Back to Journals » Cancer Management and Research » Volume 12
Comprehensive Analysis of PD-1 Gene Expression, Immune Characteristics and Prognostic Significance in 1396 Glioma Patients
Authors Liu C, Zhang Z, Ping Y, Qin G, Zhang K, Maimela NR , Huang L, Yang S, Zhang Y
Received 11 November 2019
Accepted for publication 30 May 2020
Published 10 June 2020 Volume 2020:12 Pages 4399—4410
DOI https://doi.org/10.2147/CMAR.S238174
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Chaojun Liu,1,* Zhen Zhang,1,* Yu Ping,1 Guohui Qin,1– 3 Kai Zhang,1 Nomathamsanqa Resegofetse Maimela,1 Lan Huang,1 Shengli Yang,1 Yi Zhang1– 3
1Biotherapy Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, People’s Republic of China; 2Cancer Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, People’s Republic of China; 3Henan Key Laboratory for Tumor Immunology and Biotherapy, Zhengzhou, Henan 450052, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yi Zhang; Shengli Yang
Biotherapy Center, The First Affiliated Hospital of Zhengzhou University, 1th Jianshe Road, Zhengzhou, Henan 450052, People’s Republic of China
+86-371-6691-5320
; +86-371-6691-2993
Email [email protected]; [email protected]
Background: Programmed cell death protein-1 (PD-1) blockade therapy is one of the most remarkable immunotherapy strategies in many solid tumors, excluding glioma. The PD-1 expression, immune characteristics, and prognosis relevance in glioma remain poorly understood.
Patients and Methods: RNA sequencing (RNA-seq) and mRNA microarray data were obtained for 325 and 301 glioma patients, respectively, from the Chinese Glioma Genome Atlas (CGGA) database. We analyzed the expression profile of PDCD1 (encoding PD-1) according to the different grade, isocitrate dehydrogenase (IDH) mutation status, and molecular subtype of glioblastoma. Gene ontology (GO) analyses were performed to explore biological processes of PD-1-related genes. Survival analysis was conducted using the Kaplan–Meier method. The findings were validated using The Cancer Genome Atlas (TCGA) RNA-seq data from 697 glioma samples. We also confirmed the PDCD1 gene expression feature and survival relevance in our own cohort of 73 glioma patients. R language was used for statistical analysis and generating figures.
Results: PDCD1 was enriched in glioblastoma (WHO, grade IV), IDH wild-type glioma and mesenchymal glioblastoma in CGGA and TCGA datasets; similar results were validated in our own patient cohort. GO analysis revealed that PDCD1-related genes were involved in inflammation immune responses and T cell-mediated immune responses in glioma. Circos plots indicated that PDCD1 was positively associated with CD28, ICOS, and the inhibitory checkpoint molecules CTLA4, HAVCR2, TIGIT, and LAG3. Patients with PDCD1 upregulation had much shorter overall survival.
Conclusion: PDCD1 upregulation was found in more malignant phenotypes of glioma and indicated a worse prognosis. Immunotherapy of targeting PD-1 or combined with other checkpoint molecules (eg, TIM-3, LAG-3, or TIGIT) blockade may represent a promising treatment strategy for glioma.
Keywords: programmed cell death 1, glioma, The Cancer Genome Atlas, survival analysis, immunotherapy, costimulatory, inhibitory T-cell receptors
Introduction
Glioma is the most frequent primary brain tumor in adults and has a poor prognosis owing to the limited therapeutic efficacy of conventional treatments including surgical resection, radiotherapy, and chemotherapy.1 Moreover, treatment strategies upon disease recurrence are limited and lack efficacy.2 Therefore, the new treatment approaches for glioma are urgently needed.
Ipilimumab, targeting the human cytotoxic lymphocyte antigen-4 (CTLA4), was the first immune checkpoint blocking antibody approved for the treatment of advanced melanoma by the United States Food and Drug Administration (FDA), thus opening a new era of anti-tumor immunotherapy. Subsequently, two monoclonal antibodies (pembrolizumab and nivolumab) against the PD-1 receptor were approved by the FDA. Since significantly longer recurrence-free survival and a lower rate of severe adverse effects have been observed for PD-1 antibody than for CTLA4 blockade therapy in the treatment of advanced melanoma,3 PD-1 blockade has recently received increasing attention. Thus, the indication for PD-1 blockade therapy in advanced solid tumors has rapidly extended from melanoma to other types of cancers, such as non-small cell lung cancer, head and neck squamous cell cancer, colorectal cancer, hepatocellular carcinoma, and renal cell carcinoma; however, this has excluded glioma to date. Although preclinical studies have reported distinctive anti-tumor effects of PD-1 blockade in gliomas4,5 and relevant clinical trials are underway (eg, NCT02852655 and NCT03925246, https://clinicaltrials.gov/), the evidence is still insufficient for the clinical application of PD-1 blockade therapy in glioma. Moreover, limited information is available regarding the expression and prognosis of PD-1 in glioma.
PD-1 Expression on T cells in malignant glioma patients reflects the exhaustion and activation of T cells.6 PD-1 signaling blockade therapy can restore T cell function and promote the anti-tumor immune response.7 Considering the importance of PD-1 in regulating the immune response in the glioma microenvironment, studies deciphering the immune characteristics of PD-1 in gliomas are urgently needed.
Although accumulating evidence indicates the upregulation of PD-1 in glioma tumor tissue,5,8 few studies have focused on the comprehensive characteristics of PD-1 expression in glioma. Therefore, we comprehensively analyzed PDCD1 gene expression, immune characteristics, and patient prognosis in diffuse glioma with, thus far, the largest sample size to date, containing 1396 patients. The CGGA database with 626 glioma patients, including RNA-sequencing (RNA-seq) data and mRNA microarray data from 325 and 301 glioma patients, respectively, was first analyzed as training datasets. Subsequently, the findings were validated in a cohort of 697 glioma patients from the TCGA database, as well as our own cohort of 73 glioma patients from the First Affiliated Hospital of Zhengzhou University. This study showed that PD-1 was obviously upregulated in the more malignant subtypes of gliomas, involved in pivotal biological processes, which were closely related to the immune response, and indicated a worse clinical prognosis. Thus, immunotherapy based on PD-1 blockade may provide a promising strategy for glioma therapy.
Patients and Methods
Data Sources
mRNA expression, DNA methylation data and clinicopathologic outcome information of glioma patients were downloaded from the CGGA (http://www.cgga.org.cn/) and the TCGA (http://cancergenome.nih.gov/) databases, according to the CGGA and TCGA data sharing agreements, and normalized for analysis. In the CGGA database, RNA-seq data from 325 samples and mRNA microarray data from 301 samples were included and analyzed as training cohorts. To validate the findings of the CGGA datasets, RNAseq data of 697 glioma samples of whole grade from the TCGA database were harnessed as a validation cohort. PD-1 transcriptional expression data of 1323 samples were evaluated and the clinicopathologic features of these patients are summarized in Table S1.
Patients and Tumor Samples
A total of 73 glioma patients were collected from the First Affiliated Hospital of Zhengzhou University from January 2016 to December 2018. Signed informed consent forms were obtained from all subjects. All patients underwent surgery and fresh tumor tissue was obtained afterwards. The clinicopathologic features, including gender, age, WHO grade, IDH mutation status, and overall survival time were collected (Table S2).
RNA Isolation and Real-Time PCR
Total RNA was extracted from 50~100mg fresh tumor tissue lysed with 1 mL RNAiso Plus (TaKaRa, Tokyo, Japan). Complementary DNA was generated by reverse transcriptase reaction with 1μg total RNA using the PrimeScript™ RT reagent Kit (TaKaRa, Tokyo, Japan) according to the manufacturer’s directions. The expression levels of PDCD1 and GAPDH were tested using ChamQ SYBR Color qPCR Master Mix (Vazyme, Jiangsu, China) in a CFX96™ Real-Time PCR Detection Systems (Bio-Rad, CA, USA). The genes were amplified under the conditions: 95 °C for 5 minutes; and 40 cycles of 95 °C for 10 seconds, and 60 °C for 30 seconds. Primers for PDCD1 were as follows: sense-5ʹ-CCAGGATGGTTCTTAGACTCCC-3ʹ, and antisense-5ʹ-TTTAGCACGAAGCTCTCCGAT-3ʹ; product length was 137 bp. Primers for GAPDH were as follows: sense-5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ, and antisense-5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ; product length was 138 bp. The house-keeping gene GAPDH was used as internal reference and PDCD1 expression level was ascertained using the 2−ΔΔCt method followed by logarithmic transformation of base 2.
Statistical Analysis
Variables with two groups were reported as mean ± standard error of the mean (SEM) followed by a comparison using a two-tailed Student’s t-test. The differences for more than two groups were analyzed using one-way ANOVA followed by a Bonferroni post hoc test. Survival differences were estimated using Kaplan–Meier analysis followed by a Log rank test. GO analysis was conducted using the database for annotation, visualization and integrated discovery (DAVID) 6.8 (http://david.abcc.ncifcrf.gov/home.jsp) for functional annotation of PD-1-related genes. Pearson correlation analysis was used to evaluated the correlation between PD-1 and other genes. R language (https://www.r-project.org/) and GraphPad Prism v7.0 software (GraphPad, USA) were employed for statistical analysis and generating figures. The R packages (pheatmap, ggplot2, pROC, and corrgram) were used for visualization of figures. A p-value of < 0.05 was considered statistically significant.
Results
PDCD1 Was Upregulated in Glioblastoma (GBM) and IDH Wild-Type Glioma
To explore an overview of PDCD1 (encoding PD-1) expression status in glioma, we first assessed PDCD1 expression in 325 RNA-sequencing samples and 301 mRNA microarray samples from the CGGA database, according to different tumor grades. Higher expression of PDCD1 was observed in GBM (WHO grade IV) than in grade II and III gliomas in the two CGGA datasets (Figure 1A), indicating that PDCD1 upregulation was associated with a greater malignant potential in glioma. DNA methylation is a common epigenetic phenomenon in glioma, and promoter methylation of PDCD1 is considered a favorable prognostic factor in diffuse lower-grade gliomas.9 Hence, we assessed promoter methylation of PDCD1 in different grades of gliomas and found that promoter methylation of PDCD1 was markedly higher in grade II and III gliomas than in GBM in the TCGA dataset, but not in the CGGA dataset (Figure S1A). Emerging evidence indicates that distinct clinical outcomes and the immune response in glioma patients depended on the IDH mutation status.10,11 Therefore, we further analyzed the association between PDCD1 expression levels and the IDH mutation status in glioma. IDH wild-type glioma displayed higher PDCD1 expression level than IDH-mutant glioma in the two CGGA datasets (Figure 1B). Additionally, receiver operating characteristic (ROC) curve analysis was harnessed to assess the performance of PDCD1 gene expression in IDH wild-type patients. The area under curves (AUCs) were 74.6%, 78.3% and 78.1% in the CGGA RNA-seq, CGGA microarray and TCGA datasets, respectively (Figure S1B). These results indicated that PDCD1 upregulation was significantly associated with the subtype of IDH wild-type glioma and PDCD1 may act as a predictor of IDH wild-type glioma. Moreover, the upregulation of PDCD1 gene expression in GBM and IDH wild-type glioma was also confirmed in the TCGA dataset (Figure 1A and B), as well as in our own cohort (Figure 1C and D). Taken together, these results indicated that PD-1 was a potential biomarker for GBM and IDH wild-type glioma.
Enrichment of PDCD1 in Mesenchymal GBM
To investigate the molecular expression profile of PDCD1, we analyzed the distribution of PDCD1 expression in different molecular subtypes of GBM. PDCD1 was significantly upregulated in the mesenchymal subtype compared with the other subtypes in the CGGA and TCGA datasets (Figure 2A), except for the classical subtype in the CGGA microarray dataset, probably due to the small sample size (Figure 2A). To further confirm these findings, ROC analysis was performed to investigate the potential diagnostic value of PDCD1 expression in mesenchymal GBM and the AUCs were 75.6%, 78.3%, and 85.1% for the CGGA mRNA-seq, CGGA microarray, and TCGA datasets, respectively (Figure 2B). These results showed that PD-1 was selectively enriched in the mesenchymal subtype, which is the most malignant subtype in GBM, and was a potential predictor for this group of GBM.
Correlation Between PDCD1 and PDCD1-Related Regulatory Genes in Glioma
Since PDCD1 was upregulated in the more malignant subtypes of gliomas, these data facilitated the investigation of the regulatory mechanism of PDCD1 in glioma. Thus, we accessed the association between PDCD1 and regulatory genes, including transforming growth factor beta 1 (TGFB1), T-box transcription factor 21 (TBX21), and special AT-rich sequence-binding protein 1 (SATB1), which are involved PDCD1 regulation in T cells under different disease conditions.12–15 Pearson’s correlation analysis was performed for both the CGGA and TCGA databases. In accord with the results in prostate cancer,12 TGFB1 was positively correlated with PDCD1 in both glioma and GBM in the CGGA and TCGA datasets (Figures S2A–2F). In virus-specific CD8+ T cells, TBX21 potentially suppresses PDCD1 expression.13,14 Nevertheless, a marginal association between PDCD1 and TBX21 was observed in glioma from the CGGA and TCGA databases (Figures S2A–2F), indicating that TBX21 had a limited effect on PDCD1 expression in glioma. In a mouse model of Lewis lung carcinoma, Satb1 was verified to suppress Pdcd1 expression during T cell activation.15 Intriguingly, in glioma and GBM, a remarkably negative relationship between SATB1 and PDCD1 was observed in the CGGA and TCGA datasets, except for GBM in the TCGA database (Figures S2A–2F). Thus, PD-1 was potentially upregulated by TGFB1 and downregulated by SATB1 in glioma, while the regulatory function of TBX21 in glioma remains obscure.
PDCD1 Was Synergistic with Immune-Related Molecules in Glioma
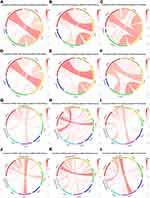
The PDCD1 checkpoint pathway is obviously associated with costimulatory and coinhibitory molecules in several solid tumors,7 thus prompting the investigation of the association between PDCD1 and immune-related genes in glioma. PDCD1 displayed a positive correlation with certain CD28 gene family members, including CD28, CTLA4, and inducible T-cell costimulatory (ICOS), both in the CGGA RNA-seq dataset (Figure 3A) and the CGGA microarray dataset (Figure 3B). In line with the CGGA datasets, the positive association between PDCD1 and these immune genes was confirmed in the TCGA database (Figure 3C). Furthermore, similar results were obtained in GBM samples from the CGGA and TCGA datasets (Figure 3D–F).
With respect to inhibitory immune checkpoints, PDCD1 displayed high concordance with T-cell immunoglobulin domain and mucin domain 3 (TIM-3), T-cell immunoglobulin and immunoreceptor tyrosine-based inhibition motif domains (TIGIT), and lymphocyte activation gene-3 (LAG3) in gliomas from the CGGA and TCGA datasets (Figure 3G–I). The obvious heterogeneity according to different grades of glioma facilitated the investigation of the associations between PDCD1 and these immune checkpoint genes in GBM, which is the most malignant grade in glioma. As expected, these immune checkpoint genes displayed an association with PDCD1 in GBM samples from the CGGA and TCGA datasets (Figure 3J–L). Furthermore, we assessed the correlation between PDCD1 and other CD28-B7 family genes. PDCD1 was closely associated with its primary ligands CD274 (also known as: programmed cell death 1 ligand 1, PD-L1), programmed cell death 1 ligand 2 (PDCD1LG2), and the costimulatory molecules CD80 and CD86 in diffuse glioma from the CGGA and TCGA datasets (Figures S3A–3C). Moreover, similar results were obtained in GBM from the CGGA and TCGA datasets (Figures S3D–3F). Together, these findings indicated that PD-1 plays an important role in costimulatory and coinhibitory immune responses in glioma, and acts as a cardinal immune regulatory molecule in glioma.
PD-1-Related Immune Responses in Glioma
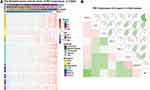
To assess the PD-1-associated immune characteristics in glioma, gene sets associated with the immune response (http://amigo.geneontology.org/amigo/landing) were refined. The genes primarily associated with PDCD1 (Pearson |r| >0.4) were identified in the CGGA RNA-seq (Figure 4A) and TCGA RNA-seq (Figure 4B) datasets, respectively. GO analysis in DAVID bioinformatics resources 6.8 was performed to decipher the biological functions of these genes. As a result, genes positively associated with PDCD1 were significantly enriched in the immune response, inflammatory response, regulation of the immune response, IFN-γ-mediated signaling pathway, T cell costimulation, and T cell activation in the CGGA datasets (Figure 4C, left). Similar outcomes were also verified in the TCGA dataset (Figure 4C, right). These results revealed that PD-1 largely participates in immune regulation in glioma, especially in T cell-mediated immune responses.
Association Between PD-1 and Inflammatory Activities
Inflammation was closely associated with the initiation, progression and metastasis in many tumors16,17 and we also found that PDCD1-related genes were closely associated with inflammatory responses in glioma (Figure 4). To further understand the association between PDCD1 and inflammatory activities, seven clusters of metagenes, serving as surrogate markers for different immune cell types and representing different types of inflammation and immune responses,18 were analyzed. PDCD1 expression was significantly associated with most of the seven clusters in the CGGA database, except for IgG which was tightly associated with the activity of B lymphocytes (Figure 5A). To validate these results, seven metagenes were generated with outcomes from gene set variation analysis (GSVA) of the corresponding gene clusters. Corrgrams were derived based on Pearson’s r values between PDCD1 and the seven metagenes (Figure 5B). PDCD1 was positively associated with LCK, MHC-II, HCK, and STAT1 and negatively associated with IgG and IFN-γ, as observed in Figure 5A. Similar results were confirmed in the TCGA database (Figures S4A–4B). Together, these findings revealed that PD-1 was deeply involved the regulation of inflammatory responses in glioma.
Highly Expressed PDCD1 Predicted Worse Survival in Glioma
To address the prognostic implication of PD-1 in all grades of gliomas, survival differences of 1323 glioma patients were analyzed via the Kaplan–Meier method. PDCD1 upregulation significantly decreased the overall survival time of glioma patients in the CGGA RNA-seq (p < 0.0001), CGGA microarray (p = 0.0012) and TCGA RNA-seq (p < 0.0001) datasets (Figure 6A–C). Similarly, high expression of PDCD1 indicated a worse prognosis in GBM patients in the CGGA RNA-seq (p = 0.0002), CGGA microarray (p = 0.0100) and TCGA RNA-seq (p = 0.017) datasets (Figure 6D–F). Additionally, patients with high expression of PDCD1 suffered a shorter survival time than patients with low expression of PDCD1 in all grades of glioma in our own cohort (p < 0.0001, Figure 6G). These findings indicated that PD-1 was a negative prognostic marker in glioma and GBM.
Discussion
Owing to the absence of improvement in glioma treatment for several decades, glioma remains the most common and fatal disease in brain tumors.1 Moreover, brain tumor associated mortality has increased slightly according to the newest epidemiological data.19 Thus, the development of novel therapeutic approaches to improve the survival of glioma patients is urgently needed. Increasing evidence indicates that the immunosuppressive tumor microenvironment results in the evasion of glioma cells from the eradication by immunologic effector cells,20 indicating that immunotherapy is a promising strategy to retard tumor progression and increases patient survival.21 Recently, encouraging results have been obtained in preclinical studies and clinical trials involving in treatment with immune checkpoint blockade in glioma,22 thus indicating the potential of immunotherapy in glioma.
PDCD1 is a member of the CD28 gene family and has become one of the most distinguished targets in anti-tumor immunotherapy for numerous advanced solid tumors, excluding glioma.23 Although aberrant PDCD1 expression in glioma has been previously reported previously, larger samples are essential to comprehensively assess the entire expression profile of PDCD1 in glioma. Therefore, we performed the largest and most comprehensive study characterizing the expression pattern and distribution of PDCD1 in 1396 glioma samples using the CGGA and TCGA datasets, as well as our own patient cohort. PDCD1 was significantly upregulated in GBM in comparison to grade II and III gliomas. PDCD1 was strongly enriched in the mesenchymal subtype of GBM, which is the most malignant subset of GBM.24 Moreover, PDCD1 was significantly upregulated in IDH wild-type glioma patients, which had a worse prognosis than IDH-mutant glioma patients, indicating that IDH potentially contributed to PDCD1 upregulation. These results indicated that PD-1 serves as a marker for more malignant subtypes in glioma, and might be a feasible therapy target for glioma.
PDCD1 promoter methylation has been considered as a beneficial prognostic factor in lower-grade glioma.9 In this study, we found that PDCD1 promoter methylation levels were much lower in GBM than in grade II and III gliomas in the TCGA database, implying that PDCD1 upregulation in GBM may partially result from PDCD1 promoter demethylation. PDCD1 promoter methylation levels in GBM marginally decreased compared to those in grade II and III gliomas in the CGGA RNAseq dataset, which might be due to a relatively small number of samples with a messy distribution. Regarding the upstream regulation of PDCD1, our findings indicated that PDCD1 expression could be positively and negatively regulated by TGFB1 and SATB1 in glioma, respectively, while the regulatory function of TBX21 remained unclear. Hence, promoter methylation, TGFB1, and SATB1 potentially regulated PDCD1 expression in glioma.
As a well-known immune checkpoint molecule, the association between PD-1 and other immune checkpoint members in glioma is still unknown. In this study, the association between PDCD1 and CD28 family genes was analyzed, and the results revealed that PDCD1 was positively associated with CD28, CTLA4, and ICOS. Since CD28 is a primary target for PD-1-mediated inhibition in the tumor milieu,25,26 the high relevance between CD28 and PDCD1 in glioma implies a promising anti-tumor effect of PD-1 blockade therapy. Unfortunately, PD-1 inhibition therapy in combination with bevacizumab had limited survival benefits in comparison with bevacizumab treatment alone in recurrent high-grade glioma.27 Moreover, a Phase III clinical trial (registration no. NCT02017717, clinicaltrials.gov) demonstrated anti-PD-1 (nivolumab) did not prolong overall survival of patients with recurrent GBM, and the trial was prematurely closed.28 There are some reasons for the unsatisfactory anti-GBM effects of anti-PD-1 therapy, such as immunosuppressive molecular characteristics of the tumor microenvironment,29 PTEN mutations,30 and additional effort is needed to dissect the tolerance mechanisms.31 Since single immune checkpoint blockade rarely obtained optimal tumor control, combinatorial treatment with different checkpoint inhibitors was rapidly developed in some tumors.32 Additionally, Wei et al has reported the distinct regulatory mechanisms between anti-CTLA4 and anti-PD-1 checkpoint inhibition in melanoma.33 A marked association between PDCD1 and CTLA4 in glioma indicated that combinatorial inhibition with these two checkpoints would potentially yield a synergetic therapeutic effect in glioma. Interestingly, it’s reported that combination therapy with anti–CTLA-4 plus anti–PD-1 cured 75% of the animals with established intracranial GL261 glioblastoma, including those with advanced-stage tumours.34 Unfortunately, instead of the inspiring results in mouse model, the combination of anti-CTLA-4 (Ipilimumab) and anti-PD-1 (nivolumab) did not acquire more improvement than anti-PD-1 alone in GBM patients in the phase III clinical trial mentioned above.28 Accordingly, combinatorial inhibition of PD-1 with CTLA-4 may be not worth considering in glioma patients. Koyama et al has reported that TIM-3 upregulation after PD-1 inhibition was a key mechanism underlying adaptive resistance to anti-PD-1 treatment in lung adenocarcinoma,35 suggesting that combinatorial treatment with TIM-3 inhibitor may overcome the resistance of anti-PD-1 therapy. Indeed, combinatorial treatment with immune checkpoint inhibitors of PD-1 and TIM-3,36 and PD-1 and TIGIT37 have shown prominent anti-tumor activity and increased the survival of glioma-bearing mice, indicating that combinatorial treatment with different immune checkpoint inhibitors based on PD-1 inhibition could be a promising therapy option for glioma patients.
The crosstalk between cancer and inflammation largely enhances tumor progression and diminishes the anti-tumor therapy efficacy.17 PD-1 predominantly causes T cell inhibition and exhaustion38 and facilitates immune evasion of glioma cells.21,39 GO analysis revealed that PDCD1-related genes were deeply involved in the inflammation response and the T cell-associated immune response, indicating that PD-1 potentially boosted tumor progression by regulating tumor-associated inflammation and T cell-related immune responses in glioma.
Furthermore, the present study showed that PDCD1 upregulation predicted a significantly worse prognosis among glioma and GBM patients. The significant prognostic value of PD-1 further implied that immunotherapy based on PD-1 inhibition could improve the prognosis of glioma patients.
The limitation of this study was that some noise in the databases would have inevitably influenced our findings and conclusions; hence, the present results require further validation through well-designed molecular biology assays.
Conclusions
In conclusion, PD-1 upregulation predicted more malignant subtypes and indicated a worse prognosis in glioma. PDCD1 was deeply involved in tumor-associated inflammation and T cell-related immune responses in glioma. Immunotherapy of targeting PD-1 or simultaneously combined with other checkpoint molecule (eg TIM-3, LAG-3 or TIGIT) inhibition represents a promising treatment strategy for glioma patients in the future.
Abbreviations
CGGA, Chinese Glioma Genome Atlas; TCGA, The Cancer Genome Atlas; OS, overall survival; GO, gene ontology; GBM, glioblastoma; GSVA, Gene Set Variation Analysis; CTLA4, cytotoxic T lymphocyte-associated antigen 4; TIM3, T-cell immunoglobulin domain and mucin domain 3; LAG3, Lymphocyte activation gene 3; TIGIT, T-cell immunoglobulin and ITIM domains.
Ethics Statement
This study was conducted according to the ethical standards of the Declaration of Helsinki and was reviewed and approved by the Ethics Committee of First Affiliated Hospital of Zhengzhou University.
Acknowledgment
We thank TCGA and CGGA project teams for their work and participation. We thank the staff and students of the Biotherapy Center for the valuable help in this study and thank Editage for English language editing.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre J-Y. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. doi:10.1016/S0140-6736(11)61346-9
2. Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma–are we there yet? Neuro Oncol. 2013;15(1):4–27. doi:10.1093/neuonc/nos273
3. Weber J, Mandala M, Del Vecchio M, et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N Engl J Med. 2017;377(19):1824–1835. doi:10.1056/NEJMoa1709030
4. Antonios JP, Soto H, Everson RG, et al. PD-1 blockade enhances the vaccination-induced immune response in glioma. JCI Insight. 2016;1(10). doi:10.1172/jci.insight.87059.
5. Dejaegher J, Verschuere T, Vercalsteren E, et al. Characterization of PD-1 upregulation on tumor-infiltrating lymphocytes in human and murine gliomas and preclinical therapeutic blockade. Int J Cancer. 2017;141(9):1891–1900. doi:10.1002/ijc.30877
6. Davidson TB, Lee A, Hsu M, et al. Expression of PD-1 by T cells in malignant glioma patients reflects exhaustion and activation. Clin Cancer Res. 2019;25(6):1913–1922. doi:10.1158/1078-0432.CCR-18-1176
7. Boussiotis VA, Longo DL. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375(18):1767–1778. doi:10.1056/NEJMra1514296
8. Hodges TR, Ott M, Xiu J, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19(8):1047–1057. doi:10.1093/neuonc/nox026
9. Rover LK, Gevensleben H, Dietrich J, et al. PD-1 (PDCD1) promoter methylation is a prognostic factor in patients with diffuse lower-grade gliomas harboring isocitrate dehydrogenase (IDH) mutations. EBioMedicine. 2018;28:97–104. doi:10.1016/j.ebiom.2018.01.016
10. Berghoff AS, Kiesel B, Widhalm G, et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 2017;19(11):1460–1468. doi:10.1093/neuonc/nox054
11. Olar A, Wani KM, Alfaro-Munoz KD, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129(4):585–596. doi:10.1007/s00401-015-1398-z
12. Donkor MK, Sarkar A, Savage PA, et al. T cell surveillance of oncogene-induced prostate cancer is impeded by T cell-derived TGF-beta1 cytokine. Immunity. 2011;35(1):123–134. doi:10.1016/j.immuni.2011.04.019
13. Kao C, Oestreich KJ, Paley MA, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12(7):663–671. doi:10.1038/ni.2046
14. Taylor A, Harker JA, Chanthong K, Stevenson PG, Zuniga EI, Rudd CE. Glycogen synthase kinase 3 inactivation drives T-bet-mediated downregulation of co-receptor PD-1 to enhance CD8(+) cytolytic t cell responses. Immunity. 2016;44(2):274–286. doi:10.1016/j.immuni.2016.01.018
15. Stephen TL, Payne KK, Chaurio RA, et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity. 2017;46(1):51–64. doi:10.1016/j.immuni.2016.12.015
16. Shalapour S, Karin M. Pas de Deux: control of anti-tumor immunity by cancer-associated inflammation. Immunity. 2019;51(1):15–26. doi:10.1016/j.immuni.2019.06.021
17. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi:10.1038/nrc3611
18. Rody A, Holtrich U, Pusztai L, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi:10.1186/bcr2234
19. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
20. Mangani D, Weller M, Roth P. The network of immunosuppressive pathways in glioblastoma. Biochem Pharmacol. 2017;130:1–9. doi:10.1016/j.bcp.2016.12.011
21. Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11(9):504–514. doi:10.1038/nrneurol.2015.139
22. Boussiotis VA, Charest A. Immunotherapies for malignant glioma. Oncogene. 2018;37(9):1121–1141. doi:10.1038/s41388-017-0024-z
23. Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer. 2018;6(1):8. doi:10.1186/s40425-018-0316-z
24. Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers. 2015;1(1):15017. doi:10.1038/nrdp.2015.17
25. Hui E, Cheung J, Zhu J, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355(6332):1428–1433. doi:10.1126/science.aaf1292
26. Kamphorst AO, Wieland A, Nasti T, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. 2017;355(6332):1423–1427. doi:10.1126/science.aaf0683
27. Kurz SC, Cabrera LP, Hastie D, et al. PD-1 inhibition has only limited clinical benefit in patients with recurrent high-grade glioma. Neurology. 2018;91(14):e1355–e1359. doi:10.1212/WNL.0000000000006283
28. Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8(53):91779–91794. doi:10.18632/oncotarget.21586
29. Wang X, Guo G, Guan H, Yu Y, Lu J, Yu J. Challenges and potential of PD-1/PD-L1 checkpoint blockade immunotherapy for glioblastoma. J Exp Clin Cancer Res. 2019;38(1):87. doi:10.1186/s13046-019-1085-3
30. Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi:10.1038/s41591-019-0349-y
31. Caccese M, Indraccolo S, Zagonel V, Lombardi G. PD-1/PD-L1 immune-checkpoint inhibitors in glioblastoma: a concise review. Crit Rev Oncol Hematol. 2019;135:128–134. doi:10.1016/j.critrevonc.2018.12.002
32. Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14(8):561–584. doi:10.1038/nrd4591
33. Wei SC, Levine JH, Cogdill AP, et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 2017;170(6):1120–1133 e1117. doi:10.1016/j.cell.2017.07.024
34. Reardon DA, Gokhale PC, Klein SR, et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol Res. 2016;4(2):124–135. doi:10.1158/2326-6066.CIR-15-0151
35. Koyama S, Akbay EA, Li YY, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7(1):10501. doi:10.1038/ncomms10501
36. Kim JE, Patel MA, Mangraviti A, et al. Combination therapy with anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin Cancer Res. 2017;23(1):124–136. doi:10.1158/1078-0432.CCR-15-1535
37. Hung AL, Maxwell R, Theodros D, et al. TIGIT and PD-1 dual checkpoint blockade enhances antitumor immunity and survival in GBM. Oncoimmunology. 2018;7(8):e1466769. doi:10.1080/2162402X.2018.1466769
38. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi:10.1038/nri3862
39. Xue S, Hu M, Iyer V, Yu J. Blocking the PD-1/PD-L1 pathway in glioma: a potential new treatment strategy. J Hematol Oncol. 2017;10(1):81. doi:10.1186/s13045-017-0455-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.