Back to Journals » Infection and Drug Resistance » Volume 14
Comparative Genome Analysis of Livestock and Human Colistin-Resistant Escherichia coli Isolates from the Same Household
Authors Kawahara R, Yamaguchi T, Yamamoto Y
Received 18 December 2020
Accepted for publication 17 February 2021
Published 3 March 2021 Volume 2021:14 Pages 841—847
DOI https://doi.org/10.2147/IDR.S298120
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Ryuji Kawahara,1 Takahiro Yamaguchi,1 Yoshimasa Yamamoto2,3
1Division of Microbiology, Osaka Institute of Public Health, Osaka, Japan; 2Graduate School of Pharmaceutical Sciences, Osaka University, Suita, Japan; 3Life Science Research Center, Gifu University, Gifu, Japan
Correspondence: Yoshimasa Yamamoto
Life Science Research Center, Gifu University, 1-1 Yanagido, Gifu, 501-1194, Japan
Tel/Fax +81-58-230-6239
Email [email protected]
Background: Emergence and dissemination of colistin-resistant bacteria that harbor mobile colistin resistance (mcr) genes pose a dire challenge for the treatment of intractable infections caused by multidrug-resistant bacteria. Current findings on colistin-resistant bacteria in both humans and livestock of the same households highlight the need to identify the dissemination mechanisms of colistin-resistant bacteria.
Methods: In this study, a comparative genome analysis of colistin-resistant Escherichia coli isolates from livestock and humans of the same household was performed to clarify the possible dissemination mechanism of mcr genes among bacteria. Pulsed-field gel electrophoresis and whole-genome sequencing followed by sequence typing of the isolates were performed for assessment of the samples.
Results: The study revealed that two colistin-resistant E. coli isolates, one each from a pig and a chicken, were phylogenetically similar but not identical to the human isolates obtained from the same household. The comparative genome analysis revealed that the chicken isolate and a human isolate shared the same IncHl2 plasmid harboring the mcr transposon (mcr-1-PAP2). The pig isolate and the other human isolate retained the mcr-1 transposon on the chromosome, with the pig isolate carrying the complete mcr transposon (ISApl1-mcr-1-PAP2-ISApl1) and the human isolate carrying the incomplete mcr transposon (ISApl1-mcr-1-PAP2).
Conclusion: The results of the study confirm the distribution of colistin-resistant bacteria and subsequent transmission of the resistance gene-carrying transposon between livestock and humans of the same household. To the best of our knowledge, this is the first report on genomic analysis of colistin-resistant E. coli isolates obtained from livestock and residents of the same household.
Keywords: colistin-resistance, multidrug resistance, comparative genome analysis, whole-genome sequencing, mcr
Introduction
Colistin, one of the few treatment options for intractable infectious diseases caused by multidrug-resistant Gram-negative bacteria,1 is a popular drug in livestock breeding; it is not only used to treat infections but also as a livestock growth promoter and infection protective agent.2 The overuse and misuse of colistin is considered responsible for the emergence and dissemination of colistin-resistant (COR) bacteria in animal husbandry. In addition, since colistin resistance can be transferred by a mobile colistin resistance gene, mcr, wide dissemination of COR bacteria can be expected in livestock as well as humans. In fact, livestock is considered a reservoir for COR bacteria.3
Our recent studies have reported wide dissemination of COR bacteria in human residents of communities where livestock carried COR bacteria.4–6 The results of these studies suggest that horizontal transmission of resistance may occur between livestock and human residents in the same community, including transmission of specific resistant isolates. However, there is a lack of direct evidence supporting the transmission of COR bacteria and/or the resistance gene, mcr, in the community.
A recent study on mcr-1-harboring plasmids obtained from COR Escherichia coli isolates from humans and livestock has reported the details of mcr-1-carrying IncI2 plasmid,7 which is one of the major replicons for the mcr-1-carrying plasmids.8 However, these studies analyzed clinical isolates from patients and livestock isolates retrospectively. In this study, we aimed to study the transmission of mcr-1 between COR E. coli isolates from humans and domestic animals in the same household by performing a comparative genomic analysis of these isolates.
Materials and Methods
Bacterial Strains
COR E. coli isolates, strains 2018–18.25PC and 2018–18.25CC, were previously isolated from rectal swab specimens of pig and chicken, respectively, in the backyard of a household in a rural community in Vietnam.4 Human COR E. coli isolates, 2017.02.01CC, 2017.02.02CC, and 2018–02.2CC, were also previously isolated from the fecal specimens of two family members of the same household as the livestock.5 The study was approved by the Ethics Committees of Osaka University (Osaka, Japan) and Thai Binh University of Medicine and Pharmacy (Thai Binh, Vietnam). The characteristics of these isolates are summarized in Table 1.
 |
Table 1 Characterization of Colistin-Resistant E. coli Isolates from Human and Livestock |
Pulsed-Field Gel Electrophoresis (PFGE)
Xbal-digested genomic DNA samples from the COR E. coli isolates were analyzed on a CHEF-DR III System (Bio-Rad, Hercules, CA, USA) following the methods reported previously.9
Genome Sequencing
In this study, we analyzed the strains 2018–18.25PC and 2018–18.25CC obtained from a pig and a chicken, respectively, by genome sequencing. Whole-genome sequencing and assembly of the isolates as well as of the plasmid harboring mcr-1, were performed using the Illumina MiSeq (Illumina Inc., CA, USA) and MinION (Oxford Nanopore Technologies, London, UK) sequencers, as described previously.10 The genome sequences were automatically annotated using the DDBJ Fast Annotation and Submission Tool Pipeline (https://dfast.nig.ac.jp). Genome analysis was performed using the Geneious R11 software (Biomatters, Ltd., NZ). Antibiotic resistance genes and replicon typing were conducted in ResFinder 4.011 and PlasmidFinder 1.3 on the Center for Genomic Epidemiology website (http://www.genomicepidemiology.org). Multilocus sequence type (MLST) analysis was performed using a MLST software (https://github.com/tseemann/mlst). The mcr-1-bearing IncHI2 plasmids were compared with the reference plasmid pECJS-59-24411 using BRIG12 and visualized with Easyfig.13
Genome analysis of the human isolates was performed previously, and the analyzed data were used in this study.14
Results
Comparison of Livestock- and Human-Derived Strains Isolated from the Same Household
The phylogenetic relation, mcr-1 location, and mcr transposon structure were assessed for five isolates. Two isolates were obtained from a chicken and a pig each, which were maintained in the backyard of the household; one isolate was obtained from a human member of the same household at the time of the isolation of the livestock strains, and two isolates were obtained from the human members of the household 10 months prior to the isolation of the livestock strains. The phylogenetic likelihood of these five isolates was assessed using PFGE, and the results are shown in Figure 1. None of the five isolates belonged to the same clone. However, the pig isolate 2018–18.25PC and the human isolate 2018–02.2CC belonged to sequence type (ST) 10, and the chicken isolate 2018–18.25CC and the human isolate 2017.02.01CC to ST48.
 |
Figure 1 Dendrogram of pulsed-field gel patterns of isolates from human and livestock. |
Comparative genomic analysis was performed to assess the genetic details of mcr-1 in these isolates. Similar to the human isolates, the livestock isolates carried mcr-1 on either the chromosome or a plasmid (Table 1). The chicken isolate 2018–18.25CC harbored mcr-1 on plasmid IncHI2 as observed in the human isolate 2017.02.01CC. Although both isolates belonged to ST48, phylogenetically, they were different. The pig isolate 2018–18.25PC belonged to ST10 and harbored mcr-1 on the chromosome similar to the human isolate 2018–02.2CC; however, these two isolates were different (Figure 1). The human isolate 2017.02.02CC was also different from the livestock isolates.
Comparative Details of mcr-1 in Human and Livestock Isolates
The results of the comparative analysis of the mcr gene showed that both the human strain 2018–02-2CC and the pig strain 2018–18.25PC retained the mcr-1 transposon on the chromosome; however, the human strain had an incomplete transposon, ISApl1-mcr-1-PAP2, whereas the pig strain possessed the complete transposon, ISApl1-mcr-1-PAP2-ISApl1 (Table 1). Furthermore, the location of mcr-1 on both chromosomes was genetically related despite the strains being different.
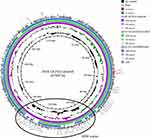
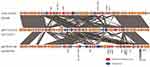
Sequence of mcr-1-plasmid IncHI2 found in the human strain 2017.02.01CC and the chicken strain 2018–18.25CC showed that both plasmids (p2017.02.01CC, and p2018-18.25CC) were similar (query cover; 88%, identity; 99.99%, Figure 2) except for some drug resistance genes in the multidrug resistance (MDR) region of p2018-18.25CC, which were inserted in the reverse direction (Figure 3). IncHI2 plasmid carries a large MDR region containing antibiotic resistance genes other than mcr-1.15 Therefore, a comparative study with the plasmids from the isolated strains and the reference plasmid pECJS-59-244 (KX084394) was conducted. As shown in Figure 3, plasmids from the human and chicken isolates were similar in terms of plasmid organization to the reference plasmid. However, the MDR region of the human and chicken isolates lacked certain resistance genes present in the reference plasmid. Interestingly, IncHI2 isolated from the human and chicken isolates harbored floR, a gene coding for resistance against the veterinary drug florfenicol; this was similar to the reference IncHI2 (KX084394).
Discussion
The results of this study showed that human members of the household harbored COR E. coli strains with the same ST as that of the livestock. In particular, the COR E. coli ST48 strains were isolated from chicken as well as humans. These isolates were genetically similar but were different clones, as assessed by PFGE analysis. This indicates that these isolates were derived from the same clone and evolved by long-term adaptation to the hosts (humans and livestock) as well as environmental conditions. Furthermore, since this study was not conducted during a short-term epidemic of COR bacteria spreading in the community, it was highly unlikely to observe the same clone in both humans and livestock. Therefore, a clonal distribution of COR strains among livestock and humans was anticipated. In addition, identification of the same IncHI2 plasmid carrying mcr-1 in these isolates might provide the initial evidence for COR bacteria dissemination and subsequent spread of plasmids in the household.
The phylogenetic similarity of human and pig ST10 COR isolates was low. The COR E. coli ST10 isolate from the pig did not possess an mcr-1-carrying plasmid but carried a mcr-1 on the chromosome akin to the human ST10 isolate. However, unlike the human ST10 isolate, the structure of the mcr-1 transposon found in the pig isolate was well conserved. Since the insertion sequences (ISs) of the mcr-1 transposon TN6330 (ISApl1-mcr-1-PAP2-ISApl1) have dropped out during stabilization of the genes across generations,16,17 it is likely that transmission of TN6330 between plasmids, chromosomes, and strains may have occurred in the household over a long period. Therefore, the ST10 clone may have spread between humans and livestock like the ST48 clone. Furthermore, the presence of floR on plasmids of both livestock and human isolates indicates the direction of this transmission. The occurrence of floR is commonly associated with mcr-1 and suggests animal origin of the isolate.8,17 However, more isolates need to be assessed to confirm this observation.
Since the discovery of mcr on plasmids, many studies have been conducted on mcr-carrying plasmids of COR bacteria isolated from various sources.7,8,18,19 Outcomes of a metadata analysis revealed that the mcr-carrying plasmids are diverse.19 Among these mcr-carrying plasmids, IncI2, IncX4, and IncHI2 plasmids are found most frequently, even in livestock isolates. The outcomes of the present study regarding mcr-1-harboring plasmid IncHI2 of the livestock COR isolate are consistent with those of previous studies.15
The IncHI2 plasmid is also known to actively acquire other foreign resistance genes.15 BLASTN and ResFinder analysis on the resistance gene database showed that the plasmids of the chicken isolate 2018–18.25CC and the human isolate 2017.02.01CC comprised MDR determinants, including floR, blaTEM, tetM, aaC3, dfrA5, and sul1. However, genes such as blaCTX-M, fosA, and aadA, reported in the reference pig isolate obtained from farms in China, were not present in our isolates.15 Addition of MDR genes to mcr-carrying plasmids and the subsequent spread of the plasmid in the microbiomes of humans and livestock poses a threat to public health worldwide. The results of the present study showed that accumulation of resistance genes was limited in the isolates. However, accumulation of resistance genes can be expected in the future. Therefore, monitoring of MDR genes besides mcr in the plasmids of human and livestock microbiomes is urgently required.
The chromosomally encoded mcr-1 is important due to its stability.13,20 The presence of chromosomal mcr-1 has been reported, but its frequency was low (3.5%).20 In this study, although only two isolates from livestock were assessed genetically, the finding of chromosomal mcr-1 in both of them is consistent with a previous study reporting the occurrence of chromosomal mcr-1 in 36.8% of the isolates from healthy residents living in the same area.13 Therefore, the monitoring of chromosomal mcr in COR bacteria it is also important.
Conclusion
To the best of our knowledge, this is the first report of a direct comparative analysis of COR E. coli isolates from livestock and humans of the same household. The initial findings of this study suggest distribution of COR E. coli and subsequent transmission of the colistin resistance gene mcr-1 transposon between livestock and humans of the same household. The findings also support that dissemination of COR bacteria in communities occurs through domestic livestock. The study warrants further genetic analysis of COR bacteria isolates in communities where COR bacteria are disseminated.
Abbreviations
COR, colistin-resistant; PFGE, pulsed-field gel electrophoresis; MDR, multidrug resistance; IS, insertion sequence; MLST, multilocus sequence type; ST, sequence type.
Data Sharing Statement
The draft genome sequences of the COR E. coli strains 2018-18.25PC and sequences of the plasmid carrying mcr-1 of strain 2018-18.25CC were deposited in DDBJ/GenBank under the accession numbers AP023286 and LC567843, respectively.
Ethics Approval and Informed Consent
The study was approved by the Ethics Committees of Osaka University (Osaka, Japan) and Thai Binh University of Medicine and Pharmacy (Thai Binh, Vietnam).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI (grants 17H01687 and 20H00561).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Giamarellou H, Poulakou G. Multidrug-resistant gram-negative infections: what are the treatment options? Drugs. 2009;69(14):1879–1901.
2. Rhouma M, Beaudry F, Letellier A. Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents. 2016;48(2):119–126.
3. Giamarellou H. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents. 2016;48(6):614–621.
4. Kawahara R, Fujiya Y, Yamaguchi T, et al. Most domestic livestock possess colistin-resistant commensal Escherichia coli harboring mcr in a rural community in Vietnam. Antimicrob Agents Chemother. 2019;63(6):e00594–19.
5. Yamamoto Y, Kawahara R, Fujiya Y, et al. Wide dissemination of colistin-resistant Escherichia coli with the mobile resistance gene mcr in healthy residents in Vietnam. J Antimicrob Chemother. 2019;74(2):523–524.
6. Yamamoto Y, Calvopina M, Izurieta R, et al. Colistin-resistant Escherichia coli with mcr genes in the livestock of rural small-scale farms in Ecuador. BMC Res Notes. 2019;12:121.
7. Yoon EJ, Hong JS, Yang JW, et al. Detection of mcr-1 plasmids in Enterobacteriaceae isolates from human specimens: comparison with those in Escherichia coli isolates from livestock in Korea. Ann Lab Med. 2018;38(6):555–562.
8. Matamoros S, van Hattem JM, Arcilla MS, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7:15364.
9. Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67.
10. Matsumoto Y, Kinjo T, Motooka D, et al. Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg Microbes Infect. 2019;8(1):1043–1053.
11. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491.
12. Alikhan NF, Petty NK, Ben Zakour NL, et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402.
13. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010.
14. Yamaguchi T, Kawahara R, Hamamoto K, et al. High prevalence of colistin-resistant Escherichia coli with chromosomally carried mcr-1 in healthy residents in Vietnam. mSphere. 2020;5(2):e00117–20.
15. Li R, Xie M, Zhang J, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393–401.
16. Snesrud E, McGann P, Chandler M. The birth and demise of the IS Apl1- mcr-1-IS Apl1 composite transposon: the vehicle for transferable colistin resistance. mBio. 2018;9(1):e02381–17.
17. Wang R, Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179.
18. Ye H, Li Y, Li Z, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio. 2016;7(2):e00177–16.
19. Lin Y, Dong X, Wu J, et al. Metadata analysis of mcr-1-bearing plasmids inspired by the sequencing evidence for horizontal transfer of antibiotic resistance genes between polluted river and wild birds. Front Microbiol. 2020;11:352.
20. Shen C, Zhong LL, Ma F, et al. Genomic patterns and characterizations of chromosomally-encoded mcr-1 in Escherichia coli populations. Gut Pathog. 2020;12:55.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.