there was no significant change in the PLK1 mRNA levels in the T-PLK1/UCNP (-L) group. However, under NIR irradiation, both the targeted T-PLK1/UCNP (+L) group and the non-targeted NT-PLK1/UCNP (+L) group exhibited a significant reduction in PLK1 mRNA levels. Notably, T-PLK1/UCNP (+L) group exhibited a more substantial reduction in PLK1 mRNA levels compared to the NT-PLK1/UCNP (+L) group, indicating that the modification of cRGD enhanced gene inhibiting effect The suppression of PLK1 protein expression was in concordance with its inhibition at the mRNA level. The above results indicated that NIR irradiation is capable of triggering siRNA-PLK1-mediated gene inhibition.
Subsequently, the MTT assay revealed that HCT116 tumor cells treated with T-SCR/UCNP (+L) or T-PLK1/UCNP (-L) exhibited high levels of viability compared to the control group (Figure 4C). In contrast, both the T-PLK1/UCNP (+L) and NT-PLK1/UCNP (+L) groups displayed a significant decrease in cell viability as siRNA-PLK1 concentrations increased. Notably, HCT116 tumor cells treated with T-PLK1/UCNP (+L) exhibited the lowest viabilities across all tested concentrations of siRNA-PLK1 (0.1–1 μg/mL). A concentration of 1 μg/mL of siRNA-PLK1 was chosen to investigate cell apoptosis. In contrast to the control group, T-SCR/UCNP (+L), and T-PLK1/UCNP (-L) groups that all caused minimal cell apoptosis, the treatments with T-PLK1/UCNP (+L) and NT-PLK1/UCNP (+L) resulted in approximately 80.6% and 48.5% apoptotic cells, respectively (Figure 4D). Similar outcomes were corroborated through live/dead-cell staining, where live cells emitted green fluorescence and dead cells emitted red fluorescence (Figure 4E). These findings confirmed that T-PLK1/UCNP exhibits NIR-controlled anti-tumor effect in vitro.
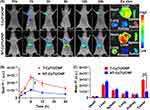
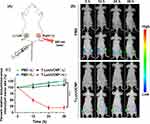
To monitor the biodistribution and tumor accumulation of nanopolyplexes, we utilized siRNA-Cy7 to fabricate T-Cy7/UCNP and NT-Cy7/UCNP. After intravenous injection, the mice harboring HCT116 xenograft tumors were subjected to an in vivo imaging system. Consequently, the fluorescence intensities of siRNA-Cy7 within tumor site peaked at 6 h after the injection of either T-Cy7/UCNP or NT-Cy7/UCNP (Figure 5A and B). Remarkably, mice receiving T-Cy7/UCNP showed the highest fluorescence intensity at the tumor, attributed to the improved tumor accumulation of nanopolyplexes facilitated by the cRGD targeting ligand. Furthermore, the outcomes of biodistribution analysis in excised organs and tumors at 24 hours after injection revealed that the Cy7 fluorescence signals in tumor site of mice treated with T-Cy7/UCNP was 2.8 times higher than in those treated with NT-Cy7/UCNP (Figure 5C). However, despite the improved siRNA accumulation in tumors achieved by the targeting nanoparticle T-Cy7/UCNP, substantial amounts of siRNAs were detected in non-tumor tissues, encompassing the liver and spleen. The findings underscored the effective delivery of siRNA into tumor tissues by T-Cy7/UCNP but also highlighted the unavoidable distribution of siRNA in normal tissues. Consequently, the desired outcome is the spatially NIR-controlled release of siRNA, enabling siRNA therapy activation within the tumor while minimizing impact on normal tissues.
To verify the in vivo NIR-controlled gene silencing potential of T-si/UCNP, we implanted recombinant HCT116 cells with constitutive luciferase (Luc) protein expression into the bilateral thighs of mice. siRNA designed to target Luc (siRNA-Luc) was complexed with the cationic micelle T-UCNP to synthesize the T-Luc/UCNP nanopolyplex. After administering PBS or T-Luc/UCNP through intravenous injection, NIR light was administered to the tumor tissue in the right thigh for a duration of 30 minutes at 6 hours post-injection, while the left thigh remained unexposed to NIR radiation (Figure 6A). We proceeded to record and analyze the bioluminescence emitted by luciferase at distinct time intervals (0, 12, 24, and 36 hours) following injection (Figure 6B). Regardless of the presence or absence of NIR irradiation, the tumor bioluminescence signal of the control group exhibited a slightly increase, attributed to tumor cell proliferation. Significantly, mice receiving T-Luc/UCNP exhibited a significant decrease of bioluminescence signal over time in the right thigh tumor exposed to NIR light, while the left thigh without NIR exposure displayed bioluminescence intensity similar to that of the control group. In comparison to the bioluminescence signal of pre-NIR exposure, the bioluminescence signal decreased by 80% at 24 hours post-injection in mice receiving T-Luc/UCNP (+L) (Figure 6C). These findings conclusively confirmed that T-Luc/UCNP can achieve NIR-controlled gene silencing in vivo.
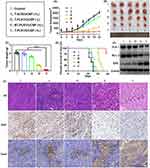
To assess the in vivo anti-tumor effect of T-PLK1/UCNP, mice harboring HCT116 xenograft tumors were allocated into 5 groups: PBS, T-SCR/UCNP (+L), T-PLK1/UCNP (-L), NT-PLK1/UCNP (+L), and T-PLK1/UCNP (+L). Subsequently, these groups underwent their respective treatments. Compared to the PBS group, the T-SCR/UCNP (+L) group exhibited no discernible impact on inhibiting tumor growth or prolonging survival rates, suggesting that both the carriers themselves and NIR irradiation had no anti-tumor effect. At the 21 days after the initial injection, the T-PLK1/UCNP (-L) group showed negligible tumor inhibitory effect, whereas the T-PLK1/UCNP (+L) group exhibited the most significant tumor inhibitory effect, leading to a reduction of tumor weight and volume by 94% and 91%, respectively, in comparison to the PBS group. (Figure 7A–C). Additionally, the treatment of T-PLK1/UCNP (+L) substantially elevated mouse survival rate and extended their overall survival durations. As shown in Figure 7D, all mice subjected to treatments of PBS, T-SCR/UCNP (+L), or T-PLK1/UCNP (-L) died within 28 days. Similarly, all mice treated with NT-PLK1/UCNP (+L) also died within 35 days. In contrast, 80% of mice subjected to T-PLK1/UCNP (+L) treatment exhibited survival beyond 40 days.
The antitumor effect resulting from PLK1 gene silencing was further elucidated by assessing the expressions of PLK1 and apoptosis-related proteins in tumor. Western blot results revealed that T-PLK1/UCNP (+L) treatment elicited the most prominent downregulation of PLK1 and the anti-apoptotic protein Bcl-2 (Figure 7E), along with the most notable upregulation of the pro-apoptotic protein BAX. In contrast, T-PLK1/UCNP (-L) treatments failed to induce the downregulation of PLK1 and Bcl-2, or the upregulation of BAX.
Furthermore, the anti-tumor efficacy of T-PLK1/UCNP was further validated through histopathological, immunohistochemical, and TUNEL assessments. As depicted in Figure 7F, treatment with T-PLK1/UCNP (+L) induced the highest levels of apoptotic tumor cells while yielding the lowest tumor cell density and Ki67 expression. Conversely, in the absence of 980 nm NIR exposure, the T-PLK1/UCNP (-L) group did not manifest a significant anti-tumor response. These results confirmed the NIR-controlled gene silencing capability of T-PLK1/UCNP, resulting in efficient anti-tumor effect in vivo.
Conclusion
In this study, we developed a nanopolyplex (T-si/UCNP) capable of efficiently targeting tumor tissue and achieving controlled release of siRNA at the tumor site upon exposure to 980 nm irradiation. By utilizing the nanopolyplex for the delivery of siRNA-PLK1, a controlled and significant inhibitory effect on HCT116 tumors in vivo was achieved under 980 nm NIR irradiation. Consequently, we believe that this nanopolyplex can not only enhance the controllability and safety of RNAi therapy for tumors but also has the potential to serve as a platform for achieving controlled release and activation of other drugs, such as mRNA and DNA.
Acknowledgments
This work was supported by Application value of 3D reconstruction of gastric vascular navigation technology in precision surgery for gastric cancer (JLY2021073) and Intestinal flora nodules in adults with diarrhea after enterostomy Structural analysis and intervention research (XYFM2020041).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Kim HJ, Kim A, Miyata K, et al. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv Drug Deliv Rev. 2016;104:61–77.
2. Chalbatani GM, Dana H, Gharagouzloo E, et al. Small interfering RNAs (siRNAs) in cancer therapy: a nano-based approach. Int J Nanomed. 2019;14:3111–3128. doi:10.2147/IJN.S200253
3. Abosalha AK, Boyajian J, Ahmad W, et al. Clinical pharmacology of siRNA therapeutics: current status and future prospects. Expert Rev Clin Phar. 2022;15(11):1327–1341. doi:10.1080/17512433.2022.2136166
4. Mehanna MM, Abla KK. siRNA nanohybrid systems: false hope or feasible answer in cancer management. Ther Deliv. 2022;13(2):109–133. doi:10.4155/tde-2021-0068
5. Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23(5):265–280. doi:10.1038/s41576-021-00439-4
6. Saw PE, Song EW. siRNA therapeutics: a clinical reality. Sci China Life Sci. 2020;63(4):485–500. doi:10.1007/s11427-018-9438-y
7. Kurakula H, Vaishnavi S, Sharif MY, et al. Emergence of small interfering RNA-based gene drugs for various diseases. ACS Omega. 2023;8(23):20234–20250. doi:10.1021/acsomega.3c01703
8. Liu S, Sun XY, Lu H, et al. Fullerene-based nanocomplex assists pulmonary delivery of siRNA for treating metastatic lung cancer. Nano Today. 2023;50:101878. doi:10.1016/j.nantod.2023.101878
9. Tarab-Ravski D, Hazan-Halevy I, Goldsmith M, et al. Delivery of therapeutic RNA to the bone marrow in multiple myeloma using CD38-targeted lipid nanoparticles. Adv Sci. 2023;10(21):e2301377. doi:10.1002/advs.202301377
10. Zhang QF, Kuang GZ, Li WZ, et al. Stimuli-responsive gene delivery nanocarriers for cancer therapy. Nano-Micro Lett. 2023;15(1):44.
11. Wang ZX, Zhang XL, Han MS, et al. An ultra pH-responsive peptide nanocarrier for cancer gene therapy. J Mater Chem B. 2023;11(37):8974–8984. doi:10.1039/D3TB01311A
12. Ye SY, Feng YL, Zhang YQ, et al. Furin enzyme-responsive siRNA delivery system for efficient anti-hypoxia-assisted cancer photodynamic therapy. CCS Chem. 2023:1–12. doi:10.31635/ccschem.023.202302777
13. Ye YH, Zhang LL, Dai YH, et al. PSMA-targeting reduction-cleavable hyperbranched polyamide-amine gene delivery system to treat the bone metastases of prostate cancer. Int J Nanomed. 2020;15:7173–7184. doi:10.2147/IJN.S268398
14. Yang YX, Ning HJ, Xia TP, et al. Electrostatic attractive self-delivery of siRNA and light-induced self-escape for synergistic gene therapy. Adv Mater. 2023;35(30):e2301409. doi:10.1002/adma.202301409
15. Mo YL, Cheng HYM, D’Elia A, et al. Light-activated siRNA endosomal release (LASER) by porphyrin lipid nanoparticles. ACS Nano. 2023;17(5):4688–4703.
16. Lin XY, Wu M, Li M, et al. Photo-responsive hollow silica nanoparticles for light-triggered genetic and photodynamic synergistic therapy. Acta Biomater. 2018;76:178–192. doi:10.1016/j.actbio.2018.07.007
17. Wang JX, He XY, Shen S, et al. ROS-sensitive cross-linked polyethylenimine for red-light-activated siRNA therapy. ACS Appl Mater Inter. 2019;11(2):1855–1863. doi:10.1021/acsami.8b18697
18. Foster AA, Greco CT, Green MD, et al. Light-mediated activation of siRNA release in diblock copolymer assemblies for controlled gene silencing. Adv Healthc Mater. 2015;4(5):760–770. doi:10.1002/adhm.201400671
19. Deng SH, Wang SY, Xiao ZC, et al. Unprotonatable and ROS-sensitive nanocarrier for NIR spatially activated siRNA therapy with synergistic drug effect. Small. 2022;18(41):e2203823.
20. Zhang MJ, Weng YH, Cao ZY, et al. ROS-activatable siRNA-engineered polyplex for NIR-triggered synergistic cancer treatment. ACS Appl Mater Inter. 2020;12(29):32289–32300. doi:10.1021/acsami.0c06614
21. Li W, Wang JS, Ren JS, et al. Near-infrared upconversion controls photocaged cell adhesion. J Am Chem Soc. 2014;136(6):2248–2251. doi:10.1021/ja412364m
22. Liang HX, Li ZH, Ren ZG, et al. Light-triggered NO-releasing nanoparticles for treating mice with liver fibrosis. Nano Res. 2020;13(8):2197–2202. doi:10.1007/s12274-020-2833-6
23. Yan ZQ, Qin HS, Ren JS, et al. Photocontrolled multidirectional differentiation of mesenchymal stem cells on an upconversion substrate. Angew Chem Int Edit. 2018;57(35):11182–11187.
24. Idris NM, Gnanasammandhan MK, Zhang J, et al. In vivo photodynamic therapy using upconversion nanoparticles as remote-controlled nanotransducers. Nat Med. 2012;18(10):1580–1585. doi:10.1038/nm.2933
25. Jayakumar MKG, Idris NM, Zhang Y. Remote activation of biomolecules in deep tissues using near-infrared-to-UV upconversion nanotransducers. Proc Natl Acad Sci USA. 2012;109(22):8483–8488. doi:10.1073/pnas.1114551109
26. Deng SH, Li XX, Liu S, et al. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci Adv. 2020;6(29):eabb4005. doi:10.1126/sciadv.abb4005
27. Yu ZL, Deng P, Chen YF, et al. Inhibition of the PLK1-coupled cell cycle machinery overcomes resistance to oxaliplatin in colorectal cancer. Adv Sci. 2021;8(23):e2100759.
28. Liu ZX, Sun QR, Wang XS. PLK1, a potential target for cancer therapy. Transl Oncol. 2017;10(1):22–32. doi:10.1016/j.tranon.2016.10.003
29. Liang S, Yang XZ, Du XJ, et al. Optimizing the size of micellar nanoparticles for efficient siRNA delivery. Adv Funct Mater. 2015;25(30):4778–4787. doi:10.1002/adfm.201501548
30. Huang XH, Li JW, Li GY, et al. Cation-free siRNA-cored nanocapsules for tumor-targeted RNAi therapy. Acta Biomater. 2023;161:226–237. doi:10.1016/j.actbio.2023.03.001
31. Degenhardt Y, Lampkin T. Targeting polo-like kinase in cancer therapy. Clin Cancer Res. 2010;16(2):384–389. doi:10.1158/1078-0432.CCR-09-1380
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.