Back to Journals » Cancer Management and Research » Volume 12
Concomitant Mutations in EGFR 19Del/L858R Mutation and Their Association with Response to EGFR-TKIs in NSCLC Patients
Authors Liang H, Li C , Zhao Y, Zhao S, Huang J, Cai X, Cheng B , Xiong S, Li J, Wang W, Zhu C, Li W, He J, Liang W
Received 28 March 2020
Accepted for publication 8 August 2020
Published 18 September 2020 Volume 2020:12 Pages 8653—8662
DOI https://doi.org/10.2147/CMAR.S255967
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Hengrui Liang,1,* Caichen Li,1,* Yi Zhao,1,* Shen Zhao,2,* Jun Huang,1 Xiuyu Cai,2,3 Bo Cheng,1 Shan Xiong,1 Jianfu Li,1 Wei Wang,1 Changbin Zhu,4 Weiwei Li,5 Jianxing He,1 Wenhua Liang1
1Department of Thoracic Surgery and Oncology, The First Affiliated Hospital of Guangzhou Medical University, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, Guangzhou, People’s Republic of China; 2Department of Medical Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, People’s Republic of China; 3Department of General Internal Medicine, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, People’s Republic of China; 4BGI Genomics, BGI-Shenzhen, Shenzhen 518083, People’s Republic of China; 5BGI-Guangzhou Medical Laboratory, BGI-Shenzhen, Guangzhou 510006, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianxing He; Wenhua Liang
Department of Thoracic Surgery and Oncology, The First Affiliated Hospital of Guangzhou Medical University, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, Guangzhou 510120, People’s Republic of China
Tel +86-20-83337792
Fax +86-20-83350363
Email [email protected]
Objective: Differences in efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKI) have been observed between non-small cell lung cancer (NSCLC) patients with 19 exon deletion (19Del) and L858R mutation. We explored whether the total number or pattern of concomitant mutations of 19Del and L858R may explain their different sensitivities.
Patients and Methods: This study contained the mutational profiles of EGFR-mutated NSCLC patients from two cohorts: Guangzhou (G1) and database (G2). Concomitant mutation status and EGFR-TKI response information were retrieved.
Results: A total of 403 patients covered 283 genes in the G1 and 803 patients with a different gene set in the G2 were included. Similar prevalence of total concomitant mutation number was observed in both G1 ( 19Del 32.48% vs L858R 30.45%; P=0.68) and G2 ( 19Del 74.9% vs L858R 73.2%; P=0.65) cohorts. Only HGF/c-Met pathway same more related to L858R mutation. EGFR-TKI response information was recorded for 134 patients in the G2 cohort. 19Del showed a higher objective response (OR) rate compared with L858R, regardless of concomitant mutations. Compared to patients with OR, non-OR patients had more concomitant mutations, both in 19Del (53.8% vs 83.3%; P=0.021) and L858R (51.4% vs 77.8%; P=0.029). In particular, total concomitant mutations (OR=0.27; P=0.03), sensitive EGFR mutations (OR=2.21; P=0.04), and T790M (OR=0.244; P=0.02) significantly affected the TKI response.
Conclusion: Concomitant mutations were widespread in 19Del and L858R and were associated with poorer OR to EGFR-TKIs. However, 19Del and L858R had similar numbers and patterns of concomitant mutations, which might not explain the different sensitivity to EGFR-TKI.
Keywords: epidermal growth factor receptor mutation, 19Del, L858R, non-small cell lung cancer, concomitant mutation
Introduction
Epidermal growth factor receptor (EGFR) mutations are a key cancerous driver in non-small cell lung cancer (NSCLC), contributing to nearly half of reported NSCLC cases in Asians and 15% in Caucasians.1 EGFR-tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib, are the mainstay of treatment for advanced patients, with an overall response rate of 70% and a median progression free survival (PFS) of 10 months.2
19 exon deletion (19Del) and L858R mutation in 21 exon (L858R) are two major subtypes of EGFR mutations and considered sensitive to EGFR-TKIs. We reported a meta-analysis confirming that EGFR 19 exon deletion was associated with better outcomes in treatments with EGFR-TKIs than L858R mutation.3 An updated comprehensive network meta-analysis from our team also revealed that the best treatment strategies differ for these two sensitive mutations.4
Hot debate persists regarding the mechanisms for the difference between these two subtypes. Some investigators have explored the structures of 19Del and L858R and suggested different binding affinity of TKIs.5 We recently reported a higher prevalence of somatic T790M in L858R (22%) than in 19Del (19%), this may be one of the mechanisms to explain the discrepancy.6 With the rapid development of multiple genetic profiling by next generation sequencing (NGS), some recent studies have suggested that other concomitant mutations might also compromise the efficacy of EGFR-TKIs.7
Thus, it is of interest to explore whether the respective concomitant mutational profiles of 19Del and L858R may explain their different sensitivities to EGFR-TKIs. In this study, we obtained individual patient data from two cohorts, the Guangzhou cohort (G1) and the public Database cohort (G2), with NGS on TKI naïve NSCLC patients, to compare the prevalence of the concomitant mutations and analyze their impact on efficacy.
Patients and Methods
Patient Selection of the Guangzhou Cohort (G1)
From May 2018 to July 2019, we consecutively collected patients with NSCLC tumors at the First Affiliated Hospital of Guangzhou Medical University. All procedures performed were in accordance with the ethical standards of the Helsinki Declaration and applicable regulatory requirements. Written informed consent was obtained from all patients to permit genetic analysis of biological samples. Inclusion criteria were: 1) single primary adenocarcinoma; 2) all resected-tissues and lymph nodes were proven by final pathology with paraffin blocks; 3) enough resected-tissue for mutational analyses; and 4) patients with EGFR sensitive mutation (19Del or L858R). Patients were excluded for: 1) multiple lung cancer, 2) non-invasive cancer (eg, adenocarcinoma in situ, minimally invasive adenocarcinoma) or non-adenocarcinoma, 3) preoperative neoadjuvant therapy, 4) local or distant metastasis, 5) patients without sensitive EGFR mutation, and 6) patients with both 19Del and L858R mutations.
Genomic driver gene mutation testing was performed in all included patients and only in resected tumor tissues with NGS. Gene alterations such as EGFR, HER2, MET, KRAS, BRAF, ALK, ROS1, and RET were tested by a targeted next-generation sequencing method, which was described in other studies.8,9 Briefly, the assay utilizes a NGS-based target enrichment panel that covers 283 genes and 300k genomic regions. The chips were loaded on a MGISEQ-500 platform and sequenced at a mean depth of over 500x, which achieved genome coverage greater than 99%.
Database Search and Collection Strategy of Database Cohort (G2)
Online electronic databases (PubMed, Medline, Web of Science, and Google Scholar) as well as abstract libraries of the American Society of Clinical Oncology (ASCO), European Society of Medical Oncology (ESMO), and World Conference on Lung Cancer (WCLC) were searched for potentially eligible studies using a combination of the following terms before August 1, 2019: lung cancer, next generation sequencing/NGS/panel, and EGFR mutation. No restriction on publication year or language was set. Reference lists of relevant articles were reviewed for supplementation. Included studies met the following criteria: 1) provided multigene profiling by NGS based on tissue or plasma samples; 2) reported the genetic profiles of both 19Del and L858R; and 3) only included TKI-naïve patients. One of the included studies from G2 is from our center,8 but the study period did not overlap with G1, avoiding patient duplication.
Data Extraction of Database Cohort (G2)
Data were extracted independently by two investigators (H.R. Liang and C.C. L), and conflicts were adjudicated by a superior investigator (W.H. Liang). For the selected studies, information on all available variables was extracted and entered into a Microsoft Excel database. Mutation status of each gene as well as EGFR-TKI response information (according to RECIST 1.1) were extracted directly from the landscape figure or table. These data were equivalent to individual patient records and, thus, pooled with our data set. Any disagreement was resolved via discussion among the authors.
Statistical Analysis
Student’s t-test was used to compare the mean of continuous variables, Chi-square test was used to compare the dichotomous variables (Fisher’s exact test was used when expected frequency was less than 5). Odds ratio of concomitant mutations were calculated, and forest plots were conducted by R software (R version 3.6.1). Logistic regression was used to measure the impact of different covariates on binary outcomes. All analyses were performed using SPSS 25.0 (Chicago, IL). Statistical significance was taken as two-sided P<0.05.
Results
Cohort Features
In the G1 cohort, 403 patients received NGS and were included in this analysis, 172 (42.68%) carried 19Del and 231 (57.32%) were L858R mutation patients, the demographic characteristics of G1 are summarized in Table 1. All patients were adenocarcinoma without any additional therapy excepted by surgery or biopsy. The average age was 59.97 years old in G1 and 75.19% patients were female. Most patients were stage I to III (73.45%) according to the 8th AJCC, only 107 (26.55%) patients were diagnosed with metastatic adenocarcinoma.
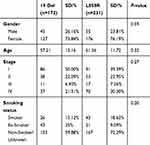 |
Table 1 Baseline Demographic Features of G1 Cohort |
In the G2 cohort, 18 eligible studies,7,8,10-25 including one publication from our team,8 were involved in this study (Table 2). A total of 803 patients with EGFR 19Del or L858R mutation were included in this study. Four hundred and fifteen patients had 19Del and 388 had L858R mutation. Five hundred and seventy-six patients were tested using tissue samples, 131 were based on plasma and 96 patients used both tissue and plasma samples. We included only TKI-naïve patients in all studies to diminish the co-founding effect after TKI resistance. As different panels were used by different studies, we analyzed T790M, TP53, and the loci reported by most studies (half of all included).
 |
Table 2 The Characteristics of Included Studies |
Mutational Profiles of EGFR 19Del and L858R of G1
The mutational landscapes of G1 are illustrated in Figure 1. The prevalence of total concomitant mutations is similar in both 19Del and L858R mutation (19Del 32.48% vs L858R 30.45%; P=0.68). The distribution of other concomitant mutations between two sensitive mutations were also similar.
 |
Figure 1 Genetic mutations landscape identified by targeted next-generation sequencing of G1: Alteration map of 403 patients with NSCLC harboring EGFR sensitive mutation. |
Mutational Profiles of EGFR 19Del and L858R of G2
The integrated mutational landscapes of G2 and percentages of EGFR 19Del and L858R are illustrated in Figure 2. We found similar prevalence of total concomitant mutations (19Del 74.9% vs L858R 73.2%; P=0.65) in 19Del and L858R mutations. The majority of other concomitant mutations were also distributed similarly between two sub-types, except for CDKN2A (19Del 12.4% vs L858R 5.5%; P=0.01), CTNNB1 (19Del 13.2% vs L858R 2.0%; P<0.001), MET (19Del 3.3% vs L858R 8.0%; P=0.03), and HRAS (19Del 7.1% vs L858R 0.0%; P=0.02). We further divided all concomitant mutations according to signaling pathway distribution and calculated their odds ratios between 19Del and L858R (Figure 3). Only HGF/c-Met pathway same more related to L858R mutation.
 |
Figure 3 Forest map of concomitant mutation in NSCLC harboring EGFR sensitive mutations according to signaling pathway. Note: Bold and italic formatting indicates P-value with significant difference. |
Factors Associated with Primary Response to EGFR-TKI of G2
One hundred and thirty-four patients in the G2 cohort reported EGFR-TKI response information (Figure 4). Compared with patients who had an objective response to TKI, those non-response or progression patients had more concomitant mutations, both in 19Del (53.8% vs 83.3%; P=0.021) and L858R (51.4% vs 77.8%; P=0.029). According to multivariate logistic regression, total concomitant mutations (Objective response: at least one 17.65% vs none 56.63%; OR=0.27; 95% CI=0.12–0.64; P=0.03), EGFR mutations (Objective response: 19Del 74.3% vs L858R 57.8%, OR=2.21; 95% CI=1.02–4.78; P=0.04), and T790M (Objective response: T790M+ 43.75% vs T790M- 73.53%, OR=0.244; 95% CI=0.100–0.590; P=0.02) significantly affected the TKI response.
Interestingly, in patients without any concomitant mutation, 19Del consistently showed better treatment outcomes (Objective response: 19Del 88.9% vs L858R 75.0%). However, the response rates in both mutation types decreased significantly when contaminated with at least one other mutation (Objective response: 19Del 65.1% vs L858R 47.5%) (Figure 5).
 |
Figure 5 The effect on response to EGFR-TKI of concomitant mutations in 19Del and L858R NSCLC patients of G2. |
Discussion
This study involved two cohorts based on NGS individual patient data and comprehensively compared the concomitant mutational profiles of EGFR 19Del and L858R in TKI naïve patients. Similar to recent studies, we observed that concomitant mutations were widespread both in 19Del and L858R mutated NSCLC patients from two cohorts, which indicated EGFR-mutant NSCLC is not a single oncogene-driven disease.8,14 We also found concomitant mutation was associated with poorer treatment outcomes both in 19Del and L858R; patients with L858R showed significantly less response to TKIs than those with 19Del regardless of the concomitant mutation status. However, we failed to conclude that the distribution of concomitant mutations was different in 19Del and L858R mutations.
Previously, it has been believed that both EGFR 19Del and L858R are sensitive to EGFR-TKIs. However, a pooled analysis of Lux-Lung 3 and Lux-Lung 6 suggested that 19Del patients might benefit more from the second-generation EGFR-TKI Afatinib than L858R.26 We also performed a meta-analysis showing a similar trend observed among first-generation TKIs, erlotinib, and gefitinib.3 A recent study found L858R had a significantly higher incidence of concomitant mutation than 19Del, and indicated the inferior treatment outcome associated with L858R could be attributed to the higher incidence of concomitant mutations.7 However, according to the present large-scale individual data in both G1 and G2, we found similar prevalence of total concomitant mutations and the majority of mutational spectrum between 19Del and L858R are the same. When dividing these concomitant mutations according to signaling pathway, no difference was found between 19Del and L858R. Furthermore, analysis of TKI treatment related outcomes suggested that both 19Del and L858R had generally similar concomitant mutation patterns, and fewer L858R patients showed an objective response to TKIs than 19Del patients, regardless of the concomitant mutation status. Thus, we claimed that the efficacy discrepancy between 19Del and L858R might not be attributed to their respective concomitant mutation status.
There are several well-known resistance markers, somatic T790M, TP53, KRAS, and so forth, which indicated a lower response rate and shorter PFS in EGFR mutant patients using TKIs.19,27 Among these alterations, somatic T790M has attracted the most attention because it is the main reason for the acquired resistance to first-generation TKIs. Our previous research showed that the presence of somatic T790m at baseline is higher in L858R than in 19Del.6 The absolute concomitant rate is 22% (L858R) and 14% (19Del), respectively, the majority of biopsy is detected by polymerase chain reaction (PCR). However, in the G2 cohort from the present study, according to pooled individual data based on NGS, concomitant somatic T790M were similar in 19Del (73.4%) and L858R (72.4%) mutations. Furthermore, the total detection rate of T790M is significantly higher than the former report and G1.
Somatic T790M variant is an indication of primary TKI resistance, the ability to identify low VAF (Variant Allele Fraction) at this locus is important in determining drug administration.28 NGS has been rapidly adopted in molecular diagnosis, and the capability to detect specific low-level mutations is important.29 Gene mutations may be detected at less than 5% VAF when performing deep sequencing of targeted NGS panels. A study showed an in-house developed cancer hotspot NGS assay at read depth of 2672X can detect T790M variant at near 2% VAF, but the real time PCR assay failed to show a positive result in the same patient,30 which indicated the NGS might had better capacity to recognize low VAF mutation. Using NGS, the current study found more somatic concomitant T790M mutations than a formal study at the baseline of EGFR mutant NSCLC patients.6 The present co-existing rate of somatic T790M in 19Del and L858R is also different compared with the former study,6 we believe the reason for the inconsistency may also be the detection method. With the use of NGS, more somatic T790M mutations of low frequency in 19Del than L858R were detected. Different concomitant mutation associated mechanisms affect the treatment response in these two sensitive mutations. The specific underlying mechanism for primary resistance and relative countermeasures in L858R and 19Del mutant patients warrants further investigation.
The total concomitant mutation rate was similar in EGFR 19Del and L858R according to G1 and G2 cohorts, however, the spectrum of concomitant mutation was different in the two sensitive mutations or different cohorts. The total concomitant mutation rate was 35% in G1 and more than 70% in G2. One of the reasons for this finding may be different screening panels and depth between G1 and G2. Another reason would be that the two sensitive mutations do not share the same concomitant mutation, but under the same screening condition, the total rate of concomitant mutation is the same. This assumption can be supported by different original studies from G2, their respective concomitant mutation spectrums were different, but the total rate was the same between 19Del and L858R. This evidence also confirmed that 19Del and L858R owned similar total numbers and patterns of concomitant mutations, which might not explain the different sensitivity to EGFR-TKI of two sub-types.
There is a concern that treatment strategies might affect the objective response, which is one of the main evaluated indices in this study. However, not all studies offered drug information. Besides, some studies included more than one EGFR TKIs, without labeled individual information of each patient. It is difficult to analyze the influence of TKI on therapeutic outcomes. We have published a Bayesian network meta-analysis, comparing every strategy that used in first-line therapy for EGFR-mutated NSCLC patients based on all published RCTs and conference abstracts.4 We found that osimertinib and gefitinib plus pemetrexed based chemotherapy were associated with the best progression free survival and overall survival benefits for patients with advanced EGFR-mutated NSCLC, compared with other first line treatments. The treatments resulting in the best progression free survival for patients with the exon 19 deletion and L858R mutations were osimertinib and gefitinib plus pemetrexed based chemotherapy, respectively, which strongly supports that the two sensitive mutations have different responses to EGFR-TKI.
There are some limitations to our study. Firstly, some heterogeneity and missing data (presented as unknown) existed in the sequencing platform from G2, which is because of not all NGS based screening panels in G2 covers the same hotspot. Furthermore, the sequencing depths are different among the included studies, which could lead to a potential bias. Secondly, the absolute number of gene mutation frequencies might affect the result, however, from the limit information of original studies that were included in our study, mutation frequencies could not be got. We only get the information of whether a patient had a mutation, thus the false discovery rates (FDRs) could not be calculated. Further large-scale study is needed to demonstrate the association between the number of samples and the mutation frequencies. Thirdly, because of the raw data limitation (Supplementary Appendix), we used objective response to evaluate the therapeutic effect rather than PFS or OS, which may not reflect the real survival situation, and G1 did not offer any therapeutic or follow-up information. Moreover, not all patients in G2 had response data, and, therefore, may not reflect the overall information for the G2 cohort. Finally, this study lacked the abundance of each mutation, thus quantitative analysis is not available.
Conclusion
Our study demonstrated that concomitant mutations were widespread both in 19Del and L858R mutated NSCLC patients and the prevalence of total concomitant mutations is similar between 19Del and L858R. Patients with L858R showed significantly less response than those with 19Del, regardless of the concomitant mutation status, indicating that some intrinsic factors, rather than concomitant mutations, might underly the varied sensitivity of 19Del and L858R to EGFR-TKIs.
Data Sharing Statement
For original deidentified individual patient data please contact [email protected]. Data will be made available for a period of 5 years after the publication date.
Ethics Approval and Consent to Participate
The study protocol has been approved by the Research Ethics Committee of the First Affiliated Hospital of Guangzhou Medical university (S-201525).
Acknowledgments
The authors thank the patients and their families who contributed to this study. We thank Ms. Linsey Hamblin for language editing. The authors would like to thank all the investigators for their contributions to this trial. Above all, we are grateful to all patients who participated in this study.
Author Contributions
All authors were involved in conception and design, provision of study materials, collection and assembly of data, data analysis and interpretation, manuscript writing, and approval. All authors agree to take responsibility and be accountable for the contents of the article. Wenhua Liang and Jianxing He provided the administrate support.
Disclosure
The authors report no conflicts of interest for this work.
References
1. da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69.
2. Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447–e459. doi:10.1016/S1470-2045(15)00246-6
3. He Q, Xin P, Zhang M, et al. The impact of epidermal growth factor receptor mutations on the prognosis of resected non-small cell lung cancer: a meta-analysis of literatures. Transl Lung Cancer Res. 2019;8(2):124–134. doi:10.21037/tlcr.2019.03.14
4. Zhao Y, Liu J, Cai X, et al. Efficacy and safety of first line treatments for patients with advanced epidermal growth factor receptor mutated, non-small cell lung cancer: systematic review and network meta-analysis. BMJ. 2019;367:l5460. doi:10.1136/bmj.l5460
5. Singh M, Jadhav HR. Targeting non-small cell lung cancer with small-molecule EGFR tyrosine kinase inhibitors. Drug Discov Today. 2018;23(3):745–753. doi:10.1016/j.drudis.2017.10.004
6. Liang H, Pan Z, Wang W, et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: a literature-based pooled analysis. J Thorac Dis. 2018;10(4):2311–2320. doi:10.21037/jtd.2018.03.150
7. Hong S, Gao F, Fu S, et al. Concomitant genetic alterations with response to treatment and epidermal growth factor receptor tyrosine kinase inhibitors in patients with EGFR-mutant advanced non-small cell lung cancer. JAMA Oncol. 2018;4(5):739–742. doi:10.1001/jamaoncol.2018.0049
8. Liu L, Liu J, Shao D, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci. 2017;108(12):2487–2494. doi:10.1111/cas.13410
9. Shao D, Lin Y, Liu J, et al. A targeted next-generation sequencing method for identifying clinically relevant mutation profiles in lung adenocarcinoma. Sci Rep. 2016;6:22338. doi:10.1038/srep22338
10. Yu JY, Yu SF, Wang SH, et al. Clinical outcomes of EGFR-TKI treatment and genetic heterogeneity in lung adenocarcinoma patients with EGFR mutations on exons 19 and 21. Chin J Cancer. 2016;35:30. doi:10.1186/s40880-016-0086-2
11. Sato S, Nagahashi M, Koike T, et al. Impact of concurrent genomic alterations detected by comprehensive genomic sequencing on clinical outcomes in east-asian patients with EGFR-mutated lung adenocarcinoma. Sci Rep. 2018;8(1):1005. doi:10.1038/s41598-017-18560-y
12. Li W, Qiu T, Guo L, et al. Primary and acquired EGFR T790M-mutant NSCLC patients identified by routine mutation testing show different characteristics but may both respond to osimertinib treatment. Cancer Lett. 2018;423:9–15. doi:10.1016/j.canlet.2018.03.005
13. Ku BM, Heo MH, Kim JH, et al. Molecular screening of small biopsy samples using next-generation sequencing in korean patients with advanced non-small cell lung cancer: Korean Lung Cancer Consortium (KLCC-13-01). J Pathol Transl Med. 2018;52(3):148–156. doi:10.4132/jptm.2018.03.12
14. Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49(12):1693–1704. doi:10.1038/ng.3990
15. Jakobsen JN, Santoni-Rugiu E, Grauslund M, Melchior L, Sorensen JB. Concomitant driver mutations in advanced EGFR-mutated non-small-cell lung cancer and their impact on erlotinib treatment. Oncotarget. 2018;9(40):26195–26208. doi:10.18632/oncotarget.25490
16. Cao L, Long L, Li M, et al. The utilization of next-generation sequencing to detect somatic mutations and predict clinical prognosis of Chinese non-small cell lung cancer patients. Onco Targets Ther. 2018;11:2637–2646. doi:10.2147/OTT.S155995
17. Jin Y, Shi X, Zhao J, et al. Mechanisms of primary resistance to EGFR targeted therapy in advanced lung adenocarcinomas. Lung Cancer. 2018;124:110–116. doi:10.1016/j.lungcan.2018.07.039
18. Liao L, Ji X, Ge M, et al. Characterization of genetic alterations in brain metastases from non-small cell lung cancer. FEBS Open Biol. 2018;8(9):1544–1552. doi:10.1002/2211-5463.12501
19. VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: correlation with clinical outcomes. Lung Cancer. 2017;106:17–21. doi:10.1016/j.lungcan.2017.01.011
20. Yu HA, Suzawa K, Jordan E, et al. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res. 2018;24(13):3108–3118. doi:10.1158/1078-0432.CCR-17-2961
21. Xu Y, Tong X, Yan J, Wu X, Shao YW, Fan Y. Short-term responders of non-small cell lung cancer patients to EGFR tyrosine kinase inhibitors display high prevalence of TP53 mutations and primary resistance mechanisms. Transl Oncol. 2018;11(6):1364–1369. doi:10.1016/j.tranon.2018.08.010
22. Tian P, Wang Y, Wang W, et al. High-throughput sequencing reveals distinct genetic features and clinical implications of NSCLC with de novo and acquired EGFR T790M mutation. Lung Cancer. 2018;124:205–210. doi:10.1016/j.lungcan.2018.08.014
23. Martin Martorell P, Huerta M, Compan Quilis A, et al. Coexistence of EGFR, KRAS, BRAF, and PIK3CA mutations and ALK rearrangement in a comprehensive cohort of 326 consecutive Spanish nonsquamous NSCLC patients. Clin Lung Cancer. 2017;18(6):e395–e402. doi:10.1016/j.cllc.2017.04.006
24. Hou H, Yang X, Zhang J, et al. Discovery of targetable genetic alterations in advanced non-small cell lung cancer using a next-generation sequencing-based circulating tumor DNA assay. Sci Rep. 2017;7(1):14605. doi:10.1038/s41598-017-14962-0
25. Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9(1):216. doi:10.1038/s41467-017-02584-z
26. Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, Phase 3 trials. Lancet Oncol. 2015;16(2):141–151. doi:10.1016/S1470-2045(14)71173-8
27. Labbe C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer. 2017;111:23–29. doi:10.1016/j.lungcan.2017.06.014
28. Reynolds JP, Zhou Y, Jakubowski MA, et al. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer Cytopathol. 2017;125(3):178–187. doi:10.1002/cncy.21812
29. Doxtader EE, Cheng YW, Zhang Y. Molecular Testing of non-small cell lung carcinoma diagnosed by endobronchial ultrasound-guided transbronchial fine-needle aspiration: the Cleveland clinic experience. Arch Pathol Lab Med. 2019;143(6):670–676. doi:10.5858/arpa.2017-0184-RA
30. Cheng YW, Stefaniuk C, Jakubowski MA. Real-time PCR and targeted next-generation sequencing in the detection of low level EGFR mutations: instructive case analyses. Respir Med Case Rep. 2019;28:100901.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


