Back to Journals » OncoTargets and Therapy » Volume 13
Comprehensive Transcriptomic Analysis and Experimental Validation Identify lncRNA HOXA-AS2/miR-184/COL6A2 as the Critical ceRNA Regulation Involved in Low-Grade Glioma Recurrence
Authors Chen PY, Li XD, Ma WN, Li H , Li MM, Yang XY, Li SY
Received 14 January 2020
Accepted for publication 16 April 2020
Published 3 June 2020 Volume 2020:13 Pages 4999—5016
DOI https://doi.org/10.2147/OTT.S245896
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Peng-Yu Chen, Xiao-Dong Li, Wei-Ning Ma, Han Li, Miao-Miao Li, Xin-Yu Yang, Shao-Yi Li
Department of Neurosurgery, Shengjing Hospital Affiliated to China Medical University, Shenyang, People’s Republic of China
Correspondence: Shao-Yi Li
Department of Neurosurgery, Shengjing Hospital Affiliated to China Medical University, 39 Hua Xiang Road, Shenyang 110022, People’s Republic of China
Email [email protected]
Purpose: The recurrence and metastasis of glioma are closely related to complex regulatory networks among protein-coding genes, lncRNAs and microRNAs. The aim of this study was to investigate core genes, lncRNAs, miRNAs and critical ceRNA regulatory mechanisms, which are involved in lower-grade glioma (LGG) recurrence.
Materials and Methods: We employed multiple datasets from Chinese Glioma Genome Atlas (CGGA) database and The Cancer Genome Atlas (TCGA) to perform comprehensive transcriptomic analysis. Further in vitro experiments including cell proliferation assay, luciferase reporter assay, and Western blot were performed to validate our results.
Results: Recurrent LGG and glioblastoma (GBM) showed different transcriptome characteristics with less overlap of differentially expressed protein-coding genes (DEPs), lncRNAs (DELs) and miRNAs (DEMs) compared with primary samples. There were no overlapping gene in ontology (GO) terms related to GBM recurrence in the TCGA and CGGA databases, but there were overlaps associated with LGG recurrence. GO analysis and protein–protein interaction (PPI) network analysis identified three core genes: TIMP1, COL1A1 and COL6A2. By hierarchical cluster analysis of them, LGGs could be clustered as Low_risk and High_risk group. The High_risk group with high expression of TIMP1, COL1A1, and COL6A2 showed worse prognosis. By coexpression networks analysis, competing endogenous RNA (ceRNA) network analysis, cell proliferation assay and luciferase reporter assay, we confirmed that lncRNA HOXA-AS2 functioned as a ceRNA for miR-184 to regulate expression of COL6A2, which induced cell proliferation of low-grade glioma.
Conclusion: In this study, we revealed a 3-hub protein-coding gene signature to improve prognostic prediction in LGG, and identified a critical ceRNA regulation involved in LGG recurrence.
Keywords: lower-grade glioma, lncRNA, miRNA, HOXA-AS2, miR-184, COL6A2
Introduction
Glioma is the most common malignant brain tumor in human adults, with a low 5-year overall survival (OS) rate (35%).1 According to the World Health Organization (WHO) grading system, gliomas are classified from grade I–IV. Grade II and III gliomas are grouped as LGGs, and GBM is regarded as grade IV. GBM, as the most invasive and aggressive glioma, shows the highest mortality despite aggressive treatment.2,3 Although LGGs have identical morphological phenotypes, the range of patient outcomes is wide.4 Because of the aggressive nature of gliomas, a residual tumor after excision is the leading cause of recurrence and disease progression, which is a major reason for the poor prognosis in glioma patients.5 Therefore, it is necessary to explore the tumor progression mechanisms and identify potential therapeutic targets in glioma.
Recently, many studies have revealed the distinctions between GBM and LGG by integrating and analyzing multilevel molecular data, including mutation, methylation, and gene expressions. Previous studies have reported that somatic intronic microsatellite loci can differentiate between GBM and LGG.6–8 Zhang et al revealed distinctions between LGG and GBM by establishing a differential mRNA-lncRNA network using mRNAs and lncRNAs differentially expressed between GBM and LGG.9 Therefore, a separate exploration of the mechanisms of LGG and GBM progression will be more appropriate.
Long noncoding RNAs (lncRNAs) and microRNAs are classic noncoding RNAs and have been reported to regulate the development of glioma.10–12 For example, Li et al found that lncRNA SNHG1 could upregulate FOXP2 and KDM5B by regulating the expression of miR-154-5p and miR-376b-3p, which contributed to the malignant behavior of glioma cells.13 Wu et al reported that lncRNA lnc-TALC increased the expression of O6-methylguanine-DNA methyltransferase by regulating the c-Met pathway by competitively binding with miR-20b-3p, and this biological process is required for temozolomide resistance and GBM recurrence.14 To date, a number of public open-access databases have helped uncover more complex protein-coding gene-lncRNA-microRNA regulatory networks. By informatics analysis using TCGA and GEO data, reciprocal regulatory networks among protein-coding genes, lncRNAs and microRNAs have been revealed to participate in cancer progression.15,16 Lou et al described a miRNA-mRNA regulatory network by using DEPs between GBM samples and normal samples that was involved in tumorigenesis and the development of GBM.17 Deng et al also revealed hub DEPs by constructing PPI networks of DEPs between primary and recurrent LGG tumor tissues in the TCGA database.18 Besides, lncRNAs, microRNAs and mRNAs could form complex ceRNA network to promote GBM growth and progression.19,20 Long et al revealed a complex lncRNA mediated ceRNA network associated with GBM progression by analyzing TCGA and GEO data.21 However, the complex regulatory networks and ceRNA networks among protein-coding genes, lncRNAs and microRNAs are still unclear in the recurrence of glioma.
In this study, DEPs, DELs and DEMs were identified, and GO analysis was performed to explore transcriptome variation in the process of LGG or GBM recurrence by cross-database analysis using CGGA and TCGA data. By intersecting the results from analyses of the CGGA and TCGA databases and constructing a PPI network, three hub genes (TIMP1, COL1A1 and COL6A2) were revealed to participate in LGG recurrence. Using the three genes, primary LGGs were clustered into two groups (High_risk and Low_risk), and the High_risk group showed poorer prognosis. Finally, protein-coding gene-lncRNA-microRNA regulatory network analysis and ceRNA network analysis were conducted to reveal hub protein-coding genes, lncRNAs and microRNAs associated with LGG recurrence.
Materials and Methods
Data Collection
We obtained transcriptomic data of GBM and LGG samples from the TCGA (https://portal.gdc.cancer.gov/) and CGGA (http://www.cgga.org.cn/) databases. The details are illustrated in Table 1. We normalized raw counts data from TCGA database by TMM using GDCRNATools package in R software. In the TCGA database, 14 LGG patients had both recurrent and primary samples, and RNA-Seq and miRNA-Seq data were available for a total of 32 samples. In addition, 6 GBM patients had both recurrent and primary samples, and RNA-Seq data were available for a total of 13 samples. RNA-Seq data of grade III glioma cell lines (SW1783, SW1088, HS683 and GOS3) were downloaded from CCLE database (https://portals.broadinstitute.org/ccle). RNA levels of COL6A2, HOXA-AS2 and miR-184 were extracted and log2-transformed.
 |
Table 1 Datasets Used in This Study |
Differentially Expressed Protein-Coding Genes, lncRNAs and microRNAs
Differentially expressed RNAs (including DEPs, DELs, and DEMs) were analyzed between primary and recurrent samples of LGG or GBM by using the “limma” package in R software. Because some cases had both primary and recurrent samples in the TCGA database, we identified DEPs, DELs, and DEMs between primary and recurrent tumors in these patients. The DEPs, DELs, and DEMs that met the criteria of P value < 0.05 and |log2 (fold change)| > 1 were selected for further analysis. Volcano plot and heatmap were plotted to show DEPs, DELs, and DEMs by using the “ggplot2” and “pheatmap” package in R software. Venn diagrams were drawn to find overlap using the Bioinformatics & Evolutionary Genomics website (http://bioinformatics.psb.ugent.be/webtools/Venn/).
GO Enrichment Analysis
GO analysis was performed using DEPs with the “clusterProfiler” package in R software. Dotplots were generated to display the most significant GO terms involved in LGG or GBM recurrence (P<0.001). Using the “enrichplot” package in R, circular diagrams were plotted to show the 5 most related GO terms (P<0.001).
Protein–Protein Interaction (PPI) Network Analysis
To better understand the interactive relationships among these enriched genes in GO terms, a PPI network was established by using the STRING database.22 According to the degree calculated using Cytoscape software (version 3.6.1), the top 10 hub genes were identified from the PPI network.
Protein Coding Gene-lncRNA-miRNA Coexpression Network Analysis
The R package “corrplot” was utilized to calculate the correlation coefficients among prognostic DEPs, DELs and DEMs. Then, node pairs with a correlation coefficient > 0.1 were selected for further analysis. A coexpression network containing protein-coding genes, lncRNAs and miRNAs was constructed, and further analysis was conducted to identify a subnetwork by using Cytoscape software (https://cytoscape.org/).
Construction of ceRNA Network
Interactions of lncRNAs with microRNAs and mRNAs were predicted by using the miRcode database. Finally, the lncRNA-miRNA-mRNA ceRNA network analysis was conducted by using Cytoscape software.
Cell Lines and Cell Culture
Two grade III glioma cell line HS683 and SW1088 were purchased from American Type Tissue Culture Collection. The cell lines were maintained in DMEM supplemented with 10% fetal bovine serum at 37°C under a humidified atmosphere of 5% CO2.
Cell Transfection
miR-184 mimics, miR-184 inhibitor, or negative control were transfected by Lipofectamine3000 (Invitrogen Inc., Carlsbad, CA, USA), which were synthesized by GenePharma (Shanghai, China). The sequences were listed as follows:23 miR-184 mimic, 5′-UGGACGGAGAACUGAUAAGGGUCCUUAUCAGUUCUCCGUCCAUU-3′; the negative control for miR-184 mimic, 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′; miR-184 inhibitor, 5′-ACCCUUAUCAGUUCUCCGUCCA-3, negative control for miR-184 inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′. siRNAs targeting COL6A2 and HOXA-AS2 were synthesized by GenePharma, and eukaryotic vectors expressing HOXA-AS2 were obtained from Genechem (Shanghai, China). The siRNAs and vectors were used to transfected to glioma cells by Lipofectamine3000. The siRNA sequences were listed as follows:24,25 siHOXA-AS2, si1: 5′-AAACCUUGUAGAUAGCUUGAGCUGG-3′, si2: 5′-CAAGCUUGACAAGUUCAGCUCAA-3′; siCOL6A2: si1: 5′-GGGCCUCCUUCAUCAAGAATT-3′; si2: 5′-GCAGGCCUGGAUUCAGCUATT-3′.
Western Blotting
Total protein was gathered by RIPA Lysis Buffer. The protein samples were separated by 8% SDS-polyacrylamide gels and transferred to PVDF membranes. After blocking with 5% BSA for 2 h, the membranes were incubated overnight with primary antibodies including mouse anti-GAPDH monoclonal antibody (Catalog number: 60,004-1-Ig, proteintech, Wuhan, China) and rabbit anti-COL6A2 monoclonal antibody (Catalog number: ab180855, Abcam, Cambridge, UK) at 4°C. After being washed in TBST for three times, membranes were incubated with HRP-conjugated anti-rabbit (Catalog number: SA00001-2, proteintech, Wuhan, China) or HRP-conjugated anti-mouse (Catalog number: SA00001-1, proteintech, Wuhan, China) secondary antibodies at a 1:5000 dilution at room temperature for 1h. Then, the binding secondary antibody was detected and recorded using the enhanced ECL Western blotting detection system.
Quantitative Real-Time PCR (qPCR)
The total RNA of glioma cells were extracted with Trizol reagent (Invitrogen, USA) according to the manufacturer’s instruction. Total RNA (1 μg) was used for cDNA synthesis using Reverse Transcription kit (TOYOBO, Osaka, Japan). The primers were: GAPDH, forward: 5′-AACGGATTTGGTCGTATTG-3′, reverse: 5′-GGAAGATGGTGATGGGATT-3′; HOXA-AS2, forward: 5′-GTTCAGCTCAAGTTGAACATA-3′, reverse: 5′-AAACCTTGTAGATAGCTTGAG-3′; COL6A2, forward: 5′- TGCTCCGTGCTCCTGCTCTG-3′, reverse: 5′- ATGGTGACGCTCTCCGAGGTG −3′. The qRT-PCR assays were carried out using SYBR Select Master Mix (Applied Biosystems, Foster City, CA, USA) on ABI 7500 Real-Time PCR system (Applied Biosystems). The PCR conditions were as follows: step 1: 95°C for 5 min; step 2: 40 cycles consisting of 95°C for 10 s, 60°C for 30 s, and 72°C for30 s; step 3: 72°C for 5 min. Relative RNA level of each gene was calculated as 2−ΔΔCt. Each sample was performed in triplicate.
Cell Counting Kit-8 (CCK-8) Proliferation Assay
The transfected HS683 and SW1088 cells were seeded into 96-well plates at a density of 5000 cells/well. After incubated at 37 °C for 2 h, the absorbance of each well was measured at a wavelength of 450 nm.
Luciferase Assay
Glioma cells were seeded in 24-well plate, and allowed to grow for 24 h. To test the binding specificity of miR-184 in COL6A2, we constructed the wild type reporter vector (COL6A2-3′UTR_WT: 5′-CCCGUCCA-3′) and the corresponding mutant (COL6A2-3′UTR_MUT: 5′-CUUAGUUA-3′). To test the binding specificity of miR-184 in HOXA-AS2, we constructed the wild type reporter vector (HOXA-AS2_WT: 5′-UCCGUCC-3′) and the corresponding mutant (HOXA-AS2_MUT: 5′- GUUAGUU −3′). Then, 100ng pGL3-COL6A2-3′UTR-luciferase plasmid (or MUT) and pGL3-HOXA-AS2-luciferase plasmid (or MUT) was transfected into glioma cells using the Lipofectamine3000 reagent. Subsequently, the cells were transfected with miR-184 mimics, miR-184 inhibitor, or negative control. The luciferase activities in cell lysates were measured at 48 h after transfection by the Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according the manufacturer’s protocol. Three independent experiments were performed.
Immunohistochemistry (IHC) and Evaluation
Four primary LGG tissues and four recurrent LGG tissues were obtained from Department of Neurosurgery of Shengjing Hospital affiliated to China Medical University and histologically diagnosed with grade II or III glioma in 2017. These experiments were approved by Human Ethics Committee of China Medical University and all patients signed the informed consent. Tissues were fixed by formalin, embedded by paraffin, and finally cut into 4μm sections. The sections were deparaffinized by three 10-min washes in xylene, and then rehydrated by 100%, 95%, 85% and 75% ethanol solutions (5min/time). Antigen retrieval was performed in 0.01 M citrate buffer (pH 6.0) at 120°C for 5 min. Endogenous peroxidase was blocked by 3% H2O2 in PBS for 15 min and washed for 2×5 min in PBS. Then, non-specific binding were blocked by 10% bovine serum albumin (BSA) for 45 min. Subsequently, sections were incubated with the primary antibody against COL6A2 (1:100) at 4 °C for overnight. After washed using PBS, the sections were incubated with HRP-conjugated goat anti-rabbit secondary antibody (1:1000) for 45 min. Finally, the sections were stained using 3,3-diaminobenzidine (DAB) for 5 min, counterstained with hematoxylin, mounted and captured representative images using an optical microscope (Olympus, Tokyo, Japan). As for the IHC scoring, protein expression was calculated by multiply the staining intensity scores and the positive cell rate scores. Staining intensity scores: 0, negative; 1, weak; 2, intense; and 3, strongly intense. Positive cell rate scores: 0, 0%; 1, 1–10%; 2, 11–25%; 3, 26–50%; 4, 51–75%; and 5, 76–100%.
Statistical Analysis
The main statistical analysis in this study is stated above. Using a 3-gene expression value, hierarchical cluster analyses were conducted with the “pheatmap” package in R. To explore the prognostic significance of DEMs, ROC curves were plotted, and optimal cutoff values were set to divide all LGG samples as low and high expression groups. The survival curves were plotted using GraphPad Prism 6.0 (La Jolla, CA, United States). When we performed differential expression analysis, only protein-coding genes, lncRNAs or miRNAs with |log2FC| > 1 and P<0.05 were considered statistically significant. Univariate Cox regression analyses were conducted using the “survival” package in R, The Student’s t-test, or the chi-squared test, was used to evaluate significant differences between groups of data. All data are represented as the means ± SD. Student’s t-test using GraphPad Prism 6.0 was used to determine significant differences between two groups. All data are represented as the means and 95% CI. A P value < 0.05 was considered statistically significant.
Results
Recurrence-Related RNA Expression Patterns Vary in GBM and LGG
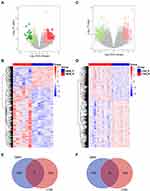
To explore whether GBM and LGG recurrences contribute to distinct mechanisms, we compared the differentially expressed RNAs between recurrent and primary samples of GBM or LGG, including protein-coding genes, pseudogenes, and long noncoding and short noncoding RNAs. In the CGGA database, there were 183 upregulated and 38 downregulated RNAs in recurrent GBM samples (|log2 (fold change)| > 1, P<0.05, Figure 1A and B, Table S1). A total of 45 upregulated and 7 downregulated RNAs were observed in recurrent LGG samples (|log2 (fold change)| > 1, P<0.05, Figure 1C and D, Table S2). Venn diagrams showed that there were only 6 overlapping RNAs among the upregulated RNAs and 1 overlapping RNA among the downregulated RNAs in recurrent LGG and GBM (Figure 1E and F). These results revealed distinct recurrence mechanisms between GBM and LGG at the transcriptional level.
To validate the above results, the same analyses were conducted in the TCGA database. There were 1332 upregulated and 466 downregulated RNAs in recurrent GBM samples (|log2 (fold change)| > 1, P<0.05, Figure 2A and B, Table S3). A total of 401 upregulated and 616 downregulated RNAs were observed in recurrent LGG samples (|log2 (fold change)| > 1, P<0.05, Figure 2C and D, Table S4). Venn diagrams showed that there were only 6 overlapping RNAs among the upregulated RNAs and 14 overlapping RNAs among the downregulated RNAs in recurrent LGG and GBM (Figure 2E and F).
Recurrent Mechanisms of GBM and LGG are Different
GO analysis was performed using DEPs. The dotplot of the top 10 enriched GO terms is displayed. In recurrent GBM, there were no identical enriched GO terms between the TCGA and CGGA databases (Figure 3A and B). However, we observed the same enriched GO terms involved in LGG recurrence (Figure 3C and D). The results showed that LGG recurrence was positively associated with GO terms involved in the extracellular matrix (such as collagens, P<0.001).
GO and PPI Network Analysis Revealed Three Core Genes Involved in LGG Recurrence
To screen critical genes that mediate LGG recurrence, we generated a circular diagram to show the top 5 related GO terms with the enriched genes. Referencing the analysis results from the CGGA and TCGA databases, a total of 20 genes were enriched in GO terms associated with the extracellular matrix (Figure 4A and B). Using these 20 genes to construct a PPI network, the top 10 hub genes were identified (Figure 4C). Among them, three genes were also intersected in both TCGA and CGGA analysis results: TIMP1, COL1A1 and COL6A2. Expression of the three genes was increased more than double in recurrent LGGs in the CGGA (Figure 4D, all P<0.001) and TCGA databases (Figure 4E, all P<0.05).
To explore the prognostic significance of the 3 genes in primary LGGs, we performed hierarchical cluster analysis. In the TCGA project and CGGA dataset (dataset 2), all primary LGG samples were clustered into two groups: High_risk and Low_risk. The High_risk group showed poorer prognosis than the Low_risk group in the CGGA (dataset 2, HR=2.97 (2.11–5.17), P<0.001, Figure 5A) and TCGA (HR=2.69 (1.86–3.75), P<0.001, Figure 5B) databases. Another two CGGA datasets (dataset 3 and dataset 4) were used as external validation, and the same results were observed (dataset 3, HR=2.20 (1.42–4.09), P=0.001, Figure 5C; dataset 4, HR=1.87 (1.08–3.01), P=0.025, Figure 5D).
Recurrence-Related miRNAs are Distinct Between LGG and GBM
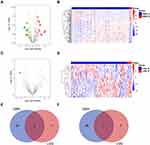
To further explore the roles of miRNAs in LGG and GBM recurrence, we used CGGA microRNA array data for the next analysis. In the CGGA dataset (dataset 6), 29 increased and 59 decreased miRNAs were observed in recurrent GBM (|log2 (fold change)| > 1, P<0.05, Figure 6A and B, Table S5). In LGG, we found 9 upregulated and 8 downregulated miRNAs in recurrent samples (|log2 (fold change)| > 1, P<0.05, Figure 6C and D, Table S6). Venn diagrams showed only 2 overlapping miRNAs among the upregulated miRNAs and 1 overlapping miRNA among the downregulated miRNAs in recurrent LGG and GBM (Figure 6E and F).
Prognostic miRNAs and lncRNAs in LGG
We further screened prognostic miRNAs in LGG patients by using TCGA miRNA-Seq data. First, we compared differentially expressed miRNAs between recurrent and primary LGG samples from 14 LGG patients. There were 23 upregulated and 20 downregulated miRNAs in recurrent LGGs (Figure 7A and B, Table S7). Among them, 8 of 23 upregulated miRNAs were significantly associated with inferior prognosis, and 11 of 20 downregulated miRNAs were positively associated with better prognosis (Figure 7C and Figure S1, P<0.01).
According to the above results on differentially expressed RNAs, we identified 35 upregulated lncRNAs and 71 downregulated lncRNAs in recurrent LGGs. By Cox univariate regression analysis, 31 of 35 upregulated lncRNAs were significantly related to worse prognosis, and 60 of 71 downregulated lncRNAs were positively associated with better prognosis (Table S8, P<0.01).
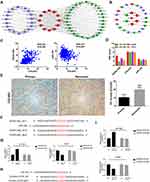
Identification and Validation of LncRNA HOXA-AS2/miR-184/COL6A2 ceRNA Regulation Associated with LGG Recurrence
The TCGA database supplies multidimensional data, which includes somatic mutations, copy number variants (CNVs), gene expression, lncRNA expression, and miRNA expression. Therefore, we could establish a protein-coding gene-lncRNA-miRNA coexpression network using these data. There were 12 protein-coding gens enriched in the GO terms involved in the extracellular matrix, 19 prognostic miRNAs and 91 prognostic lncRNAs in the TCGA database. Using the protein-coding genes and prognostic lncRNAs and miRNAs, the node pairs with correlation coefficients > 0.1 and P value < 0.05 were selected for further analysis. Accordingly, we constituted a complex co-expression network containing 12 protein coding genes, 18 miRNAs and 29 lncRNAs (Figure 8A). To better understand whether the above 29 hub lncRNAs regulated 12 protein coding genes via ceRNA regulatory mechanism, we performed ceRNA network analysis by using miRcode database. Finally, we constructed a ceRNA network including 9 protein coding genes, 6 miRNAs and 12 lncRNAs. As shown in Figure 8B, HOXA-AS2 may act as a competing endogenous RNA (ceRNA) for miR-184 to regulate the hub gene COL6A2 expression. COL6A2 expression was positively related to LncRNA HOXA-AS2 expression, and negatively related to miR-184 expression in glioma in TCGA database (Figure 8C). In CCLE database, the four grade III glioma cell lines showed different levels of expression of LncRNA HOXA-AS2, COL6A2 and miR-184 (Figure 8D). Because HS683 showed high expression of them and SW-1088 showed low levels of them, we further selected these two cell lines for the next experiments. We also detected the COL6A2 protein expression by IHC, and COL6A2 expression was significantly higher in recurrent LGG tissues (n=4) than in primary LGG tissues (n=4) (P<0.01, Figure 8E). By referencing to miRcode database, we obtained two potential binding site of miR-184 in LncRNA HOXA-AS2 mRNA (Figure 8F). In order to further validate the interaction, luciferase reporter assays were performed. The miR-184 inhibitors enhanced the luciferase activity of HOXA-AS2_WT reporter systems in SW1088, but not HOXA-AS2_MUT (Figure 8G). The miR-184 mimics inhibited the luciferase activity of HOXA-AS2_WT reporter systems in HS683, but not HOXA-AS2_MUT (Figure 8G). Furthermore, we also obtained the binding site of miR-184 in 3′UTR of COL6A2 mRNA (Figure 8H). Similarly, the miR-184 inhibitors enhanced the luciferase activity of COL6A2-3′UTR_WT reporter systems in SW1088, but not COL6A2-3′UTR_MUT (Figure 8I). The miR-184 mimics inhibited the luciferase activity of COL6A2-3′UTR_WT reporter systems in HS683, but not COL6A2-3′UTR_MUT (Figure 8I).
LncRNA HOXA-AS2/miR-184/COL6A2 ceRNA Regulation Promoted Glioma Cell Proliferation
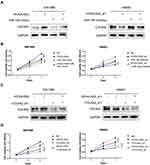
In order to explore whether LncRNA HOXA-AS2/miR-184/COL6A2 ceRNA regulation involved in glioma cell proliferation, rescue experiments were performed. We first knocked down (Figure S2A) or overexpressed HOXA-AS2 (Figure S2B), and knocked down COL6A2 (Figure S2C) in glioma cells. The more effective siRNA sequence was selected for the further experiments. HOXA-AS2 upregulation increased COL6A2 expression in SW1088 cells, which was partly reversed by miR-184 mimics (Figure 9A). HOXA-AS2 downregulation suppressed COL6A2 expression in HS683, which was restored by miR-184 inhibitors to some extent (Figure 9A). Forcing expression of HOXA-AS2 enhanced cell proliferation ability of SW1088 cells, and the effect was attenuated by miR-184 mimics (Figure 9B). In HS683 cells, HOXA-AS2 knockdown mediated cell proliferation inhibition was reversed by miR-184 inhibitors (Figure 9B). HOXA-AS2-mediated COL6A2 upregulation could be partly blocked by COL6A2 knockdown in SW1088 cells (Figure 9C). Double knockdown of HOXA-AS2 and COL12A1 more apparently suppressed expression of COL6A2 in HS683 cells than single knockdown of HOXA-AS2 and COL6A2 (Figure 9C). Cell proliferation ability of SW1088 was enhanced by HOXA-AS2, which was attenuated by COL6A2 downregulation (Figure 9D). Double knockdown of HOXA-AS2 and COL6A2 more apparently inhibited cell proliferation in HS683 than single knockdown of them (Figure 9D).
Discussion
Although glioma patients receive aggressive treatment including surgery, chemotherapy and radiotherapy, some patients suffer tumor recurrence.26,27 Therefore, exploring the recurrence mechanisms of glioma will be conducive to improving the diagnosis and treatment of glioma. An increasing number of studies have inferred the distinctions between GBM and LGG.6–9 Hence, we explored the recurrence mechanisms of GBM and LGG separately. By comparing them to primary samples, we found that recurrent samples of GBM and LGG showed distinct changes in transcriptomic characteristics due to less overlapping DEPs, DELs and DEMs. Further GO analysis using DEPs also indicated that the recurrent mechanisms of GBM were different from those of LGG. However, in GBM, we did not obtain similar results of GO analysis from the CGGA and TCGA databases, which may be due to the large heterogeneity of GBM.28 For example, the markedly enriched GO terms (MF) in recurrent GBMs were protein heterodimerization activity and structural constituent of ribosome in CGGA database and channel activity and passive transmembrane transporter activity in the TCGA database, although these GO terms have been reported to contribute to GBM progression.28–31 Glioma stem cells play critical roles in glioblastoma multiforme chemotherapy resistance and recurrence, such as CD133 cells.32 Glioma stem cells can self-renew and differentiate into other specific GBM subpopulations, which is involved in activation of Wnt/β-catenin pathway.33 Maintenance of stem cell characteristics may be involved in activation of the above pathway related GBM recurrence.33,34 Therefore, they have been held responsible for malignant relapse after primary standard therapy and the poor prognosis of recurrent GBM. Nevertheless, the enriched GO terms in recurrent LGG in the CGGA database were similar to those in the TCGA database, which were involved in the extracellular matrix (mainly including collagens). The extracellular matrix plays pivotal roles in multiple biological processes in cancers, including metastasis, immune evasion and drug resistance.35–37
Of the genes in the top 5 most relevant GO terms involved in LGG recurrence, TIMP1, COL1A1 and COL6A2 were upregulated in recurrent samples in both the CGGA and TCGA databases. In pancreatic cancer, high expression of TIMP1 can attenuate the gemcitabine therapy response and accelerate tumor growth and liver metastasis.38 COL1A1 is a member of the collagen I family and has been reported to be associated with tumor cell proliferation and invasion in many cancers, such as breast, lung and renal cancers.39 As a member of the Collagen VI family, COL6A2 is widely expressed in a variety of cancers and favors cancer progression.40 These three core genes were found to interact with each other and with other genes involved in the extracellular matrix as demonstrated using the STRING database. Furthermore, hierarchical cluster analysis using the three genes indicated that there were two clusters of LGGs, and the High_risk group with high expression of these genes showed poor prognosis in primary LGGs. These results indicated the critical roles of these genes in LGG recurrence. These three genes have been reported to be significantly associated with poor prognosis in other cancers, such as bladder cancer and colorectal cancer.41,42 Many previous studies have been designed to acquire prognostic signatures of microRNAs, pseudogenes and lncRNAs, and the studies screened the genes by prognostic analyses.43–45 However, in this study, we screened the hub genes by analyzing DEPs between recurrent and primary samples. Deng et al also screened DEPs between recurrent and primary samples using TCGA data and identified hub genes (APOA2, COL3A1, COL1A1, TYR, COL1A2, COL5A1, PAPOLB, IGF2BP1 and ANHX) by PPI analysis, but did not intersect these data with those from other databases.18 Our screened hub genes were validated in another dataset. Because some cases had both primary and recurrent lesions in the TCGA database, in order to obtain more reliable results, we analyzed DEPs, DELs and DEMs using mRNA-Seq and miRNA-Seq data from these cases. LGG patients were Chinese in CGGA and Westerners in TCGA. In this study, TIMP1, COL1A1 and COL6A2 were considered critical genes in the process of LGG recurrence and were identical in different geographic locations.
It is well known that cancer progression is orchestrated by a complex regulatory network.46,47 Recently, many studies have reported that miRNAs and lncRNAs are involved in gene expression regulation. Song et al revealed a regulatory network of CLDN4 in gastric cancer, which was mediated by miRNAs and lncRNAs, resulting in cancer cell proliferation and invasion.48 In GBM, AGAP2-AS1 epigenetically decreases TFPI2 expression by binding to EZH2 and LSD1, and this biological process promotes cancer cell proliferation and invasion.49 Therefore, exploring complex regulatory networks among protein-coding genes, lncRNAs and miRNAs will contribute to clarifying the recurrence mechanisms of cancer. In this study, we constructed a coexpression networks among 12 protein coding genes, 12 miRNAs and 29 lncRNAs. Of the lncRNAs, HOXA10-AS was upregulated in LUAD tissues and correlated with poor prognosis in lung cancer patients, and HOXA-AS2 could promote cancer progression via complicated regulatory mechanisms, including induction of EMT, direct inhibition of gene expression and methylation.50,51 For the screened miRNAs, miR-139-3p was identified as a key miRNA related to hepatocellular carcinoma, and miR-326 could inhibit cell proliferation, migration, and invasion by targeting oncogenic CCND1.52,53 In skin fibroblasts, lncRNA AC067945.2 overexpression inhibited the expression of COL1A1, COL1A2, COL3A1.54 Cao et al demonstrated that lncRNA PVT1 promotes atrial fibrillation by increasing collagen expression via the miR-128-3p-SP1-TGF-β1-Smad axis in atrial fibroblasts.55 In this study, we identified a complex regulatory network of extracellular matrix-related genes, lncRNAs and miRNAs in the process of LGG recurrence.
The ceRNA network is also a critical regulatory mechanism between lncRNAs and Mrna56,57 We identified a complex ceRNA network among 9 protein coding genes, 6 miRNAs and 12 lncRNAs. The results indicated that HOXA-AS2 could act as a competing endogenous RNA (ceRNA) for miR-184 to regulate the hub gene COL6A2 expression. In osteosarcoma cells, LncRNA HOXA-AS2 could promote cancer cell migration and invasion by acting as a ceRNA of miR-520c-3p.58 Although the research is not innovative enough, we confirmed that LncRNA HOXA-AS2 sponge miR-184 with complementary binding sites to increase expression of COL6A2, which resulted in LGG proliferation.
Conclusion
We demonstrated that the recurrence mechanisms of GBM and LGG are different at the transcriptional level, including protein coding genes, lncRNAs and miRNAs. Further analysis revealed three critical genes, TIMP1, COL1A1 and COL6A2, and the 3-gene signature was identified to improve prognosis prediction in LGG. Finally, LncRNA HOXA-AS2 acted as a ceRNA for miR-184 to regulate COL6A2 expression, and this ceRNA regulation induced cell proliferation in LGG.
Acknowledgments
Financial support from National Natural Science Foundation of China (Number A344). We acknowledge the open databases of CCLE, CGGA and TCGA.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl_4):iv1–iv86. doi:10.1093/neuonc/noy131
2. Lim M, Xia Y, Bettegowda C, et al. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442. doi:10.1038/s41571-018-0003-5
3. Newman JP, Wang GY, Arima K, et al. Interleukin-13 receptor alpha 2 cooperates with EGFRvIII signaling to promote glioblastoma multiforme. Nat Commun. 2017;8:1913. doi:10.1038/s41467-017-01392-9
4. Broen MPG, Smits M, Wijnenga MMJ, et al. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncol. 2018;20:1393–1399. doi:10.1093/neuonc/noy048
5. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498.
6. Karunasena E, McIver LJ, Rood BR, et al. Somatic intronic microsatellite loci differentiate glioblastoma from lower-grade gliomas. Oncotarget. 2014;5:6003–6014. doi:10.18632/oncotarget.2076
7. Reon BJ, Anaya J, Zhang Y, et al. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med. 2016;13:e1002192. doi:10.1371/journal.pmed.1002192
8. Paul Y, Mondal B, Patil V, et al. DNA methylation signatures for 2016 WHO classification subtypes of diffuse gliomas. Clin Epigenetics. 2017;9(1):32. doi:10.1186/s13148-017-0331-9
9. Zhang X, Lu X, Liu Z, et al. Integrating multiple-level molecular data to infer the distinctions between glioblastoma and lower-grade glioma. Int J Cancer. 2019;145:952–961. doi:10.1002/ijc.32174
10. Teng H, Wang P, Xue Y, et al. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther. 2016;24:1806–1822. doi:10.1038/mt.2016.103
11. Cai H, Liu X, Zheng J, et al. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi:10.1038/onc.2016.212
12. Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8. doi:10.1016/j.canlet.2017.08.015
13. Li H, Xue Y, Ma J, et al. SNHG1 promotes malignant biological behaviors of glioma cells via microRNA-154-5p/miR-376b-3p- FOXP2- KDM5B participating positive feedback loop. J Exp Clin Cancer Res. 2019;38:59. doi:10.1186/s13046-019-1063-9
14. Wu P, Cai J, Chen Q, et al. Lnc-TALC promotes O(6)-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat Commun. 2019;10:2045. doi:10.1038/s41467-019-10025-2
15. He K, Li WX, Guan D, et al. Regulatory network reconstruction of five essential microRNAs for survival analysis in breast cancer by integrating miRNA and mRNA expression datasets. Funct Integr Genomics. 2019;19:645–658. doi:10.1007/s10142-019-00670-7
16. Song J, Ye A, Jiang E, et al. Reconstruction and analysis of the aberrant lncRNA-miRNA-mRNA network based on competitive endogenous RNA in CESC. J Cell Biochem. 2018;119:6665–6673. doi:10.1002/jcb.26850
17. Lou W, Ding B, Xu L, et al. Construction of potential glioblastoma multiforme-related miRNA-mRNA regulatory network. Front Mol Neurosci. 2019;12:66. doi:10.3389/fnmol.2019.00066
18. Deng T, Gong YZ, Wang XK, et al. Use of genome-scale integrated analysis to identify key genes and potential molecular mechanisms in recurrence of lower-grade brain glioma. Med Sci Monit. 2019;25:3716–3727. doi:10.12659/MSM.913602
19. Wu W, Yu T, Wu Y, et al. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res. 2019;38(1):133. doi:10.1186/s13046-019-1132-0
20. Wang Q, Cai J, Fang C, et al. Mesenchymal glioblastoma constitutes a major ceRNA signature in the TGF-beta pathway. Theranostics. 2018;8:4733–4749. doi:10.7150/thno.26550
21. Long S, Li G. Comprehensive analysis of a long non-coding RNA-mediated competitive endogenous RNA network in glioblastoma multiforme. Exp Ther Med. 2019;18:1081–1090. doi:10.3892/etm.2019.7647
22. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi:10.1093/nar/gkw937
23. Cheng Z, Wang HZ, Li X, et al. MicroRNA-184 inhibits cell proliferation and invasion, and specifically targets TNFAIP2 in glioma. J Exp Clin Cancer Res. 2015;34:27. doi:10.1186/s13046-015-0142-9
24. Xiang Z, Li J, Song S, et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res. 2019;38:314. doi:10.1186/s13046-019-1318-5
25. Lian Y, Li Z, Fan Y, et al. The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex promotes cell proliferation in pancreatic cancer. Am J Transl Res. 2017;9:5496–5506.
26. Pala A, Coburger J, Scherer M, et al. To treat or not to treat? A retrospective multicenter assessment of survival in patients with IDH-mutant low-grade glioma based on adjuvant treatment. J Neurosurg;2019. 1–8. doi:10.3171/2019.4.JNS183395
27. Shergalis A, Bankhead A
28. Dirkse A, Golebiewska A, Buder T, et al. Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1787. doi:10.1038/s41467-019-09853-z
29. Morelli MB, Amantini C, Tomassoni D, et al. Transient receptor potential mucolipin-1 channels in glioblastoma: role in patient’s survival. Cancers (Basel). 2019;11:525. doi:10.3390/cancers11040525
30. Chakravarty D, Pedraza AM, Cotari J, et al. EGFR and PDGFRA co-expression and heterodimerization in glioblastoma tumor sphere lines. Sci Rep. 2017;7:9043. doi:10.1038/s41598-017-08940-9
31. Ruan M, Liu J, Ren X, et al. Whole transcriptome sequencing analyses of DHA treated glioblastoma cells. J Neurol Sci. 2019;396:247–253. doi:10.1016/j.jns.2018.11.027
32. Auffinger B, Spencer D, Pytel P, et al. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev Neurother. 2015;15(7):741–752. doi:10.1586/14737175.2015.1051968
33. Janda CY, Dang LT, You C, et al. Surrogate Wnt agonists that phenocopy canonical Wnt and beta-catenin signalling. Nature. 2017;545:234–237. doi:10.1038/nature22306
34. Klumpp L, Sezgin EC, Skardelly M, et al. KCa3.1 channels and glioblastoma: in vitro studies. Curr Neuropharmacol. 2018;16(5):627–635. doi:10.2174/1570159X15666170808115821
35. Hanker AB, Estrada MV, Bianchini G, et al. Extracellular matrix/integrin signaling promotes resistance to combined inhibition of HER2 and PI3K in HER2(+) breast cancer. Cancer Res. 2017;77:3280–3292. doi:10.1158/0008-5472.CAN-16-2808
36. Chakravarthy A, Khan L, Bensler NP, et al. TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun. 2018;9:4692. doi:10.1038/s41467-018-06654-8
37. Yuzhalin AE, Gordon-Weeks AN, Tognoli ML, et al. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat Commun. 2018;9:4783. doi:10.1038/s41467-018-07306-7
38. D’Costa Z, Jones K, Azad A, et al. Gemcitabine-induced TIMP1 attenuates therapy response and promotes tumor growth and liver metastasis in pancreatic cancer. Cancer Res. 2017;77:5952–5962. doi:10.1158/0008-5472.CAN-16-2833
39. Shi Y, Duan Z, Zhang X, et al. Down-regulation of the let-7i facilitates gastric cancer invasion and metastasis by targeting COL1A1. Protein Cell. 2019;10(2):143–148. doi:10.1007/s13238-018-0550-7
40. Chen P, Cescon M, Bonaldo P. Collagen VI in cancer and its biological mechanisms. Trends Mol Med. 2013;19(7):410–417. doi:10.1016/j.molmed.2013.04.001
41. Chen L, Lu D, Sun K, et al. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene. 2019;692:119–125. doi:10.1016/j.gene.2019.01.001
42. Zhu H, Chen H, Wang J, et al. Collagen stiffness promoted non-muscle-invasive bladder cancer progression to muscle-invasive bladder cancer. Onco Targets Ther. 2019;12:3441–3457. doi:10.2147/OTT.S194568
43. Qian Z, Li Y, Fan X, et al. Prognostic value of a microRNA signature as a novel biomarker in patients with lower-grade gliomas. J Neurooncol. 2018;137:127–137. doi:10.1007/s11060-017-2704-5
44. Liu B, Liu J, Liu K, et al. A prognostic signature of five pseudogenes for predicting lower-grade gliomas. Biomed Pharmacother. 2019;117:109116. doi:10.1016/j.biopha.2019.109116
45. Kiran M, Chatrath A, Tang X, et al. A prognostic signature for lower grade gliomas based on expression of long non-coding RNAs. Mol Neurobiol. 2019;56(7):4786–4798. doi:10.1007/s12035-018-1416-y
46. Syafruddin SE, Rodrigues P, Vojtasova E, et al. A KLF6-driven transcriptional network links lipid homeostasis and tumour growth in renal carcinoma. Nat Commun. 2019;10:1152. doi:10.1038/s41467-019-09116-x
47. Takaku M, Grimm SA, Roberts JD, et al. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat Commun. 2018;9:1059. doi:10.1038/s41467-018-03478-4
48. Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi:10.1038/s41467-017-00304-1
49. Luo W, Li X, Song Z, et al. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging (Albany NY). 2019;11(11):3811–3823. doi:10.18632/aging.102018
50. Li L, Wang Y, Song G, et al. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019;450:14–21. doi:10.1016/j.canlet.2019.02.036
51. Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi:10.1016/j.cca.2018.07.004
52. Wang X, Gao J, Zhou B, et al. Identification of prognostic markers for hepatocellular carcinoma based on miRNA expression profiles. Life Sci. 2019;232:116596. doi:10.1016/j.lfs.2019.116596
53. Sun C, Huang C, Li S, et al. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7:8341–8359. doi:10.18632/oncotarget.7071
54. Chen L, Li J, Li Q, et al. Overexpression of LncRNA AC067945.2 down-regulates collagen expression in skin fibroblasts and possibly correlates with the VEGF and Wnt signalling pathways. Cell Physiol Biochem. 2018;45:761–771. doi:10.1159/000487167
55. Cao F, Li Z, Ding WM, et al. LncRNA PVT1 regulates atrial fibrosis via miR-128-3p-SP1-TGF-beta1-Smad axis in atrial fibrillation. Mol Med. 2019;25:7. doi:10.1186/s10020-019-0074-5
56. Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234:18837–18848. doi:10.1002/jcp.28522
57. Li XY, Zhou LY, Luo H, et al. The long noncoding RNA MIR210HG promotes tumor metastasis by acting as a ceRNA of miR-1226-3p to regulate mucin-1c expression in invasive breast cancer. Aging (Albany NY). 2019;11(15):5646.
58. Wang Y, Zhang R, Cheng G, et al. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle. 2018;17(13):1637–1648. doi:10.1080/15384101.2018.1489174
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.