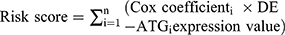Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
Competing Endogenous RNA (ceRNA) Network Analysis of Autophagy-Related Genes in Hepatocellular Carcinoma
Authors Yang C , Wang Y, Xue W, Xie Y, Dong Q, Zhu C
Received 13 June 2020
Accepted for publication 7 September 2020
Published 13 October 2020 Volume 2020:13 Pages 445—462
DOI https://doi.org/10.2147/PGPM.S267563
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Chenyu Yang,1,2,* Yixiu Wang,3 Weijie Xue,4 Yuwei Xie,3 Qian Dong,1,2,* Chengzhan Zhu2,3,*
1Department of Pediatric Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266003, People’s Republic of China; 2Shandong Provincial Key Laboratory of Digital Medicine and Computer-Assisted Surgery, Qingdao 266003, People’s Republic of China; 3Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266003, People’s Republic of China; 4Department of Gastrointestinal Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266003, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qian Dong
Department of Pediatric Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266003, People’s Republic of China
Email [email protected]
Chengzhan Zhu
Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266003, People’s Republic of China
Email [email protected]
Purpose: Autophagy plays an important role in the occurrence and development of hepatocellular carcinoma (HCC). We aimed to develop an autophagy-related genes signature predicting the prognosis of HCC and to depict a competing endogenous RNA (ceRNA) network.
Methods: Differentially expressed autophagy-related genes (DE-ATGs), miRNAs and lncRNAs and clinical data of HCC patients were extracted from TCGA. The GO and KEGG analysis were performed to investigate the gene function. Univariate and multivariate Cox regression analysis were used to identify a prognostic signature with the DE-ATGs. And a nomogram, adapted to the clinical characteristics, was established. Then, we established a ceRNA network related to autophagy genes.
Results: We screened out 27 differentially expressed genes which were enriched in GO and KEGG pathways related to autophagy and cancers. In univariate and multivariate Cox regression analysis, BIRC5, HSPB8, and SQSTM1 were screened out to establish a prognostic risk score model (AUC=0.749, p< 0.01). Kaplan–Meier survival analysis showed that the overall survival of high-risk patients was significantly worse. Furthermore, the signature was validated in the other two independent databases. The nomogram, including the autophagy-related risk signature, gender, stage and TNM, was constructed and validated (C-index=0.736). Finally, the ceRNA network was established based on DE-ATGs, differentially expressed miRNAs and lncRNAs.
Conclusion: We constructed a reliable prognostic model of HCC with autophagy-related genes and depicted a ceRNA network of DE-ATGs in HCC which provides a basis for the study of post-transcriptional modification and regulation of autophagy-related genes in HCC.
Keywords: competing endogenous RNA, ceRNA, autophagy-related genes, hepatocellular carcinoma, HCC, TCGA
Introduction
Hepatocellular carcinoma (HCC) is one of the most common liver tumors and has been the fourth most common cause of cancer-related deaths in the world.1–3 Although great efforts have been taken to prevent, diagnose and treat HCC, but the morbidity and mortality of HCC are still high.1,4,5 When a series of genetic and epigenetic events occurs in chronic liver disease, normal hepatocyte will gradually acquire the abilities of proliferation, migration, and invasion, becoming cancer cells.4,6 The molecular mechanism of developing HCC involves many classical cancer-related pathways, including WNT pathway, p53 pathway, and apoptosis signal pathway, etc.7 In recent years, autophagy has been found to play important roles in the occurrence, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma.8
Autophagy is a highly conservative biological process in eukaryote evolution that plays an important role in regulating metabolism, recycling of intracellular substances and maintaining homeostasis of the internal environment.9 The vesicles in the cytoplasm wraps and isolates the proteins to be degraded, and the destroyed or aged organelles form autophagosomes, which eventually fuse with lysosomes, leading to the degradation of isolated components.9,10 Disorders of autophagy can lead to a variety of diseases, including cancer as HCC,8 breast cancer11 and lung cancer.12 Autophagy is not only a constitutive reaction, but also a result of adaptability.9 The main function of the former is to remove damaged or aged organelles and maintain basic material and energy balance but the latter is characterized by the mobilization of the intracellular material cycle to meet material or energy requirements when nutrients are deficient.9,10 Thus, autophagy, like a double-edged sword, plays different roles in different stages of cancer. Basic autophagy inhibits tumor initiation and early stage development by maintaining the cell genomic stability and removing heterogeneous cells. In contrast, activated autophagy contributes to the survival of cancer cells and promotes the development of cancer.8,10,13 Previous studies have explored the role of some autophagy-related genes (ATGs) in the occurrence and development of HCC.14–17 But there are a lack of studies dealing with the mechanism of autophagy in the occurrence and development of cancer from a global view. So, we not only studied the relationship between autophagy and HCC from the perspective of DE-ATGs, but also predicted ceRNA to explain the relationship between autophagy and HCC.
Competing endogenous RNA (ceRNA) is a general term for a class of RNA that can bind with shared miRNAs and cross-regulate each other at post-transcription level.18 CeRNAs are a group of non-coding RNA, such as lncRNAs, pseudogenic RNAs, and circular RNAs.18,19 At present, mRNA-miRNA-lncRNA network is mostly studied in the ceRNA network. MiRNA is an important factor in regulating gene expression which promotes the degradation of mRNA to prevent translation, and then inhibiting gene expression.20 CeRNA, like a sponge, can competitively bind and enrich miRNA.18,19 Thus, ceRNA promotes gene expression by reducing the binding of miRNA and mRNA to protect mRNA from degradation. Some studies have focused on the relationship between ceRNAs and autophagy in HCC.21–24 Therefore, we were committed to building a ceRNA network related to autophagy in order to better understand the role of autophagy in HCC.
Differently expressed genes have been demonstrated to have advantages to use for the early detection of HCC and predicting prognosis.1 Many single genes have been reported as predictors of HCC, but a gene as a predictor cannot fully describe the characteristics of tumors. Cancer, as a disease involving multi-gene changes, compared with a single gene, means the multi-gene expression pattern can be used as a good molecular biomarker in HCC, and could provide potential treatment targets.
Here, we explored the DE-ATGs and differentially expressed miRNA and lncRNA in HCC. Three signature genes were screened from DE-ATGs to develop a prognostic model and construct a nomogram with clinicopathologic features. Finally, we selected the differentially expressed miRNA and lncRNA which were related to the DE-ATGs to construct a ceRNA network to better explore the role and position of autophagy in the occurrence and development of HCC.
Materials and Methods
Acquisition and Collation of Datasets
The 424 gene expression datasets (374 tumor tissues and 50 non-tumor samples) and clinical information of 377 HCC patients were obtained from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and the gene expression datasets and clinical information of 231 HCC patients were obtained from the International Cancer Genome Consortium (ICGC) (https://icgc.org/). A total of 232 autophagy-related genes were identified in the Human Autophagy Database (HADb, http://www.autophagy.lu/index.html).
High Throughput Sequencing of Tissue Samples and Patient Follow-Up
A total of 39 patients diagnosed as HCC underwent hepatectomy between May 2017 and May 2018 at the Department of Hepatobiliary and Pancreatic Surgery of the Affiliated Hospital of Qingdao University were enrolled.
Total RNAs of samples were extracted from fresh frozen tissues using RNeasy Mini kit (QIAGEN’s RNeasy kit; Qiagen, Valencia, CA) and isolated using NEBNext rRNA Depletion Kit (NEB, Ipswich, MA, USA). Complementary DNA (cDNA) was synthesized from RNA by reverse transcription and frozen to −20 °C. Libraries were constructed using KAPA HyperPrep Kit (KAPA Biosystems, Roche, USA) and then sequenced by a NovaSeq 6000 system (Illumina, San Diego, CA, USA).
We conducted this study in strict compliance with the Helsinki Declaration principles. And the study was allowed and approved by the Institutional Ethics Committee of the Affiliated Hospital of Qingdao University (Ethics ID: QYFYWZLL25880; Qingdao, China). Informed consent was obtained from patients and their families before surgery and agreements were signed by them.
Functional Enrichment Analysis of Differently Expressed Autophagy-Related Genes (DE-ATGs)
The expression level of the 232 ATGs was obtained from gene expression datasets, and the DE-ATGs were analyzed between HCC tumor and non-tumor samples. DE-ATGs were screened with the Limma package by R software (https://www.r-project.org/). In this study false discovery rate (FDR) < 0.05 and a log2 |fold change| > 1.5 were set as the cutoff values. 27 DE-ATGs were screened out, including two down-expression genes and 25 up-expression genes. After that, gene functional enrichment analyses were performed to identify the major biological function, including the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. The DOSE, clusterProfiler and enrichplot packages were used to visualize the enrichment terms. The Z-score of both GO analysis and KEGG were >0, indicating that the expression of the DE-ATGs enriched in these biological functions and pathways were mainly up-regulated, and <0 indicating down-regulated.
Construction of the Autophagy-Related Prognostic Risk Model in the Training Cohort
The data obtained from TCGA was taken as the training cohort. The survival times and status of the patients were extracted and combined with the DE-ATGs for analysis by univariate Cox regression analysis and multivariate Cox regression analysis. HR >1 was defined as a high risk gene and HR <1 was defined as low risk gene, p<0.05. Three DE-ATGs, BIRC5, HSPB8, and SQSTM1, were identified as an autophagy-related risk signature and multivariate Cox regression coefficient was used to weight the expression values of the three DE-ATGs correspondingly. The prognosis index (PI) or risk score for every HCC patient was calculated with a weighted Sum Method. According to the PI, HCC patients were divided into a high-risk group and a low-risk group for follow-up analysis and study.
Development and Validation of the Nomogram
Based on the results of multivariate analysis, six independent prognostic parameters, including autophagy-related risk signature, gender, stage and TNM stages, were enrolled in the nomogram model to assess the 1-, 3- and 5-year survival for TCGA patients by rms R package (https://cran.r-project.org/web/packages/rms/). The prediction ability of the nomogram was measured by concordance index (C-index) and the discriminant ability of the nomogram was evaluated graphically by calibration curve.
Depiction of ceRNA Network
Differentially expressed miRNA and lncRNA were screened from TCGA, and |log FC|≥1.5 and FDR<0.05 were the cutoff values. The target miRNAs of differential lncRNAs were predicted in the miRcode (http://www.mircode.org/). The target mRNAs of the target miRNAs were predicted in three miRNA databases, miRDB (http://www.mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) and TargetScan (http://www.targetscan.org/). The target mRNA was considered a valid target only if it appears in all three databases simultaneously. Then, screened autophagy-related mRNA from target mRNAs. Finally, the lncRNA-miRNA and miRNA-mRNA relationship pairs were input into open-source Cyotoscape 3.6.0 (http://www.cytoscape.org/) to establish a ceRNA network.
Statistical Analyses
Statistical analysis was performed by R programming language of R software and SPSS 23.0, and p < 0.05 was considered statistically significant. Student’s t-test was used to assess differences between variables. Kaplan–Meier analysis and Log rank tests were used to assess differences in patient overall survival. We used the online tool Kaplan–Meier Plotter (http://kmplot.com) to analyze the survival of a single gene which auto selects the best cutoff to perform the analysis.25 To identify the independent prognostic factor, univariate and multivariate prognostic analyses were done by using Cox regression analysis. The accuracy of the prognostic risk model of autophagy-related signature was evaluated with ROC analysis. The area under the curve (AUC) of the survival ROC curve was calculated via the survival ROC package of R. The diagnostic ability of ROC was lower when AUC was 0.5~0.7, moderate at 0.7~0.9 and higher when it was greater than 0.9.
Result
Differentially Expressed Autophagy-Related Genes (DE-ATGs) and Function Enrichment Analysis in HCC
The gene expression profiles of 424 samples were downloaded from the TCGA database, composed of 50 normal samples and 374 tumor samples. R language was used to extract the expression values of the 232 autophagy-related genes in each expression profile. Comparing the expression between tumor and normal tissue, 205 DE-ATGs were obtained (Figure 1A). Then we set the cutoff values as FDR < 0.05 and logFC ≥ ±1.5, only 27 DE-ATGs were screened out. Among them, two genes were down-regulated and 25 genes were up-regulated (Figure 1B and C).
Functional enrichment of the 27 DE-ATGs was performed by GO terms and KEGG pathway analysis. In GO terms analysis, except autophagy and autophagy-related events, the apoptotic signaling pathway was mostly enriched (Figure 2). In KEEG analysis, the genes shown were notably associated with the pathways in apoptosis, platinum resistance, P53 signaling pathway and PI3K-Akt signaling pathway, etc. (Figure 3).
Identification of the Candidate DE-ATGs and Single Gene Prognostic Analysis
We explored the prognosis related DE-ATGs in the 377 HCC patients from TCGA. Twelve DE-ATGs related to prognosis were screened out from the 27 DE-ATGs by univariate Cox regression analysis (p < 0.05) (Figure 4A). The interrelationships among these 12 genes were explored, demonstrating that these genes interact in platinum drug resistance, pathways in cancer, hepatitis B, etc. (Figure 4B). Further, 3 genes (BIRC5, HSPB8, SQSTM1) were screened out as independent prognostic factors using multivariate Cox regression analysis (Figure 4C).
Single Gene Prognostic Analysis of the Three DE-ATGs
In order to rule out that a single gene can be used as an independent prognostic factor and have better predictive ability than our model, we decided to conduct a single gene analysis of three differential genes before constructing a multi-gene prognostic model. Significantly different expressions of the three genes existed between non-tumor tissue and tumor tissue (Figure 5A).
 |
Figure 5 The three genes expression in tumor and non-tumor tissues and survival analysis. (A) The three genes expression level in tumor and normal tissues. (B) The K–M curves of the three genes. |
Kaplan–Meier survival analyses showed that after automatically matching the best cutoff value there was a significant negative correlation between each of the three genes and survival rate, respectively (Figure 5B). Multivariate Cox regression analysis was performed according to the three genes and 7 clinicopathologic features (age, gender, grade, stage, T stage, N stage and M stage) respectively. Results of the analysis indicated that only BIRC5 and SQSTM1 could be used as independent predictors of the prognosis of patients with HCC (Figure 6A–C).
 |
Figure 6 BIRC5 and SQSTM1 could be used as independent predictors. (A–C) The forest plot of BIRC5 and SQSTM1. (D, E) The ROC curves of BIRC5 and SQSTM1. |
By combining clinical data with gene expression, 370 eligible HCC patients were selected from TCGA. According to the survival time from low to high, and with the median survival time as the demarcation value, this group was divided into two subgroups: high risk group (n = 185) and low risk group (n = 185). The predictive effect of BIRC5 and SQSTM1 on HCC was evaluated by the ROC curve, and the AUC of BIRC5 and SQSTM1 was 0.597 (p = 0.0013) and 0.590 (p = 0.0028), respectively (Figure 6D and E).
We further investigated the correlation of BIRC5 and SQSTM1 expression with the clinical characteristics of patients. BIRC5 was related to survival status, grade, staging, and T staging (Figure 7A–D). And the expression level of SQSTM1 was significantly different in patients of different ages and genders, and affected the survival outcome (Figure 7E–G). We thought that the BIRC5 expression of StageIV is lower than those of StageI–III because the number of patients at StageIV is very little, which may be related to the fact that patients at StageIV are not suitable to operate and fewer samples of tissues are obtained. A similar situation might exist in T4.
An Autophagy-Related Risk Signature for the Prognosis Model of HCC
Then we further explore the role of BIRC5, HSPB8 and SQSTM1 as a risk signature to predict the prognosis of HCC. According to the multivariate Cox regression analysis, we obtained the expression coefficients of the three genes and constructed a Prognosis Index (PI) formula: PI= (0.117212775×BIRC5 expression value) + (0.260475987 × HSPB8 expression value) + (0.306471973 × SQSTM1 expression value) (Figure 4C). Patients were divided into two groups according to the PI. The OS of the high-risk group was shorter than that of the low-risk group, the 5-year survival rate of which were 42% and 52.6%, respectively (Figure 8A). The expression of these 3 DE-ATGs was significantly increased in the high-risk group (Figure 8B). The number of deaths in the high-risk group was higher than that in the low-risk group (Figure 8C). Thus, a rising curve could be obtained by sorting patients according to the risk scores from low to high (Figure 8D). Univariate analysis indicated that the PI, stage, T and M were risk factors of prognosis (Figure 8E). Multivariate Cox regression analysis found that only the PI was an independent prognostic predictor (Figure 8F). And the AUC (0.749,p<0.01) of the PI was higher than that of the other clinicopathologic features (Figure 8G). Meanwhile, the PI also obtained a prominent AUC compared to BIRC5 or SQSTM1(Figures 8G and 6D and E).
Relationship Between Autophagy-Related Risk Signature and Clinicopathologic Features in Patients with HCC
We selected 240 patients with complete clinical data to investigate the correlation of autophagy-related risk signature with clinicopathologic features. The results showed that the autophagy-related risk signature was closely related to survival status, overall survival time, sex, grade, T stage and distant metastasis (Figure 9).
Validation of the Autophagy-Related Risk Signature via Two Independent Datasets
We firstly validated the risk score in our patient cohort (n = 39) obtained from the Affiliated Hospital of Qingdao University. Kaplan–Meier analysis showed that the high-risk patients had a shorter DFS (p = 0.022) and tended to have a shorter OS (p = 0.072) (Figure 10A and B). Because of the small amount of our data and no statistical difference in OS, we then validated the risk score in a patient cohort from the International Cancer Genome Consortium (ICGC) including 224 patients. Kaplan–Meier analysis showed that the high-risk patients had a shorter OS (p = 0.034) (Figure 10C).
Development and Validation of the Nomogram
We established a nomogram to predict the survival probability at 1-, 3-, and 5-years which could be a clinically applicable method for predicting the prognosis of HCC patients. Six independent prognostic parameters, including the autophagy-related risk signature, gender, stage and TNM stages, were enrolled in the nomogram (Figure 11A). The survival probability of 1-, 3- and 5-years can be obtained by querying the points corresponding to different prognostic parameters and calculating the total points. The C index of the nomogram model was 0.736 (95% CI, 0.68 to 0.80; p < 0.05). The calibration plots showed good and acceptable agreement between actual situations and the nomogram model predictions of the 1-, 3- and 5-years overall survival rates (Figure 11B–D).
Depiction of a ceRNA Network of Autophagy in HCC
We screened the differentially expressed miRNA and lncRNA from TCGA by |log FC|≥1.5 and FDR<0.05. A total of 172 miRNAs had been found differently expressed, 166 up- and 6 down-regulated (Figure 12A). And 1587 lncRNAs were differently expressed, 1444 up- and 143 down- regulated (Figure 12B). The target miRNAs of differently expressed lncRNAs and the target genes of differently expressed miRNAs were predicted by comparing with the database, including miRcode, miRDB, miRTarBase and TargetScan. We observed 35 DE-ATGs were targeted by the differently expressed miRNAs. And 131 lncRNAs interacted with 22 miRNAs, forming 658 lncRNA-miRNA relationship pairs. Finally, a lncRNA-miRNA-mRNA (DE-ATGs) network was constructed after intersection (Figure 12C).
Discussion
Autophagy has been demonstrated to associate with the occurrence, metastasis, targeted therapy and drug resistance of HCC.8 In this study, we explored the DE-ATGs and differentially expressed miRNA and lncRNA in HCC. Three signature genes were screened from DE-ATGs to develop a prognostic model and construct a nomogram with clinicopathologic features. Finally, we selected the differentially expressed miRNA and lncRNA which were related to the DE-ATGs to construct a ceRNA network to better explore the role and position of autophagy in the occurrence and development of HCC.
In the GO and KEGG functional analyses, biological function including apoptosis, insulin-like factor binding, death domain binding platinum drug resistance, and lung cancer were indicated other than autophagy. All of the DE-ATGs enriched in these biological functions and pathways were associated with cancer and mainly up-regulated in HCC. Univariate Cox regression analysis and multivariate Cox regression analysis identified 3 DE-ATGs (BIRC5, HSPB8 and SQSTM1) to be a prognostic signature used as an independent prognostic indicator for HCC patients. In order to better prove that the three ATGs as a gene signature are more advantageous to predict the prognosis of HCC, we performed single gene analysis of the three genes, respectively. Single gene analysis showed that BIRC5 and SQSTM1 could independently predict the prognosis of HCC. When compared with single gene analysis, the gene signature established by BIRC5, HSPB8, and SQSTM1 had a better predictive ability than single genes (the AUC of the gene signature = 0.749; the AUC of BIRC5 = 0.594; the AUC of SQSTM1 = 0.590). This was also confirmed by multivariate Cox regression analysis.
The validation of the autophagy-related risk signature in our own clinical cohort indicated that the high-risk patients had a shorter DFS. And the signature was related with a shorter OS in ICGC patients. Therefore, we have reason to believe that the gene signature composed of BIRC5, HSPB8 and SQSTM1 can be used as an independent predictor of HCC. We developed a nomogram with six independent prognostic parameters, including autophagy-related risk signature, gender, stage and T, N, M stages. The survival probability of 1-, 3- and 5-years can be obtained by querying the points corresponding to different prognostic parameters and calculating the total points.
BIRC5 encodes Survivin, which is an evolutionarily conserved eukaryotic protein and plays a key role in cell mitosis.26 BIRC5/Survivin may act as a bridge between apoptosis and autophagy, inhibiting cell death and protecting cells from apoptosis and autophagic death.26–28 Usually it only expresses in actively proliferating cells, but up-regulated in most cancers.26 The Survivin and ATG7 were negatively correlated, and it is conjectured that Survivin could inhibit the basic autophagic level of cells, promoting normal cells transforming to cancer cells.27 In HCC and prostate cancer, activation of the PI3K/AKT signaling pathway will promote the expression of BIRC5, leading to the inhibition of autophagic death.29,30 BIRC5 expression could also be induced by IGF-1 signaling to promote epithelial-mesenchymal transition.31,32 YM155, an inhibitor of Survivin, could induce autophagy-dependent apoptosis of cancer cells and could be a candidate drug of HCC.33,34 BIRC5 is also involved in drugs resistance. Melatonin could overcome drug resistance in HCC cells by suppressing Survivin via the COX-2/PI3K/AKT pathway.35,36 In addition, the expression of Survivin is not regulated only on a transcription level, but also on a post-transcriptional level.37,38 Therefore, Survivin can be used as a new serological biomarker of HCC.
SQSTM1, a conserved encoding gene of p62, is associated with HCC.39 During the autophagy process, p62 is an autophagy receptor in the form of ubiquitin - ready to participate in selective autophagy which is essential for removing damaged or excess macromolecules or organelles efficiently.39–41 The accumulation of p62 caused by autophagy damage could promote glycolysis and proliferation of HCC cells.42 Autophagy on a low-level leads to the accumulation of p62, while activated autophagy can also lead to the increase and accumulation of p62 and promote the selective autophagy of some molecules. Low level autophagy and the deletion or decreased expression of the SQSTM1 led to the inhibition of cyclin D1 degradation, which promoted the development of HCC.16 And cellular thyroid hormone (TH) promotes selective autophagy through SQSTM1, thereby protecting hepatocytes from diethylnitrosamine-induced hepatotoxicity or carcinogenesis.43 However, high protein levels of SQSTM1 have also been reported to promote the development of HCC. DDX5 (DEADboxprotein5) can induce autophagy, also promoting the degradation of p62, reducing the ubiquitination level of mTOR, and thus blocking the signaling pathway and inhibiting tumor formation.44 In addition, some studies have demonstrated that SQSTM1 could influence cancers beyond autophagy.41 Therefore, more study should be performed to clarify the function of SQSTM1 in tumorigenesis.
There were few reports about HSPB8 in HCC. In breast cancer, HSPB8 plays a role as a generalist in promoting cancer and drug resistance. HSPB8 is highly expressed in McF-7 cells, and down-regulation of HSPB8leads to decreased ability of cells to pass through restriction points and migrate.45 However, another study reported that HSPB8 could protect MCF7 cells from tamoxifen and block autophagy.46,47 So, from promoting autophagy to blocking autophagy, the mechanism regulating HSPB8 function is not yet clear, which may be through the mTOR pathway.47 HSPB8 forms complexes with BAG3, subsequently forming a chaperon complex through recruiting HSP70 and CHIP (C-terminus of Hsc70 interacting protein) and then interacts with SQSTM1/p62 promoting the autophagy process.48–50 HSPB8 could also be involved in the development and migration of cancer in a non-autophagy dependent form.51,52
All of the three genes seem to have two-sided effects, promoting and inhibiting cancer. It is more interesting and significant to study the role of a cluster of ATGs involved in cancer than a single gene. Our results indicate that the three DE-ATGs may influence the development of cancer in a special way, and provides a new method for clinicians to predict the prognosis of HCC patients, as well as a new target for the role of autophagy in HCC.
The central dogma reveals the synthesis process of protein, but some post-transcriptional modifications in DNA-RNA-protein affect the final protein expression. MiRNA can block mRNA and promote degradation of mRNA to induce diseases, including cancer.20 CeRNA is a general term for a class of RNA including lncRNAs, pseudogenic RNAs, and circular RNAs that could bind with shared miRNAs and cross-regulate with each other at a post-transcription level.18–20 In the ceRNA network, including lncRNAs, miRNAs, and mRNAs, lncRNAs as sponges can competitively bind miRNAs to indirectly regulate mRNA expression levels. Now, some studies have focused on the relationship between ceRNAs and autophagy in HCC. In cancer, lncRNA is the most common and studied ceRNA. Many lncRNAs have been identified to be related to the occurrence and development of HCC and involved in autophagy, also that lncRNAs promote the proliferation, metastasis and drug resistance of HCC cells by activating autophagy. For example, lncRNA HULC triggers autophagy and attenuates the chemosensitivity of HCC, lncRNA HULC accelerates HCC by inhibiting PTEN via autophagy cooperation to miR15a and lncRNA NEAT1 promotes autophagy and enhanced HCC cell resistance to Sorafenib.53–55 Although some studies have studied the relationship between the lncRNA-miRNA axis and HCC, there is not a good ceRNA network to explain the status and role of autophagy in HCC. So, we depicted a lncRNA-miRNA-mRNA (DE-ATGs) network including 35 DE-ATGs, 131 lncRNAs and 22 miRNAs. We hope the network could help us to better understand the role and status of autophagy in HCC.
However, this study was limited to retrospective analysis, and we did not understand the changes in autophagy flux in these patients with HCC. Although the molecular mechanism is a cause, the position and role of genes in different pathways, the interaction between pathways, epigenetics and so on will affect the final outcome. At the same time, based on the contradictory role of autophagy, both enhancement and attenuation may promote the development of cancer, so determining the balance point of autophagy is also a problem. Therefore, more fundamental and prospective studies are needed to further elucidate the role of autophagy networks in HCC.
Conclusion
In summary, molecular mechanisms play an important role in the relationship between autophagy and HCC. Our results are expected to be used to predict the prognosis of HCC patients in clinical practice and provide new ideas based on the autophagy-related risk signature and the ceRNA network for the treatment of HCC. It also provided a direction for further research. On the basis of public data, we revealed autophagy-related genes and autophagy may influence the outcome of HCC.
Data Sharing Statement
The RNA-seq data and clinical information of HCC analyzed in this study can be downloaded from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC).
Acknowledgments
The authors thank The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), and The Human Autophagy Database (HADb, http://www.autophagy.lu/index.html). The authors also want to thank Yixiu Wang and Weijie Xue for their assistance in data processing. This work was supported by the Shandong Higher Education Young Science and Technology Support Program(grant number 2020KJL005); China Postdoctoral Science Foundation (2016M602098); Young Taishan Scholars Program of Shandong Province (No.2019010668); and Qingdao Postdoctoral Science Foundation (2016046).
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
2. Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 Cancer Groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3(4):524–548. doi:10.1001/jamaoncol.2016.5688
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70.
4. Villanueva A, Longo DL. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
5. Fitzmaurice C, Akinyemiju TF, Al Lami FH, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 Cancer Groups, 1990 to 2016: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4(11):1553–1568. doi:10.1001/jamaoncol.2018.2706
6. Torrecilla S, Sia D, Harrington AN, et al. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol. 2017;67(6):1222–1231. doi:10.1016/j.jhep.2017.08.013
7. El–Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi:10.1053/j.gastro.2007.04.061
8. Huang F, Wang B-R, Wang Y-G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24(41):4643–4651. doi:10.3748/wjg.v24.i41.4643
9. Kim KH, Lee M-S. Autophagy–a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10:322–337.
10. Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124(16):3307–3318. doi:10.1002/cncr.31335
11. Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun. 2018;9(1):1944. doi:10.1038/s41467-018-04070-6
12. Yao C, Ni Z, Gong C, et al. Rocaglamide enhances NK cell-mediated killing of non-small cell lung cancer cells by inhibiting autophagy. Autophagy. 2018;14(10):1831–1844. doi:10.1080/15548627.2018.1489946
13. Li Y-J, Lei Y-H, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36(1):52. doi:10.1186/s40880-017-0219-2
14. Zhang Z, Guo M, Li Y, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16(8):1482–1505.
15. Pant K, Saraya A, Venugopal SK. Oxidative stress plays a key role in butyrate-mediated autophagy via Akt/mTOR pathway in hepatoma cells. Chem Biol Interact. 2017;273:99–106. doi:10.1016/j.cbi.2017.06.001
16. Wu S-Y, Lan S-H, Wu S-R, et al. Hepatocellular carcinoma-related cyclin D1 is selectively regulated by autophagy degradation system. Hepatology. 2018;68(1):141–154. doi:10.1002/hep.29781
17. Chen D-P, Ning W-R, Li X-F, et al. Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma. Autophagy. 2018;14(8):1335–1346. doi:10.1080/15548627.2018.1474994
18. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
19. Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
20. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi:10.1038/nrc3932
21. Yang L, Peng X, Jin H, Liu J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene. 2019;697:94–102. doi:10.1016/j.gene.2019.02.036
22. Wei H, Hu J, Pu J, et al. Long noncoding RNA HAGLROS promotes cell proliferation, inhibits apoptosis and enhances autophagy via regulating miR-5095/ATG12 axis in hepatocellular carcinoma cells. Int Immunopharmacol. 2019;73:72–80. doi:10.1016/j.intimp.2019.04.049
23. Liu Z, Wei X, Zhang A, et al. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473(4):1268–1275. doi:10.1016/j.bbrc.2016.04.054
24. Shi Y, Yang X, Xue X, et al. HANR enhances autophagy-associated sorafenib resistance through miR-29b/ATG9A axis in hepatocellular carcinoma. Onco Targets Ther. 2020;13:2127–2137. doi:10.2147/OTT.S229913
25. Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
26. Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci. 2019;132(7):jcs223826. doi:10.1242/jcs.223826
27. Lin T-Y, Chan HH, Chen SH, et al. BIRC5/survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy. 2020;16(7):1296–1313
28. Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3(1):46–54. doi:10.1038/nrc968
29. Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2011;146(1):130–147. doi:10.1002/hep.30795
30. Roca H, Varsos Z, Pienta KJ. CCL2 protects prostate cancer PC3 cells from autophagic death via phosphatidylinositol 3-kinase/AKT-dependent survivin up-regulation. J Biol Chem. 2008;283(36):25057–25073. doi:10.1074/jbc.M801073200
31. Li C, Li J, Wu D, Han G. The involvement of survivin in insulin-like growth factor 1-induced epithelial-mesenchymal transition in gastric cancer. Tumour Biol. 2016;37(1):1091–1096. doi:10.1007/s13277-015-3909-x
32. Liu F, Sun Y, Liu B, et al. Insulin-like growth factor-1 induces epithelial-mesenchymal transition in hepatocellular carcinoma by activating survivin. Oncol Rep. 2018;40:952–958.
33. Wang Q, Chen Z, Diao X, Huang S. Induction of autophagy-dependent apoptosis by the survivin suppressant YM155 in prostate cancer cells. Cancer Lett. 2011;302(1):29–36. doi:10.1016/j.canlet.2010.12.007
34. Zhang B, Wang N, Zhang C, et al. Novel multi-substituted benzyl acridone derivatives as survivin inhibitors for hepatocellular carcinoma treatment. Eur J Med Chem. 2017;129:337–348. doi:10.1016/j.ejmech.2017.02.027
35. Fan L, Sun G, Ma T, Zhong F, Wei W. Melatonin overcomes apoptosis resistance in human hepatocellular carcinoma by targeting survivin and XIAP. J Pineal Res. 2013;55(2):174–183. doi:10.1111/jpi.12060
36. Zhou J, Bi C, Janakakumara JV, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113(17):4052–4062. doi:10.1182/blood-2008-05-156422
37. Yang R, Liu M, Liang H, et al. miR-138-5p contributes to cell proliferation and invasion by targeting survivin in bladder cancer cells. Mol Cancer. 2016;15(1):82. doi:10.1186/s12943-016-0569-4
38. Jaiswal PK, Goel A, Mittal RD. Survivin: a molecular biomarker in cancer. Indian J Med Res. 2015;141:389–397.
39. Sánchez-Martín P, Saito T, Komatsu M. p62/ SQSTM 1: ‘jack of all trades’ in health and cancer. FEBS J. 2019;286(1):8–23. doi:10.1111/febs.14712
40. Lamark T, Svenning S, Johansen T, Lane JD, Korolchuk VI, Murray JT. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61(6):609–624. doi:10.1042/EBC20170035
41. Moscat J, Karin M, Diaz-Meco MT. p62 in cancer: signaling adaptor beyond autophagy. Cell. 2016;167(3):606–609. doi:10.1016/j.cell.2016.09.030
42. Jiao L, Zhang H-L, Li -D-D, et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy. 2018;14(4):671–684. doi:10.1080/15548627.2017.1381804
43. Chi H-C, Chen S-L, Tsai C-Y, et al. Thyroid hormone suppresses hepatocarcinogenesis via DAPK2 and SQSTM1-dependent selective autophagy. Autophagy. 2016;12(12):2271–2285. doi:10.1080/15548627.2016.1230583
44. Zhang H, Zhang Y, Zhu X, et al. DEAD box protein 5 inhibits liver tumorigenesis by stimulating autophagy via interaction with p62/SQSTM1. Hepatology. 2019;69(3):1046–1063. doi:10.1002/hep.30300
45. Piccolella M, Crippa V, Cristofani R, et al. The small heat shock protein B8 (HSPB8) modulates proliferation and migration of breast cancer cells. Oncotarget. 2017;8(6):10400–10415. doi:10.18632/oncotarget.14422
46. Gonzalez-Malerva L, Park J, Zou L, et al. High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc Natl Acad Sci U S A. 2011;108(5):2058–2063. doi:10.1073/pnas.1018157108
47. Shi -J-J, Chen S-M, Guo C-L, et al. The mTOR inhibitor AZD8055 overcomes tamoxifen resistance in breast cancer cells by down-regulating HSPB8. Acta Pharmacol Sin. 2018;39(8):1338–1346. doi:10.1038/aps.2017.181
48. Li F, Xiao H, Hu Z, Zhou F, Yang B. Exploring the multifaceted roles of heat shock protein B8 (HSPB8) in diseases. Eur J Cell Biol. 2018;97(3):216–229. doi:10.1016/j.ejcb.2018.03.003
49. Fuchs M, Luthold C, Guilbert SM, et al. A role for the chaperone complex BAG3-HSPB8 in actin dynamics, spindle orientation and proper chromosome segregation during mitosis. PLoS Genet. 2015;11(10):e1005582. doi:10.1371/journal.pgen.1005582
50. Gamerdinger M, Carra S, Behl C. Emerging roles of molecular chaperones and co-chaperones in selective autophagy: focus on BAG proteins. J Mol Med. 2011;89(12):1175–1182. doi:10.1007/s00109-011-0795-6
51. Shen J, Li M, Min L. HSPB8 promotes cancer cell growth by activating the ERK‑CREB pathway and is indicative of a poor prognosis in gastric cancer patients. Oncol Rep. 2018;39:2978–2986.
52. Suzuki M, Matsushima-Nishiwaki R, Kuroyanagi G, et al. Regulation by heat shock protein 22 (HSPB8) of transforming growth factor-α-induced ovary cancer cell migration. Arch Biochem Biophys. 2015;571:40–49. doi:10.1016/j.abb.2015.02.030
53. Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540.
54. Xin X, Wu M, Meng Q, et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17(1):94. doi:10.1186/s12943-018-0843-8
55. Li X, Zhou Y, Yang L, et al. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. 2020;235(4):3402–3413. doi:10.1002/jcp.29230
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.