Back to Journals » Cancer Management and Research » Volume 13
Comprehensive Analysis of Peritoneal Metastasis Sequencing Data to Identify LINC00924 as a Prognostic Biomarker in Gastric Cancer
Authors Fang Y, Huang S, Han L, Wang S, Xiong B
Received 5 May 2021
Accepted for publication 29 June 2021
Published 12 July 2021 Volume 2021:13 Pages 5599—5611
DOI https://doi.org/10.2147/CMAR.S318704
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Yan Fang,1– 4 Sihao Huang,1– 3 Lei Han,1– 3 Shuyi Wang,1– 3 Bin Xiong1– 3
1Department of Gastrointestinal Surgery & Department of Gastric and Colorectal Surgical Oncology, Zhongnan Hospital of Wuhan University, Wuhan, 430071, People’s Republic of China; 2Hubei Key Laboratory of Tumor Biological Behaviors, Zhongnan Hospital of Wuhan university, Wuhan, 430071, People’s Republic of China; 3Hubei Cancer Clinical Study Center, Zhongnan Hospital of Wuhan university, Wuhan, 430071, People’s Republic of China; 4Department of Obstetrics and Gynecology, Guangzhou Women and Children’s Medical Center, Guangzhou, 510620, People’s Republic of China
Correspondence: Bin Xiong
Department of Gastrointestinal Surgery & Department of Gastric and Colorectal Surgical Oncology, Zhongnan Hospital of Wuhan University, Hubei Key Laboratory of Tumor Biological Behaviors & Hubei Cancer Clinical Study Center, No. 169 Donghu Road, Wuchang District, Wuhan, 430071, Hubei Province, People’s Republic of China
Tel +86 027 6781 3152
Fax +86 027 6781 2829
Email [email protected]
Background: Gastric cancer peritoneal metastasis has high mortality. At present, there is no effective way to cure the patients diagnosed with gastric cancer peritoneal metastasis due to its indistinct molecular mechanism. Therefore, to understand the pathogenesis and help for further target therapy, we conduct comprehensive analysis of peritoneal metastasis by bioinformatics in gastric cancer.
Methods: Microarray sequencing was used to find differential mRNAs and long non-coding RNAs (lncRNAs) expression between primary foci and peritoneal metastases lesion in gastric cancer. RT-qPCR was used to verify the expression levels of lncRNAs in gastric cancer cells after co-culture with adipocytes. TCGA, Cytoscape, lnCAR, cBioPoratal and R packages (ggrisk, survival, survminer, timeROC, forestplot, immunedeconv, ggplot2, pheatmap and ggpubr) were applied in this work.
Results: Adipocytes co-culture model was used to mimic the peritoneal microenvironment and found that LINC01151 (NR_126348), FAM27B (NR_027422) and LINC00924 (NR_027133) were up-regulated in co-culture group. Increased LINC00924 expression was significantly associated with reduced overall survival and up-regulated percentage abundance of tumor-infiltrating CD8+ T, B, macrophage and NK immune cells; moreover, immune checkpoint blockers (ICBs) had a worse effect on the LINC00924 high expression group. Furthermore, through univariate and multivariate Cox regression analysis, we found that LINC00924-related PEX5L in CNC network was an independent prognostic factor in gastric cancer progression.
Conclusion: LINC00924 expression was associated with poor survival, immune infiltrations and worse response to ICBs. LINC00924 might be immunotherapeutic targets of advanced gastric cancer.
Keywords: gastric cancer, LINC00924, immune infiltrations, target therapy
Introduction
Gastric cancer peritoneal metastasis was less sensitive to chemotherapy, and the patients diagnosed with peritoneal metastasis had a poor clinical prognosis and high mortality.1 Cytoreductive surgery combined with hyperthermic intraperitoneal chemotherapy could delay tumor progression; however, the current medical treatment was still limited for prolonging patients’ survival.2 Therefore, it is necessary to study the molecular mechanism of gastric cancer peritoneal metastasis which could improve early diagnosis and provide basis for targeted therapy.
Long non-coding RNAs (lncRNAs) were a type of RNAs longer than 200 nucleotides and could not be translated into protein.3 lncRNA-involved in gene regulation were associated with many biological processes including tumor growth, cancer metastasis and immune escape.4–6 Huang reported that LncRNA AK023391 promoted gastric cancer invasion through PI3K/Akt signaling pathway.7 LncRNA SNHG11 activated Wnt/beta-Catenin axis to facilitate cell migration and proliferation in gastric cancer.8 However, the function of lncRNAs in gastric cancer peritoneal metastasis was still unclear.
Immune checkpoint therapy – as a promising anti-cancer therapy method – had benefited many patients and extended overall survival.9 LncRNA was associated with immunity closely.10 Li’s research showed that the positive feedback loop of LncRNA SNHG14/miR-5990-3P/ZEB1 promoted diffuse large B cell lymphoma progression through regulating PD-1/PD-L1 checkpoint.11 However, due to individual differences in gene expression, most cancer patients did not respond to immunotherapy.12 Therefore, the study of gastric cancer peritoneal metastasis gene expression profile and the relationship between different genes and immune infiltrations was beneficial to improve the curative effect of immunotherapy.
Our study results showed that compared with primary foci, there were 3433 lncRNAs and 1210 mRNAs up-regulated, 1341 lncRNAs and 3192 mRNAs down-regulated in peritoneal metastasis lesion. Thirty-eight up-regulated lncRNAs and 9 down-regulated lncRNAs with known function and fold change >2 were selected to construct CNC network. Adipocytes were a major cell type of peritoneal microenvironment13 and we used adipocytes to co-culture with tumor cells to mimic the tumor’s internal environment and exclude the influence of cell diversity on sequencing results. Our results showed that LINC01151, FAM27B and LINC00924 were up-regulated in co-culture group and LINC00924 was associated with poor survival. Furthermore, immune checkpoint blockers (ICBs) had a worse effect on the LINC00924 high expression group. We also found that LINC00924-related PEX5L in CNC network was an independent prognostic factor by univariate and multivariate Cox regression analysis in gastric cancer progression.
Materials and Methods
Patient Tissue Samples
In this study, we collected three paired gastric cancer with peritoneal metastasis tissue samples at the Zhongnan Hospital of Wuhan University (Additional File, Table 1). The patient had not experienced any chemotherapy or radiotherapy. All samples were obtained with informed patients’ consent before collection, and approved by the Zhongnan Hospital of Wuhan University Ethics Committee. Moreover, the collected samples need to be fixed in formalin and embedded in paraffin for further study.
Microarray of Primary Lesion and Peritoneal Metastasis Lesion
Biopulverizer was used for cryopulverization of three paired tissue samples, then Mini-Bead-Beater-16 was used for homogenization. Quick Amp Labeling Kit, One-Color (Agilent p/n 5190–0442) was used for labeling reaction, RNeasy Mini Kit (Qiagen p/n 74104) for RNA purifying, NanoDrop ND-1000 for labeled cDNA QC. After hybridization and microarray wash, Agilent Microarray Scanner (Agilent p/n G2565BA) was used for scanning. All data were extracted using Agilent Feature Extraction Software.
Coding-Non-Coding (CNC) Gene Co-Expression Network
The normalized data of mRNA was taken to calculate the medium value of different transcripts of the same gene to represent expression value of gene. Then, the Pearson’s correlation coefficient (PCC) was calculated between selected lncRNAs and all mRNAs according to normalized data. The mRNAs with abs (PCC) ≥0.95, P-value ≤0.05 and FDR ≤0.1 were recorded. Cytoscape was used to draw the CNC gene co-expression network of lncRNAs and mRNAs.
Cell Culture and Adipocyte Differentiation Induction
The human gastric cancer cell lines (AGS, BGC823) culturing in RPMI 1640 medium (Gibco, USA) and mouse preadipocyte cell line (3T3-L1) culturing in DMEM (Gibco, USA) were purchased from Chinese Academy of Sciences in Shanghai. For 3T3-L1 induction, the medium of 3T3-L1 cells was changed with DMEM containing 10% FBS, 10µg/mL insulin, 1µmol/L dexamethasone, 0.5mmol/L 3-isobutyl-1-methylxanthine (IBMX, HY-12318; MCE) for 2 days cultivation after 2 days of contact inhibition, and subsequently, the medium was replaced with high glucose DMEM containing 10ug/mL insulin and 10% FBS for 2 days, then, the medium was changed every day with DMEM supplemented with 10% FBS until 3T3-L1 was induced into mature adipocyte.14 The gastric cancer lines were cocultured with the supernatant of adipocytes to mimic peritoneal microenvironment.15
RNA Isolation and Quantitative Real-Time PCR (RT-qPCR)
According to the manufacturer’s instructions, the Trizol Reagent was used to isolate the total RNA from gastric cancer cells. RNA concentration was measured and 1µg RNA was quantified for reverse transcription using the PrimeScriptTM RT reagent kit (Toyobo, Osaka), then RT-qPCR was run on the Bio-Rad IQ5 Real PCR machine (Bio-Rad, USA) in a 96-well plate containing reaction mixture of pre-primer, post-primer, cDNA, SYBR-Green PCR Master Mix (Takara, Osaka). The 2−ΔΔCt method was used to calculate the relative expression. The primer sequences used in the study are presented in Additional File, Table 2.
Prognostic Analysis
Gastric cancer sequencing data and corresponding clinical information were obtained from the Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.com). Log rank was used to verify KM survival analysis to compare the differences in survival and time-ROC analysis was performed to compare the predictive accuracy and risk score. Univariate and multivariate Cox regression analyses through survival R package were performed to identify the proper terms to build the nomogram. The forest was used to show the P value, HR and 95% CI of each variable through forestplot R package.
Immunity Analysis
According to gastric cancer sequencing data of TCGA, we divided the overall sample into high expression group and low expression group of LINC00924. Immunedeconv R package was used to assess the percentage abundance of tumor-infiltrating immune cells in each sample. TIDE algorithm was applied to predict potential ICB response.
lnCAR
lnCAR (https://lncar.renlab.org/) was a cancer-related microarray data interactive tool and used for lncRNAs differential expression, co-expression network, KEGG pathway and survival analysis.16
cBioPortal
cBioPortal (https://www.cbioportal.org/), an open-source platform, could be used to analyze and visualize cancer genomics and clinical data.17 In this study, cBioPortal was used for mRNAs survival analysis.
Statistics Analysis
Results expressed as mean ± standard error (m ± SE) were analyzed by two-tailed Student’s t-test. All experiments were performed in triplicate and statistical significance was defined as p < 0.05 using the GraphPad Prism software (version 6.0, GraphPad Software, USA). Wilcox test was used for immune checkpoint analysis.
Results
Microarray Data of lncRNAs and mRNAs
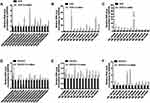
To distinguish the differential expression of lncRNAs and mRNAs, we performed microarray assay between peritoneal metastasis lesion and primary gastric cancer foci from three patients without any treatment who were diagnosed with gastric cancer peritoneal metastasis. Compared with primary foci, there was a total of 26,793 lncRNAs and 18,985 mRNAs, of which 3433 lncRNAs and 1210 mRNAs up-regulated significantly, 1341 lncRNAs and 3192 mRNAs down-regulated significantly (Figure 1A–D). Among up-regulated lncRNAs, 38 up-regulated and 9 down-regulated lncRNAs with fold change >2 had known structural and functional studies (Figure 1E). According to the sequencing results of mRNAs and lncRNAs, we further used 38 up-regulated and 9 down-regulated lncRNAs to construct CNC network between lncRNAs and mRNAs (Figure 2A–G). To analyze the function of lncRNAs and mRNAs, GO analysis were used. The GO analysis results of up-regulated lncRNA-related mRNAs showed that most mRNAs were located at cytosol and associated with positive regulation of transcription from RNA polymerase II promoter, positive regulation of gene expression, regulation of apoptotic process, response to drug and negative regulation of gene expression during the biological process. Furthermore, the molecular function of most mRNAs was protein binding, protein homodimerization activity, ubiquitin protein ligase binding, protease binding and methyltransferase activity (Figure 3A–C). The GO analysis results of down-regulated lncRNA-related mRNAs demonstrated that most mRNAs were in cytoplasm and had protease binding and protein homodimerization activity function. The top six enriched GO terms of the biological process in down-regulated lncRNA-related transcripts were positive regulation of transcription from RNA polymerase II promoter, positive regulation of gene expression, response to drug, positive regulation of sequence-specific DNA binding transcription factor activity, positive regulation of I-kappaB kinase/NF-kappaB signaling and extracellular matrix organization (Figure 3D–F).
LINC00924 Was Upregulated Significantly in Simulated Peritoneal Microenvironment
Due to the diversity of cell types in the microenvironment, microarray assay of peritoneal metastasis lesion and primary gastric cancer foci did not represent the expression level of tumor genes accurately. Adipocytes, the most abundant cell type of peritoneum, were used to coculture with gastric cancer cell lines to perform in vivo environment simulation. The results of RT-qPCR showed that NR_126348 (LINC01151), NR_027422 (FAM27B) and NR_027133 (LINC00924) had a significant increase in AGS, BGC823 co-culture group (Figure 4A–F). Furthermore, we used lnCAR website to analyze and visualize the expression, co-expression and pathway analysis. The results showed that LINC00924 was up-regulated in gastric cancer and the related genes were associated with metabolic pathways according to GSE65801 dataset (Figure 5A–C). GSE30727 and GSE13911 analysis showed that LINC01151 and FAM27B were also up-regulated and related-mRNAs involved in metabolic pathways (Figure 5D–I); however, LINC01151 and FAM27B had no effect on the survival of patients with gastric cancer surprisingly (data was not shown). Above all, the LINC00924 gene expression profile was peritoneal metastases > primary foci > normal tissue and high LINC00924 expression was closely associated with low overall survival.
LINC00924-Related PEX5L in CNC Network Was an Independent Prognostic Factor and Associated with Poor Survival
According to the CNC network based on our sequencing data, we took the intersection of LINC00924 and FAM27B-related mRNAs. CD28, NHLRC1, CYREN (C7orf49), COLQ, PEX5L, PRMT8, SNX32, HAX1, E2F1, AADACL4, PITX1, ATG10, ELOF1, RGR, SYP, ISLR2, CDS1, TAZ and RP1 were in the intersection. Furthermore, we studied the effects of lncRNA-related mRNAs, age, gender and p-TNM stage on prognosis of gastric cancer. In univariate Cox regression analysis, PEX5L, AADACL-4 and TNM stage were high-risk factors. The hazard ratio (95% CI) of PEX5L, AADACL-4 and TNM stage was 1.91265 (1.30176,2.81020), 2.60071 (1.34318, 5.03559) and 1.27432 (1.06347, 1.52698), respectively (Figure 6A–D). For multivariate Cox regression analysis, the hazard ratio (95% CI) of PEX5L, AADACL-4 and TNM stage was 1.91455 (1.24433, 2.94577), 2.56854 (1.32327, 4.98567) and 1.40575 (1.16321, 1.69886), respectively (Figure 6E–H). Furthermore, we used stomach adenocarcinoma (TCGA, Firehose Legacy) clinical and genomic data in cBioPortal website and found that the patients with high PEX5L expression had a poor survival (Figure 6I–L). The above results demonstrated that LINC00924-related PEX5L was an independent prognostic factor in gastric cancer progression.
LINC00924 Was Associated with a Worse Response Rate to ICB Treatment
According to the median of expression, we divided 375 gastric cancer samples into two groups: high LINC00924 expression group and low LINC00924 expression group. To explore the relationship between LINC00924 and immunity, we used EPIC algorithms to make immune infiltration estimations. In high LINC00924 expression group, the percentage abundance of tumor-infiltrating B cells, CD4+ T cells, endothelial cells, macrophages and NK cells were up-regulated (Figure 7A). CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT and SIGLEC15 were immune checkpoints. In high LINC00924 expression group, CD274, CTLA4, HAVCR2, PDCD1, PDCD1LG2 and TIGIT were up-regulated significantly (Figure 7B), so we further used TIDE algorithm to analyze ICB response. In our study, high LINC00924 expression was associated with high TIDE score; moreover, there were 48 responders in high expression group and 92 responders in low expression group (Figure 7C). To further explored the effect of LINC00924 on gastric cancer prognosis, we plotted a scatter plot of gene expression and the corresponding survival time in different samples (Figure 8A). The prognosis of LINC00924 gene was significant (Log rank P =0.0989), and the higher expression was related to the worse prognosis (Figure 8B). Time-ROC analysis showed that 1-Year, AUC=0.584, 3-Year, AUC=0.594 and 5-Year, AUC=0.676 (Figure 8C). All results showed that LINC00924 was associated with a worse response rate to ICB treatment and LINC00924 could be used to predict the prognosis of gastric cancer.
Discussion
In this study, we found the differential lncRNAs and mRNAs of peritoneal metastases foci versus primary foci through microarray sequencing. LINC00924 played an important role in gastric cancer progression. LINC00924 was associated with poor survival and immune escape. LINC00924-related PEX5L was an independent prognostic factor. Our study was helpful to provide the basis for molecular targeted therapy of gastric cancer patients with peritoneal metastasis.
Adipocytes was major cell components of peritoneum and adipocyte-derived cytokines and fatty acids promoted cancer progression in varieties of molecular mechanisms. On the one hand, FABP4 (fatty acid binding protein 4)18 and PITPNC1 (Phosphatidylinositol transfer protein, cytoplasmic 1)19 were up-regulated after induction of adipocytes which accelerated fatty acids transport to satisfy energy demands of rapid tumor growth; On the other hand, adipokines derived from adipocytes activated the intracellular signaling pathway to promote oncogenes expression to increase the malignant potential.20 In our results, lncRNAs were up-regulated after coculture with adipocytes, but restricted by many factors, we could not find the upstream molecular of these lncRNAs secreted by adipocytes.
Gastric cancer peritoneal metastasis was a complex biological process concerned with abnormal expression of many coding and non-coding RNAs. In our study, 38 lncRNAs were upregulated. Xiaorong Lin’s study showed that lncRNA BDNF-AS (NR_033312) promoted RNH1 ubiquitination degradation in TRIM21-dependent manner to induce poor outcomes and endocrine therapy resistance of breast cancer, thereby leading to malignant progression.21 FGD5-AS1 (NR_046252) enhanced non-small cell lung cancer cells’ proliferation through regulating FGD5-AS1/has-miR-107/FGFRL1 axis,22 and Fu et al’s study also demonstrated that FGD5-AS1 improved cisplatin resistance of non-small cell lung cancer cells through miR-140-5p/WEE1 signaling pathway.23 Moreover, other malignant tumors have also been reported to correlate with high FGD5-AS1 expression. FGD5-AS1/miR-383/SP1 axis promoted esophageal squamous cell carcinoma progression and FGD5-AS1/ miR-153-3p-CITED2 enhanced gastric cancer cell malignant characteristics both in a competing endogenous RNA mechanism.24,25 Our results showed that many lncRNA-related mRNAs participated in positive regulation of gene expression. PITX1 (paired like homeodomain 1), IKZF1 (IKAROS family zinc finger 1), CREB3L4 (cAMP-responsive element-binding protein 3 like 4) and E2F1 (E2F transcription factor 1) encoded a transcription factor to function as a regulator of gene. PITX1 as a known tumor suppressor gene cis-regulated PDCD5 expression to promote tumor cell apoptosis in gastric cancer.26 IKZF1 was a hub gene of pediatric acute lymphoblastic leukemia and its effect on cancer was still obscure.27 High CREB3L4 expression inhibited preadipocytes 3T3-L1 differentiation into mature adipocytes by reducing expression of C/EBPα and PPARγ2,28 so we speculated that low CREB3L4 expression in peritoneal metastasis lesion was associated with omentum adipogenesis and energy supply of cancer cells. E2F1-a tumor promoter gene-derived breast cancer progression by regulating FGF13.29 Summarily, lncRNAs and lncRNA-related mRNAs promoted tumor development through metabolic programming change induced by microenvironmental remodeling and proto-oncogene activation.
Tumor immune escape referred to the phenomenon that tumor cells survived and proliferated in vivo by escaping from the recognition and attack of the body’s immune system through a variety of mechanisms. The immune system of the body had the function of immune monitoring. When the malignant cells appeared in the body, the immune system could recognize and eliminate these “non-self” cells specifically through the immune mechanism to resist the occurrence and development of tumors. However, malignant cells could evade the immune surveillance of the body through a variety of mechanisms in some cases, and proliferate rapidly in the body to form tumors.30 Cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) was constitutively expressed on T cells. When bound to CD80 or CD86 on the surface of antigen-presenting cells, it acted as a “off” switch to downregulate the immune response.31 In LINC00924 high expression group, the immune checkpoints were also up-regulated which meant the immunosuppressive status of gastric cancer. CD4+T cells not only killed tumor cells directly through IFN-gamma-dependent mechanisms but also played an important role in the generation of cytotoxic T lymphocytes (CTL).32 However, the ratio of CD4+ T cells was upregulated in LINC00924 high expression group in our results. The abnormal results might be due to the fact that tumor was a heterogeneous mass and cancer progression was the result of competition between antitumor cells and suppressor cells.33
Conclusion
In conclusion, LINC00924 was associated with poor survival, immune escape and the related-mRNA, PEX5L was a high-risk factor for gastric cancer progression. LINC00924 might be the therapeutic targets for gastric cancer.
Ethics Approval
This study was approved by the Zhongnan Hospital of Wuhan University ethics committee and the patient tissue samples were conducted in accordance with the Declaration of Helsinki.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81572874), National Natural Science Fund Youth Fund of China (81702411), Zhongnan Hospital of Wuhan University, Technology and Innovation Seed Found (znpy 2016058).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Alyami M, Hubner M, Grass F, et al. Pressurised intraperitoneal aerosol chemotherapy: rationale, evidence, and potential indications. Lancet Oncol. 2019;20(7):e368–e377. doi:10.1016/S1470-2045(19)30318-3
2. Bonnot PE, Piessen G, Kepenekian V, et al. Cytoreductive surgery with or without hyperthermic intraperitoneal chemotherapy for gastric cancer with peritoneal metastases (CYTO-CHIP study): a propensity score analysis. J Clin Oncol. 2019;37(23):2028–2040. doi:10.1200/JCO.18.01688
3. Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol. 2020;21(3):123–136. doi:10.1038/s41580-019-0209-0
4. Srirangam Nadhamuni V, Korbonits M. Novel insights into pituitary tumorigenesis: genetic and epigenetic mechanisms. Endocr Rev. 2020;41(6).
5. Wang CJ, Zhu CC, Xu J, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18(1):115. doi:10.1186/s12943-019-1032-0
6. Zhuo W, Liu Y, Li S, et al. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156(3):676–691 e611. doi:10.1053/j.gastro.2018.10.054
7. Huang Y, Zhang J, Hou L, et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36(1):194.
8. Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. lncRNA SNHG11 promotes gastric cancer progression by activating the wnt/beta-catenin pathway and oncogenic autophagy. Mol Ther. 2021;29(3):1258–1278. doi:10.1016/j.ymthe.2020.10.011
9. Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27(8):1492–1504. doi:10.1093/annonc/mdw217
10. Li Y, Jiang T, Zhou W, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun. 2020;11(1):1000. doi:10.1038/s41467-020-14802-2
11. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10(10):731. doi:10.1038/s41419-019-1886-5
12. Gil Del Alcazar CR, Aleckovic M, Polyak K. Immune escape during breast tumor progression. Cancer Immunol Res. 2020;8(4):422–427. doi:10.1158/2326-6066.CIR-19-0786
13. Thibault B, Castells M, Delord JP, Couderc B. Ovarian cancer microenvironment: implications for cancer dissemination and chemoresistance acquisition. Cancer Metastasis Rev. 2014;33(1):17–39.
14. Yang W, Yang C, Luo J, Wei Y, Wang W, Zhong Y. Adiponectin promotes preadipocyte differentiation via the PPARgamma pathway. Mol Med Rep. 2018;17(1):428–435. doi:10.3892/mmr.2017.7881
15. Ladanyi A, Mukherjee A, Kenny HA, et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene. 2018;37(17):2285–2301. doi:10.1038/s41388-017-0093-z
16. Zheng Y, Xu Q, Liu M, et al. lnCAR: a comprehensive resource for lncRNAs from cancer arrays. Cancer Res. 2019;79(8):2076–2083. doi:10.1158/0008-5472.CAN-18-2169
17. Wu P, Heins ZJ, Muller JT, et al. Integration and analysis of CPTAC proteomics data in the context of cancer genomics in the cBioPortal. Mol Cell Proteomics. 2019;18(9):1893–1898. doi:10.1074/mcp.TIR119.001673
18. Huang M, Narita S, Inoue T, et al. Fatty acid binding protein 4 enhances prostate cancer progression by upregulating matrix metalloproteinases and stromal cell cytokine production. Oncotarget. 2017;8(67):111780–111794. doi:10.18632/oncotarget.22908
19. Tan Y, Lin K, Zhao Y, et al. Adipocytes fuel gastric cancer omental metastasis via PITPNC1-mediated fatty acid metabolic reprogramming. Theranostics. 2018;8(19):5452–5468. doi:10.7150/thno.28219
20. He JY, Wei XH, Li SJ, et al. Adipocyte-derived IL-6 and leptin promote breast cancer metastasis via upregulation of Lysyl hydroxylase-2 expression. Cell Commun Signal. 2018;16(1):100. doi:10.1186/s12964-018-0309-z
21. Lin X, Dinglin X, Cao S, et al. Enhancer-driven lncRNA BDNF-AS induces endocrine resistance and malignant progression of breast cancer through the RNH1/TRIM21/mTOR cascade. Cell Rep. 2020;31(10):107753. doi:10.1016/j.celrep.2020.107753
22. Fan Y, Li H, Yu Z, et al. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci Rep. 2020;40(1). doi:10.1042/BSR20193309
23. Fu J, Cai H, Wu Y, Fang S, Wang D. Elevation of FGD5-AS1 contributes to cell progression by improving cisplatin resistance against non-small cell lung cancer cells through regulating miR-140-5p/WEE1 axis. Gene. 2020;755:144886. doi:10.1016/j.gene.2020.144886
24. Gao J, Zhang Z, Su H, Zong L, Li Y. Long noncoding RNA FGD5-AS1 acts as a competing endogenous RNA on microRNA-383 to enhance the malignant characteristics of esophageal squamous cell carcinoma by increasing SP1 expression. Cancer Manag Res. 2020;12:2265–2278. doi:10.2147/CMAR.S236576
25. Gao Y, Xie M, Guo Y, Yang Q, Hu S, Li Z. Long non-coding RNA FGD5-AS1 regulates cancer cell proliferation and chemoresistance in gastric cancer through miR-153-3p/CITED2 axis. Front Genet. 2020;11:715. doi:10.3389/fgene.2020.00715
26. Qiao F, Gong P, Song Y, et al. Downregulated PITX1 modulated by MiR-19a-3p promotes cell malignancy and predicts a poor prognosis of gastric cancer by affecting transcriptionally activated PDCD5. Cell Physiol Biochem. 2018;46(6):2215–2231. doi:10.1159/000489590
27. Stanulla M, Cave H, Moorman AV. IKZF1 deletions in pediatric acute lymphoblastic leukemia: still a poor prognostic marker? Blood. 2020;135(4):252–260. doi:10.1182/blood.2019000813
28. Kim TH, Jo SH, Choi H, et al. Identification of Creb3l4 as an essential negative regulator of adipogenesis. Cell Death Dis. 2014;5:e1527. doi:10.1038/cddis.2014.490
29. Hollern DP, Swiatnicki MR, Rennhack JP, et al. E2F1 drives breast cancer metastasis by regulating the target gene FGF13 and altering cell migration. Sci Rep. 2019;9(1):10718. doi:10.1038/s41598-019-47218-0
30. Garrido F, Aptsiauri N. Cancer immune escape: MHC expression in primary tumours versus metastases. Immunology. 2019;158(4):255–266. doi:10.1111/imm.13114
31. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi:10.1038/nrc3239
32. Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18(10):635–647. doi:10.1038/s41577-018-0044-0
33. Tian X, Shen H, Li Z, Wang T, Wang S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12(1):84. doi:10.1186/s13045-019-0772-z
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.