Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 15
Construction of Potential miRNA–mRNA Regulatory Network in COPD Plasma by Bioinformatics Analysis
Authors Zhu M, Ye M , Wang J, Ye L , Jin M
Received 31 March 2020
Accepted for publication 18 August 2020
Published 10 September 2020 Volume 2020:15 Pages 2135—2145
DOI https://doi.org/10.2147/COPD.S255262
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Mengchan Zhu,* Maosong Ye,* Jian Wang, Ling Ye, Meiling Jin
Department of Pulmonary and Critical Care Medicine, Zhongshan Hospital, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Meiling Jin; Ling Ye Email [email protected]; [email protected]
Background: Chronic obstructive pulmonary disease (COPD) has become a major cause of morbidity and mortality worldwide. Increasing evidence indicates that aberrantly expressed microRNAs (miRNAs) are involved in the pathogenesis of COPD. However, an integrative exploration of miRNA–mRNA regulatory network in COPD plasma remains lacking.
Methods: The microarray datasets GSE24709, GSE61741, and GSE31568 were downloaded from the GEO database and analyzed using GEO2R tool to identify differentially expressed miRNAs (DEMs) between COPD and normal plasma. The consistently changing miRNAs in the three datasets were screened out as candidate DEMs. Potential upstream transcription factors and downstream target genes of candidate DEMs were predicted by FunRich and miRNet, respectively. Next, GO annotation and KEGG pathway enrichment analysis for target genes were performed using DAVID. Then, PPI and DEM-hub gene network were constructed using the STRING database and Cytoscape software. Finally, GSE56768 was used to evaluate the hub gene expressions.
Results: A total of nine (six upregulated and three downregulated) DEMs were screened out in the above three datasets. SP1 was predicted to potentially regulate most of the downregulated DEMs, while YY1 and E2F1 could regulate both upregulated and downregulated DEMs. 1139 target genes were then predicted, including 596 upregulated DEM target genes and 543 downregulated DEM target genes. Target genes of DEMs were mainly enriched in PI3K/Akt signaling pathway, mTOR signaling pathway, and autophagy. Through the DEM-hub gene network construction, most of the hub genes were found to be potentially modulated by miR-497-5p, miR-130b-5p, and miR-126-5p. Among the top 12 hub genes, MYC and FOXO1 expressions were consistent with that in the GSE56768 dataset.
Conclusion: In the study, potential miRNA–mRNA regulatory network was firstly constructed in COPD plasma, which may provide a new insight into the pathogenesis and treatment of COPD.
Keywords: microRNAs (miRNAs), chronic obstructive pulmonary disease (COPD), bioinformatics analysis, miRNA–mRNA regulatory network
Introduction
Chronic obstructive pulmonary disease (COPD), characterized by persistent respiratory symptoms, progressive airflow limitation, and lack of effective treatment, is one of the leading cause of morbidity and mortality all over the world.1,2 In China, two national surveys of COPD showed that the overall prevalence of COPD in people aged 40 years or older has increased from 8.2% during 2002–20043 to 13.7% during 2014–2015,4 which has almost doubled, putting increasing economic pressure on the patients and society. As we all know, exposure to cigarette smoke (CS) is the largest risk factor for COPD. However, most studies have shown that no more than 80% of patients with COPD are caused by CS,5,6 that is to say, there are many other factors that contribute to COPD susceptibility.
MicroRNAs (miRNAs) are a class of endogenous small noncoding RNA molecules, consisting of 18–25 nucleotides. They can regulate gene expression by directly degrading target messenger RNAs (mRNAs) or blocking protein translation,7,8 and thus participate in many biological processes such as cell proliferation, differentiation, apoptosis, disease occurrence and progression.9,10 Studies have shown that dysregulated miRNAs are related to the onset and development of multiple diseases, including COPD.11–13 Since miRNAs can also be stably expressed in body fluids, miRNAs (especially circulating miRNAs) may be an integrated part of the COPD screening programs during future preventive treatment. Hu et al14 identified that plasma miR-125b could distinguish acute exacerbation of COPD from stable COPD and healthy controls, and correlated with inflammatory cytokine expressions, including tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-8, and leukotriene B4 (LTB-4). Tang et al15 showed that compared with healthy non-smokers, the plasma levels of miR-29b was reduced in smokers and COPD patients, with a positive correlation with decreased pulmonary function. Moreover, a recent study performed by Sundar et al16 for the first time reported that plasma-derived exosome miRNAs functioned as novel COPD biomarkers, which represents a greater translational potential for miRNAs in the diagnosis and treatment of COPD.
Despite many studies on the expression and function of miRNAs in COPD have been carried out, however, little study on the role of miRNA–mRNA regulatory network in COPD plasma has been conducted to date. In this study, we for the first time screened DEMs in COPD plasma compared with normal plasma by analyzing three datasets (GSE24709, GSE61741, and GSE31568) from the GEO database. Transcription factor-DEM, DEM-target gene, functional analysis of target genes, and DEM-hub gene network were then performed. In addition, the plasma levels of hub genes were further verified in the GSE56768 dataset. Herein, we aimed to construct potential miRNA–mRNA regulatory network in COPD plasma, and to provide promising targets for COPD diagnosis and treatment.
Methods
miRNA Microarray
According to the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), the following words, that “COPD” (study keywords), “noncoding RNA profiles by array” (study type), “Home sapiens” (organism), and “plasma” (attribute name) were used to screen the miRNA datasets related to COPD plasma. Finally, three miRNA datasets (GSE24709, GSE6174117 and GSE31568) were chosen for subsequent analysis, which were all based on the GPL9040 platform (febit Homo Sapiens miRBase 13.0). Detailed information of the three datasets is shown in Table 1, and a flow chart of the study design is shown in Figure 1.
 |
Table 1 Details for GEO COPD Data |
 |
Figure 1 Flow chart of constructing the miRNA–mRNA regulatory network in COPD plasma. |
Screening of DEMs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), a R-based web application that helps to analyze GEO data, was applied to compare and screen DEMs between COPD and normal plasma. The adjusted P value using the Benjamini and Hochberg false discovery rate (FDR) method was applied to correct for the occurrence of false positive results. Here, an adjusted P value <0.05 and |Log fold change (FC)|>1.5 were set as the threshold for identifying DEMs. Venn diagram was utilized to analyze the overlapping of DEMs in the three datasets (GSE24709, GSE61741, and GSE31568), and these consistently changed miRNAs were selected as candidate DEMs. Heatmaps of DEMs and candidate DEMs were generated using the TBtools, which is a toolkit integrating various biological data handling tools.
Prediction of Upstream Transcription Factors of DEMs
FunRich (http://www.funrich.org/), a stand-alone software tool mainly for analyzing functional enrichment and interaction network analysis of genes and proteins, was used to predict the potential upstream transcription factors of candidate DEMs. The P value < 0.05 was considered statistically significant.
Prediction of Downstream Target Genes of DEMs
miRNet (https://www.mirnet.ca/), an integrated tool kit that contains information about miRNA-target interactions and displays the association in a visual network, was used to predict the potential downstream target genes of candidate DEMs.
GO and KEGG Analysis of Target Genes
To investigate the biological function of DEM target genes, the database for annotation, visualization, and integrated discovery (DAVID) online tool was utilized to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The adjusted P value < 0.05 was statistically significant.
Construction of PPI Network and Screening of Hub Genes
The protein-protein interaction (PPI) network of DEM target genes was performed by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online database (http://stringdb.org/) and then visualized in Cytoscape software (version 3.6.1). CytoHubba, a plugin of Cytoscape, was used to rank nodes in a preloaded PPI network by its features through several topological algorithms. Here, the hub genes were selected out as the top 30 nodes of the PPI network using the Maximal Clique Centrality (MCC) method, which has a better performance on the precision of predicting essential proteins. The score for a node v by MCC is defined as MCC(v) = ∑C∈S(v)(|C| - 1)!, where S(v) is the collection of maximal cliques which contain v, and (|C|-1)! is the product of all positive integers less than |C|.
Expression Analysis of Hub Genes by GSE56768 Dataset
GSE56768, downloaded from the GEO database, was introduced to analyze the expressions of hub genes due to no other data on mRNA expressions in COPD plasma. The dataset was based on the platform of GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array), in which 49 COPD and 29 normal plasma were selected. Student’s t-test was used to identify the differentially expressed genes (DEGs) between COPD and normal plasma. The significantly hub genes needed to meet two criteria as follows: one was that upregulated DEM target genes were downregulated or downregulated DEM target genes were upregulated; the second was the P value should be less than 0.05.
Results
Identification of DEMs in COPD Plasma
After screening with the threshold of an adjusted Pvalue <0.05 and |logFC|>1.5, 148 DEMs (78 upregulated and 70 downregulated) were identified in the GSE24709 dataset, 26 DEMs (19 upregulated and seven downregulated) were identified in GSE61741, and 71 DEMs (18 upregulated and 53 downregulated) were identified in GSE31568. The heatmaps of DEMs in the above three datasets are shown in Figure 2A–C, and the detailed information about DEMs is listed in Supplementary Table S1–3. Among these DEMs, six upregulated (miR-1246, miR-1258, miR-556-3p, miR-1468-5p, miR-126-5p, and miR-130b-5p) and three downregulated (miR-182-3p, miR-497-5p, and miR-492) DEMs were consistently changed in the three datasets (Figure 2D and E).
 |
Figure 2 Identification of the candidate DEMs. |
Prediction of Upstream Transcription Factors of DEMs
In the study, FunRich software was used to predict the upstream transcription factors of candidate DEMs. The transcription factors for upregulated and downregulated DEMs are presented in Figure 3A and B, respectively. For upregulated DEMs, the transcription factors were YY1 and E2F1. For downregulated DEMs, the transcription factors were SP1, YY1, EGR1, E2F1, ZFP161, ELF1, SP4, and MYC.
 |
Figure 3 Potential transcription factors of DEMs predicted by FunRich. |
Prediction of Downstream Target Genes of DEMs
A total of 1139 genes targeted by candidate DEMs were predicted by the miRNet database, among which there were 596 target genes for upregulated DEMs and 543 target genes for downregulated DEMs. For better visualization, upregulated and downregulated DEMs with their target genes were displayed in a DEM-target gene network in Figure 4A and B, respectively. In addition, the number of target genes for each DEM is listed in Table 2, and all predicted target genes are listed in Supplementary Table S4.
 |
Table 2 Potential Target Genes of the Significantly Upregulated and Downregulated DEMs |
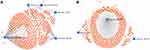 |
Figure 4 Potential target genes of DEMs predicted by miRNet. |
GO and KEGG Analysis of Target Genes
Then, we performed GO and KEGG analysis on the 1139 target genes of DEMs. As shown in Figure 5A and B, biological process (BP) analysis showed that DEM target genes were particularly enriched in muscle tissue development, striated muscle tissue development, and activation of protein kinase activity; cellular component (CC) analysis revealed that DEM target genes were mainly enriched in nuclear speck, transferase complex, transferring phosphorus-containing groups, and protein kinase complex; molecular function (MF) analysis demonstrated that DEM target genes were significantly enriched in DNA-binding transcription activator activity, RNA polymerase II specific, cell adhesion molecule binding, and protein serine/threonine kinase activity.
 |
Figure 5 GO annotation analysis for the target genes of DEMs in the biological process, cellular component, and molecular function. |
Furtherly, KEGG pathway analysis showed that DEM target genes were significantly enriched in PI3K-Akt signaling pathway, human papillomavirus infection, focal adhesion, breast cancer, human T-cell leukemia virus 1 infection and cellular senescence (Figure 6A and B).
 |
Figure 6 KEGG pathway analysis for the target genes of DEMs. |
Construction of PPI and DEM-Hub Gene Network
The PPI network of genes targeted by upregulated and downregulated DEMs was established by the STRING database, respectively. Then we uploaded the above PPI network to Cytoscape and used its cytohubba plugin to screen for hub genes. The top 30 hub genes for upregulated and downregulated DEMs are shown in Figure 7A and B, respectively. For upregulated DEMs, the top 10 hub genes were MYC, VEGFA, MAPK8, STAT5A, FOXO3, FOXO1, IGF1, NR3C1, HSPA8, and EP300; For downregulated DEMs, the top 10 hub genes were FBXW7, CUL3, BTRC, ZBTB16, ANAPC13, CDC34, FBXL18, FBXL20, SMURF1, and UBE2V1 (Table 3).
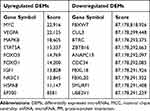 |
Table 3 Top 10 Hub Genes of the Significantly Upregulated and Downregulated DEMs in the PPI Network Ranked by MCC |
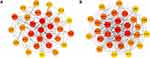 |
Figure 7 Identification of the hub genes for DEMs in the PPI network. |
To better investigate the molecular mechanisms of these DEMs in COPD plasma, the DEM-hub gene network was constructed by Cytoscape software. The interactions between upregulated DEMs and hub genes were as follows: miR-130b-5p interacted with five hub genes, including EP300, HSPA8, STAT5A, IGF1, and FOXO1; miR-126-5p interacted with five hub genes, including MYC, MAPK8, VEGFA, NR3C1, and FOXO3; The interactions between downregulated DEMs and hub genes were as follows: miR-497-5p interacted with eight hub genes, including SMURF1, BTRC, ZBTB16, FBXL20, CUL3, UBE2V1, FBXL18 and ANAPC13; miR-182-3p interacted with FBXW7; and miR-492 interacted with CDC34 (Figure 8).
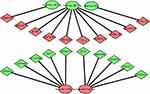 |
Figure 8 The miRNA-hub gene regulatory network. |
Validation of Hub Genes Expression
According to the DEM-hub gene network, plasma levels of the top 12 hub genes (MYC, VEGFA, MAPK8, STAT5A, FOXO3, FOXO1, FBXW7, CUL3, BTRC, ZBTB16, ANAPC13, and CDC34), were then identified using the GSE56768 dataset. For upregulated DEMs, compared with normal plasma, only the expressions of MYC and FOXO1 were consistently decreased in COPD plasma, while VEGFA expression was increased on the contrary (Figure 9A–F). For downregulated DEMs, no miRNAs were significantly changed (Figure 9G–L). Therefore, miR-126-5p-MYC and miR-130b-5p-FOXO1 were identified as two potential regulatory pathways in COPD plasma.
 |
Figure 9 The mRNA expression of the top 12 hub genes was determined from the GSE56768 dataset. |
Discussion
During the past few years, increasing number of studies have been conducted in COPD diagnosis and treatment, however, the prognosis of these patients is still poor partly due to the limited understanding of COPD pathogenesis. Recently, with microarray technology, it is easier to reveal thousands of genetic alterations in the progression of various diseases. In the present study, data from three datasets, namely GSE24709, GSE61741, and GSE31568, were extracted to identify DEMs between COPD and normal plasma. A total of six upregulated DEMs (miR-1246, miR-1258, miR-556-3p, miR-1468-5p, miR-126-5p, and miR-130b-5p) and three downregulated DEMs (miR-182-3p, miR-497-5p, and miR-492), which were consistently changed in the three datasets, were identified as candidate DEMs to be further analyzed. Among upregulated DEMs, detection of plasma and plasma exosome miR-1246 and miR-130b-5p can be used as diagnosis biomarkers for various cancers. Despite that, anti-miR-1246 alleviated lipopolysaccharide (LPS)-induced inflammation and permeability in vivo and vitro.18 Anti-miR-130b-5p prevented hepatic lipid accumulation and insulin resistance in nonalcoholic fatty liver disease.19 For downregulated DEMs, overexpression of miR-497-5p inhibited IL-1β-induced cartilage matrix degradation in chondrocytes, providing a potential therapeutic target for osteoarthritis.20 Notably, many of these miRNAs were reported in cancer biology, and need further studies for their roles in COPD.
Recent studies have shown that the expression of miRNA can be modulated by transcription factors,21,22 and thus we predicted transcription factors that can potentially regulate these candidate DEMs. Specificity protein 1 (SP1), a C2H2-type zinc-finger transcription factor,23 was predicted to account for the highest percentage of downregulated DEMs, and has been widely documented in COPD. In the year of 2012, Di et al24 showed that CS induced MUC5AC gene transcription predominantly through increased SP1 nuclear protein levels and increased SP1 binding to its promoter region. Following that, more experiments suggested that ERK-SP1 may be a promising target for inhibiting COPD-related inflammation and mucus hypersecretion.25–27 Yin Yang 1 (YY1) and E2F1 were predicted to regulate the expressions of both upregulated and downregulated DEMs. In the study of YY1, a transcriptional repressor that inhibits muscle gene expression and myogenesis, Natanek et al28 found that YY1 expression was related to smaller quadriceps fiber cross-sectional area (CSA), and its nuclear localization was in muscle of COPD patients but not controls, suggesting an important role of YY1 in COPD-related muscle atrophy. E2F1, a key proliferative factor,29,30 was found to be targeted by miR-197 and participate in the vascular remodeling of COPD.12 In fact, despite of ZFP161 and SP4, the remaining transcription factors have been reported in COPD,31–33 which in turn supports the importance of these candidate DEMs in the pathogenesis of COPD.
KEGG analysis showed that the DEMs were mainly enriched in PI3K-Akt signaling pathway, mTOR signaling pathway, and autophagy. In the previous studies, the activation of PI3K-Akt signaling pathway has been amply documented to have an association with COPD-related inflammation. A study performed by Horiguchi et al34 showed that pulmonary administration of Wortmannin, an inhibitor of PI3K, was capable of inducing the repair of alveolar injury and a significant recovery of the respiratory function. Conversely, the activation of mTOR signaling pathway was reported to suppresses CS-induced inflammation and emphysema development, likely through modulation of autophagy, apoptosis, and necroptosis.35 Wu et al36 identified that increased levels of autophagy in peripheral blood mononuclear cells (PBMCs) of COPD patients were negatively correlated with FEV1% predicted, and positively correlated with circulating expressions of inflammatory cytokines. Therefore, inhibition of PI3K-Akt, activation of mTOR and inhibition of autophagy may represent novel therapeutic strategies for COPD. Although the numbers of genes targeted by either upregulated or downregulated DEMs were similar, however, many of the target genes of upregulated DEMs were not involved in GO and KEGG pathways (results were not shown), including the transcription factors described above. This may hint that downregulated DEMs may play a more critical role in the pathogenesis of COPD.
By constructing DEM-hub gene network, we found that most of hub genes could be potentially targeted by miR-497-5p, miR-130b-5p, and miR-126-5p. Among the top 12 hub genes, only two genes (MYC, FOXO1) expressions were in accordance with that in the GSE56768 dataset, which may result from different sample sources. MYC is well known to play a vital role in the pathogenesis of squamous cell lung cancer (SCLC). In COPD, highly expressed c-MYC in lung tissue was reported to induce the pulmonary vessels structural remodeling.37 However, in our results, the expression level of MYC in plasma was opposite to that in lung tissue. Volckaert et al38 showed that conditional deletion of c-Myc from parabronchial smooth muscle cells (PSMCs) inhibited airway epithelial repair, and thus plasma MYC downregulation may explain the dysregulation of lung repair in COPD. FOXO1, a member of the forkhead-box O family, can act as a homeostasis regulator and coordinate responses to various environmental changes, including oxidative stress.39,40 Liu et al41 identified that AMPK alleviated endoplasmic reticulum stress in COPD through activation of FOXO1. In agreement with the study of Hueso et al,42 FOXO1 expression of our analysis was decreased in COPD plasma compared with normal plasma, suggesting that miR-130b-5p-FOXO1 may participate in the pathogenesis of COPD through excessive oxidative stress.
Although we for the first time investigated the potential miRNA–mRNA regulatory network in COPD plasma by integrating multiple microarray datasets, however, there were several limitations existing in our present study. Firstly, we only focused on miRNAs and mRNAs between COPD and normal plasma, however, some of these may vary in different stages of COPD, which need to be further identified. Secondly, the sample size of each dataset used here was not enough, compared with that usually required for biomarker analysis. Thirdly, the miRNA–mRNA interactions were only based on predictions from public databases, and further studies with experimental validations in vivo and in vitro are still needed to validate our analysis.
Conclusion
In summary, based on the GEO database and bioinformatics analysis, we firstly constructed two potential miRNA–mRNA pathways (miR-126-5p-MYC, miR-130b-5p-FOXO1) in COPD plasma as potential biomarkers for the diagnosis and treatment of COPD patients. We hope that these findings can contribute to future in-depth studies and achieve the goal of improving the prognosis of patients with COPD.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Feigin V. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388(10053):1545–1602. doi:10.1016/S0140-6736(16)31678-6
2. Soriano JB, Abajobir AA, Abate KH. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet Respir Med. 2017;5(9):691–706. doi:10.1016/S2213-2600(17)30293-X
3. Zhong N, Wang C, Yao W, et al. Prevalence of chronic obstructive pulmonary disease in China: a large, population-based survey. Am J Respir Crit Care Med. 2007;176(8):753–760. doi:10.1164/rccm.200612-1749OC
4. Wang C, Xu J, Yang L, et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China pulmonary health [CPH] study): a national cross-sectional study. Lancet. 2018;391(10131):1706–1717. doi:10.1016/S0140-6736(18)30841-9
5. Lindberg A, Eriksson B, Larsson LG, Ronmark E, Sandstrom T, Lundback B. Seven-year cumulative incidence of COPD in an age-stratified general population sample. Chest. 2006;129(4):879–885. doi:10.1378/chest.129.4.879
6. Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J. Developing COPD: a 25 year follow up study of the general population. Thorax. 2006;61(11):935–939. doi:10.1136/thx.2006.062802
7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
8. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
9. Wu B, Liu G, Jin Y, et al. miR-15b-5p promotes growth and metastasis in breast cancer by targeting HPSE2. Front Oncol. 2020;10:108. doi:10.3389/fonc.2020.00108
10. Wittstatt J, Weider M, Wegner M, Reiprich S. MicroRNA miR-204 regulates proliferation and differentiation of oligodendroglia in culture. Glia. 2020;68(10):2015–2027. doi:10.1002/glia.23821
11. Conickx G, Mestdagh P, Avila CF, et al. MicroRNA profiling reveals a role for microRNA-218-5p in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;195(1):43–56. doi:10.1164/rccm.201506-1182OC
12. Musri MM, Coll-Bonfill N, Maron BA, et al. MicroRNA dysregulation in pulmonary arteries from chronic obstructive pulmonary disease. Relationships with vascular remodeling. Am J Respir Cell Mol Biol. 2018;59(4):490–499. doi:10.1165/rcmb.2017-0040OC
13. Mohamed A, Kunda NK, Ross K, Hutcheon GA, Saleem IY. Polymeric nanoparticles for the delivery of miRNA to treat chronic obstructive pulmonary disease (COPD). Eur J Pharm Biopharm. 2019;136:1–8. doi:10.1016/j.ejpb.2019.01.002
14. Hu HL, Nie ZQ, Lu Y, et al. Circulating miR-125b but not miR-125a correlates with acute exacerbations of chronic obstructive pulmonary disease and the expressions of inflammatory cytokines. Medicine. 2017;96(51):e9059. doi:10.1097/MD.0000000000009059
15. Tang K, Zhao J, Xie J, Wang J. Decreased miR-29b expression is associated with airway inflammation in chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 2019;316(4):L621–L629. doi:10.1152/ajplung.00436.2018
16. Sundar IK, Li D, Rahman RI. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J Extracell Vesicles. 2019;8(1):1684816. doi:10.1080/20013078.2019.1684816
17. Wu C, Zhao Y, Liu Y, et al. Identifying miRNA-mRNA regulation network of major depressive disorder in ovarian cancer patients. Oncol Lett. 2018;16(4):5375–5382. doi:10.3892/ol.2018.9243
18. Fang Y, Gao F, Hao J, Liu Z. microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. Am J Transl Res. 2017;9(3):1287–1296.
19. Liu X, Chen S, Zhang L. Downregulated microRNA-130b-5p prevents lipid accumulation and insulin resistance in a murine model of nonalcoholic fatty liver disease. Am J Physiol Endocrinol Metab. 2020;319(1):E34–E42. doi:10.1152/ajpendo.00528.2019
20. Hou L, Shi H, Wang M, Liu J, Liu G. MicroRNA-497-5p attenuates IL-1β-induced cartilage matrix degradation in chondrocytes via Wnt/β-catenin signal pathway. Int J Clin Exp Pathol. 2019;12(8):3108–3118.
21. Mullany LE, Herrick JS, Wolff RK, Stevens JR, Samowitz W, Slattery ML. MicroRNA-transcription factor interactions and their combined effect on target gene expression in colon cancer cases. Genes Chromosomes Cancer. 2018;57(4):192–202. doi:10.1002/gcc.22520
22. Uddin MN, Li M, Wang X. Identification of transcriptional markers and microRNA-mRNA regulatory networks in colon cancer by integrative analysis of mRNA and microRNA expression profiles in colon tumor stroma. Cells-Basel. 2019;8(9):1054. doi:10.3390/cells8091054
23. Chu S. Transcriptional regulation by post-transcriptional modification–role of phosphorylation in Sp1 transcriptional activity. Gene. 2012;508(1):1–8. doi:10.1016/j.gene.2012.07.022
24. Di YP, Zhao J, Harper R. Cigarette smoke induces MUC5AC protein expression through the activation of Sp1. J Biol Chem. 2012;287(33):27948–27958. doi:10.1074/jbc.M111.334375
25. Shin IS, Shin NR, Park JW, et al. Melatonin attenuates neutrophil inflammation and mucus secretion in cigarette smoke-induced chronic obstructive pulmonary diseases via the suppression of Erk-Sp1 signaling. J Pineal Res. 2015;58(1):50–60. doi:10.1111/jpi.12192
26. Park JW, Shin NR, Shin IS, et al. Silibinin inhibits neutrophilic inflammation and mucus secretion induced by cigarette smoke via suppression of ERK-SP1 pathway. Phytother Res. 2016;30(12):1926–1936. doi:10.1002/ptr.5686
27. Shin NR, Ryu HW, Ko JW, et al. A standardized bark extract of Pinus pinaster Aiton (Pycnogenol((R))) attenuated chronic obstructive pulmonary disease via Erk-sp1 signaling pathway. J Ethnopharmacol. 2016;194:412–420. doi:10.1016/j.jep.2016.10.029
28. Natanek SA, Riddoch-Contreras J, Marsh GS, et al. Yin Yang 1 expression and localisation in quadriceps muscle in COPD. Arch Bronconeumol. 2011;47(6):296–302. doi:10.1016/j.arbres.2011.02.015
29. Attwooll C, Denchi EL, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23(24):4709–4716. doi:10.1038/sj.emboj.7600481
30. Crosby ME, Almasan A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol Ther. 2004;3(12):1208–1211. doi:10.4161/cbt.3.12.1494
31. Savarimuthu FS, Larsen JE, Pavey SJ, et al. Genes and gene ontologies common to airflow obstruction and emphysema in the lungs of patients with COPD. PLoS One. 2011;6(3):e17442. doi:10.1371/journal.pone.0017442
32. Hou HH, Wang HC, Cheng SL, Chen YF, Lu KZ, Yu CJ. MMP-12 activates protease-activated receptor-1, upregulates placenta growth factor, and leads to pulmonary emphysema. Am J Physiol Lung Cell Mol Physiol. 2018;315(3):L432–L442. doi:10.1152/ajplung.00216.2017
33. Sun X, Shang J, Wu A, Xia J, Xu F. Identification of dynamic signatures associated with smoking-related squamous cell lung cancer and chronic obstructive pulmonary disease. J Cell Mol Med. 2020;24(2):1614–1625. doi:10.1111/jcmm.14852
34. Horiguchi M, Oiso Y, Sakai H, Motomura T, Yamashita C. Pulmonary administration of phosphoinositide 3-kinase inhibitor is a curative treatment for chronic obstructive pulmonary disease by alveolar regeneration. J Control Release. 2015;213:112–119. doi:10.1016/j.jconrel.2015.07.004
35. Houssaini A, Breau M, Kebe K, et al. mTOR pathway activation drives lung cell senescence and emphysema. JCI Insight. 2018;3(3). doi:10.1172/jci.insight.93203
36. Wu Y, Xu B, He X, et al. Correlation between autophagy levels in peripheral blood mononuclear cells and clinical parameters in patients with chronic obstructive pulmonary disease. Mol Med Rep. 2018;17(6):8003–8009. doi:10.3892/mmr.2018.8831
37. Li R, Xu F, Wu X, Ji S, Xia R. CUL1-mediated organelle fission pathway inhibits the development of chronic obstructive pulmonary disease. Comput Math Methods Med. 2020;2020:5390107. doi:10.1155/2020/5390107
38. Volckaert T, Campbell A, De Langhe S. c-Myc regulates proliferation and Fgf10 expression in airway smooth muscle after airway epithelial injury in mouse. PLoS One. 2013;8(8):e71426. doi:10.1371/journal.pone.0071426
39. Taka C, Hayashi R, Shimokawa K, et al. SIRT1 and FOXO1 mRNA expression in PBMC correlates to physical activity in COPD patients. Int J Chron Obstruct Pulmon Dis. 2017;12:3237–3244. doi:10.2147/COPD.S144969
40. Xing YQ, Li A, Yang Y, Li XX, Zhang LN, Guo HC. The regulation of FOXO1 and its role in disease progression. Life Sci. 2018;193:124–131. doi:10.1016/j.lfs.2017.11.030
41. Liu JQ, Zhang L, Yao J, Yao S, Yuan T. AMPK alleviates endoplasmic reticulum stress by inducing the ER-chaperone ORP150 via FOXO1 to protect human bronchial cells from apoptosis. Biochem Biophys Res Commun. 2018;497(2):564–570. doi:10.1016/j.bbrc.2018.02.095
42. Hueso M, Mallen A, Casas A, et al. Integrated miRNA/mRNA counter-expression analysis highlights oxidative stress-related genes CCR7 and FOXO1 as blood markers of coronary arterial disease. Int J Mol Sci. 2020;21(6):1943. doi:10.3390/ijms21061943
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
