Back to Journals » Infection and Drug Resistance » Volume 14
Comparative Evaluation of Seven Tigecycline Susceptibility Testing Methods for Carbapenem-Resistant Enterobacteriaceae
Authors Li H, Zhou M, Chen X, Zhang Y, Jian Z , Yan Q, Liu WE
Received 31 December 2020
Accepted for publication 9 March 2021
Published 20 April 2021 Volume 2021:14 Pages 1511—1516
DOI https://doi.org/10.2147/IDR.S289499
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Hongling Li, Mao Zhou, Xia Chen, Yawen Zhang, Zijuan Jian, Qun Yan, Wen-En Liu
Department of Clinical Laboratory, Xiangya Hospital, Central South University, Changsha, People’s Republic of China
Correspondence: Wen-En Liu
Department of Clinical Laboratory, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha, Hunan 410008, People’s Republic of China
Tel +86-731-84327440
Fax +86-731-84327332
Email [email protected]
Purpose: Carbapenem-resistant Enterobacteriaceae (CRE) strains are extensively resistant to most antibiotics. Tigecycline is one of the few effective drugs that can be used to treat infections caused by CRE. The aim of this study was to evaluate the accuracy of different methods for detecting the susceptibility of CRE to tigecycline.
Methods: Seven commonly used drug susceptibility testing methods were compared and evaluated for the ability to determine CRE tigecycline susceptibility: broth microdilution (BMD), agar dilution method (ADM), disk diffusion method, Etest, MicroScan, Vitek2 COMPACT, and BD Phoenix 100.
Results: The minimum inhibitory concentration (MIC) of tigecycline to inhibit 50% and 90% of CRE growth (MIC50 and MIC90, respectively) assessed by ADM and BD Phoenix 100 was the same as that determined by the reference method, BMD. The MIC50 was 2 μg/mL, and the MIC90 was 4 μg/mL. The highest number of susceptible strains was detected by MicroScan, followed by BMD, Etest, ADM, BD Phoenix 100, Vitek2 COMPACT, and disk diffusion method, in descending order. No significant differences were observed among the tigecycline susceptibility results (P > 0.05) obtained from MicroScan, Etest, BD Phoenix 100, and BMD. BMD confirmed that 82.0% of strains were susceptible to tigecycline. ADM, MicroScan, and BD Phoenix 100 yielded the categorical agreement of 96%, 92%, and 93%, respectively. No method was found to present any very major errors (VMEs), and only the Vitek2 COMPACT yielded major errors (MEs) greater than 3%.
Conclusion: Among the seven methods tested, the ADM, MicroScan, and BD Phoenix 100 methods were accurate for determining the tigecycline susceptibility of CRE. MicroScan was acceptable with better performance than other methods.
Keywords: tigecycline, antibiotic susceptibility test, carbapenem-resistant Enterobacteriaceae, CRE
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) has become an urgent and serious threat to public health due to the lack of effective alternative treatment options.1 According to the Centers for Disease Control and Prevention (CDC), CRE refers to any member of Enterobacteriaceae when developing resistance to the carbapenem antibiotics.2 Bloodstream infections caused by CRE are associated with high mortality rates of up to 24% to 43%, according to some studies.3–5 Tigecycline is one of the few remaining options for the treatment of multidrug-resistant gram-negative bacteria, including CRE.6 As the use of tigecycline increases, an increasing number of tigecycline-resistant strains are being reported.7,8 Commonly used methods for determining tigecycline susceptibility include broth microdilution (BMD), agar dilution method (ADM), disk diffusion method, Etest, and automated microbial identification and drug susceptibility systems, such as the Vitek2 COMPACT and the BD Phoenix 100. Previous studies have shown that the outcomes of in vitro tigecycline susceptibility testing can be affected by the testing method used.9
Clinical microbiologists require accurate tigecycline susceptibility results to guide clinical drug use, which can have an important impact on the treatment strategies used for CRE. The Clinical and Laboratory Standards Institute (CLSI) offers no breakpoints or recommended methods for the tigecycline susceptibility testing of Enterobacteriaceae, and few studies have examined the reliability of the various methods for testing the tigecycline susceptibility of Enterobacteriales. Recently, Yin et al evaluated several methods for testing the tigecycline susceptibility of carbapenem-resistant Klebsiella pneumoniae (CRKP) and carbapenem-resistant Acinetobacter baumannii (CRAB). The authors clarified that a modified Kirby-Bauer (K-B) disk diffusion method was a simple, accurate, and inexpensive method for testing tigecycline susceptibility in CRKP and CRAB.10 However, they did not include any tigecycline-resistant isolates in their study. Furthermore, the CRE isolates were limited to CRKP. The aim of the present study was to examine a variety of CRE, species including tigecycline-resistant isolates, to evaluate the currently available methods for determining tigecycline susceptibility, including BMD, ADM, Etest (bioMérieux, France), disk diffusion method, MicroScan (Beckman Coulter, America), Vitek2 COMPACT (bioMérieux, France), and BD Phoenix 100, to guide the choice of suitable, convenient, and accurate methods for use in clinical laboratories.
Materials and Methods
Bacterial Isolates
In this study, a total of 100 CRE isolates obtained from sputum (48%), blood (18%), ascites (13%), urine (13%), bile samples (6%), and catheter tip (2%) were collected from 2017 to 2018 at Xiangya Hospital, including 78 Klebsiella pneumoniae isolate, 10 Escherichia coli isolates, 7 Enterobacter cloacae isolates, 2 Serratia marcescens isolates, 2 Klebsiella oxytoca isolates, and 1 Enterobacter aerogenes isolate. All of the strains were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF; Bruker, Germany), and carbapenem antimicrobial susceptibility was determined using an AST CN16 panel (VITEK Compact, bioMérieux, France).
CRE isolates with minimum inhibitory concentrations (MICs) of ≥2 µg/mL for ertapenem or ≥4 µg/mL for meropenem or imipenem were included in this study. The E. coli strain ATCC 25922 was used as a quality control strain for tigecycline susceptibility testing according to CLSI recommendations.11
Antimicrobial Susceptibility Testing
The MICs of tigecycline against all strains were detected using six methods, including BMD, ADM, and Etest (bioMérieux, France), and three automated microbial identification and drug susceptibility systems: MicroScan (Beckman Coulter, America), using the NUC61 type card; Vitek2 COMPACT (bioMérieux, France), using the AST-GN16 card; and BD Phoenix 100 (Becton Dickinson, America), using the NMIC-413 type card. The disk diffusion method (Oxoid, UK) was used to detect the inhibition zone diameter for MIC evaluation. The size of the inhibition zone is inversely proportional to the MIC. All tests were performed in accordance with CLSI recommendations.12–14
For the BMD method, the strains were grown in 0.5 McFarland Standard (McF) bacterial suspension using Mueller–Hinton broth (Oxoid, UK) and diluted 100 times. Next, bacterial suspensions were added to broth containing serial 2-fold dilutions of tigecycline in a 96-well flat-bottom cell culture plate (Costar, America). For all other methods, the strains were grown in 0.5 McF bacterial suspension using normal saline. ADM was performed by adding various concentrations of tigecycline-containing solutions into Mueller–Hinton agar (Oxoid, UK), 2 µL of the bacterial suspension was diluted 10 times and dropped onto the agar surface. The 96-well culture plates and agar plates were incubated at 35°C ± 2°C for 16–20 hours.
To perform the disk diffusion method and Etest, the bacterial suspension was applied to the Mueller–Hinton agar surface with a sterile cotton swab. A disk containing 15 µg tigecycline or an Etest strip was placed onto the agar surface. For the MicroScan, Vitek2 COMPACT, and BD Phoenix 100 analyses, diluted suspensions were added to the respective gram-negative bacilli susceptibility identification cards for culture and identification, according to the manufacturer’s instructions and the standard operating procedures provided for the instrument.
Data Analysis
The tigecycline breakpoint interpretations for Enterobacteriaceae issued by the United States Food and Drug Administration (FDA) were as follows: susceptible (S): MIC ≤ 2 µg/mL; intermediate (I): MIC = 4 µg/mL; and resistant (R) MIC ≥ 8 µg/mL.15 For the disk diffusion method, an inhibition zone diameter ≤14 mm was classified as R, a diameter ≥ 19 mm was classified as S, and a diameter of 15–18 mm was classified as I. According to the interpretation criteria, the proportions of susceptible, intermediate, and resistant isolates were determined using each method. All data analysis was performed using SPSS software (version 25.0) with the McNemar test. A P-value < 0.05 was considered significant.
The consistency of the results determined by all methods was compared against those for the BMD method, which was set as the standard method. The misclassification of a resistant strain as susceptible was considered to be a very major error (VME), whereas the reporting of a susceptible strain as resistant was classified as a major error (ME). The interpretive categories of either susceptible or resistant strains as intermediate or vice versa was considered a minor error (mE). Categorical agreement (CA) was evaluated as the percentage of isolate characterizations produced by each method that was consistent with the results (R, S, or I) reported for the BMD method. Essential agreement (EA) was established when the method reported an MIC of no more than one dilution different from that determined by the BMD method. When CA or EA ≥ 90% (if reporting MIC), VME ≤ 1.5%, and ME ≤ 3%, the tested method was considered acceptable according to CLSI criteria.16
Ethics
This study was reviewed and approved by the Ethics Committee of Xiangya Hospital of Central South University and was performed in accordance with the Declaration of Helsinki and its amendments or comparable ethical standards. The requirement for patient informed consent was waived because this study only focused on the susceptibility testing methods, and no patient information was used.
Results
In this study, we evaluated seven tigecycline susceptibility testing methods for 100 CRE strains. The MIC50 and MIC90 values, tigecycline susceptibility categories, and the rates of CA, EA, VME, ME, and mE relative to those for the BMD method are shown in Table 1. The MIC50 and MIC90 values for the ADM and BD Phoenix 100 analyses were the same as the reference method, BMD, which resulted in an MIC50 of 2 µg/mL and an MIC90 of 4 µg/mL. Due to the MicroScan drug susceptibility card that we used only included two concentrations of tigecycline, the MIC50 was documented as ≤2 µg/mL. The MIC50 of the Etest was 1.5 µg/mL below that of the BMD evaluation. The MIC90 of Vitek2 COMPACT was 8 µg/mL, which was higher than that evaluated by the BMD method. The percentage of resistance among the tested strains is shown in Supplementary Table 1.
 |
Table 1 Tigecycline Susceptibility Results Against 100 Enterobacteriaceae Strains Using Seven Different Testing Methods |
As shown in Table 1, for all seven methods, the MicroScan reported the highest number of susceptible strains, followed by BMD, Etest, ADM, BD Phoenix 100, Vitek2 COMPACT, and the disk diffusion method, in descending order. After paired analysis, no significant difference was detected between the MicroScan, Etest, BD Phoenix 100, and BMD test results (P > 0.05) for CRE susceptibility to tigecycline. The ADM, MicroScan, and BD Phoenix 100 yielded CA values of 96%, 92%, and 93%, respectively. No methods resulted in VME, and only the Vitek2 COMPACT yielded an ME rate greater than 3%.
In contrast, the proportions of resistant and intermediate strains measured by ADM and the disk diffusion method were higher than those identified by the BMD method. Strains that were identified as susceptible and intermediate by BMD were identified as resistant by ADM, which increased the resistance rate of the strains. In contrast, the disk diffusion method identified multiple susceptible strains as being intermediate strains. The Vitek2 COMPACT also returned higher resistance rates than the BMD method. The results for all methods are shown in Table 2.
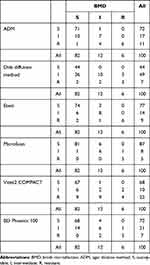 |
Table 2 Comparison of Results Between the Reference Method, BMD, and Six Other Testing Methods |
With reference to previous CLSI standards, the acceptable methods were identified as ADM, MicroScan, and BD Phoenix 100. However, the ADM and BD Phoenix 100 both had high probabilities of returning mEs, at 15% and 20%, respectively.
Discussion
This study was conducted to compare and evaluate seven common tigecycline susceptibility testing methods for 100 CRE strains collected over a 12-month period from 2017 to 2018. Because no convenient and accurate method has been established for tigecycline susceptibility testing in clinical, microbiological laboratories, the identification of an appropriate method for clinical use remains urgent.
The BMD method was used as the reference standard for the comparison and analysis of the results returned for the other six methods. However, the BMD method is time-consuming and requires an experienced professional to perform accurately. Previous studies have reported that some factors associated with the culture medium could affect the performance of tigecycline susceptibility testing; therefore, fresh (≤12 hours) broth was used in this experiment.17 The different methods examined in this study resulted in different MIC values for tigecycline against CRE.
Studies have shown that for severe multidrug-resistant Acinetobacter baumannii infections, the tigecycline MIC values are more reliably determined by reference tests, such as ADM or BMD.18 Similar to other studies, in our study, the ADM showed better stability than other tests, resulting in the same MICs as BMD; however, the ADM wrongly reported 10 susceptible strains as being intermediate, 1 intermediate strain as being susceptible, and 1 susceptible and 4 intermediate strains as being resistant,19 which were inconsistent with the test results obtained using the BMD method. Although the ADM was identified as an acceptable method for tigecycline susceptibility testing, and the mE rate was 15%, indicating that this test still returns errors. In addition, the operation and interpretation of results for the ADM can be complicated, making it less commonly used in clinical practice.
Compared with the BMD, the results of the disk diffusion method had a relatively high and significantly different intermediate rate of 49%. The disk diffusion method also had the highest probability of mEs among all tested methods, as a large number of susceptible strains were reported as intermediate. The disk diffusion method is cheap, simple, and fast, which has resulted in its wide adoption by most clinical laboratories in China. Intermediate results returned by this method may require the use of additional methods for further confirmation.
Zarkotou et al considered the Etest to be consistent with the BMD method in an experiment comparing the BMD, Etest, MTS strip, and Vitek2 system for the detection of tigecycline susceptibility against CRE.20 In our study, we also found that compared with the Vitek2 system, the Etest was more consistent with the BMD method. Casal et al reported that when interpreting the tigecycline susceptibility results against Acinetobacter baumannii assessed by Etest, for MICs was higher than 2 µg/mL, especially those in the 2–4 µg/mL range, some intermediate results were reported as susceptible when using the BMD method.21 Our experiment identified a similar phenomenon, suggesting that this issue is worthy of attention. However, the Etest is associated with high costs and is not suitable for large-scale testing.
Automated microbial identification and drug susceptibility systems are increasingly being used in clinical practice, but these systems remain unreliable for some drug susceptibility tests. A previous study reported that the Vitek2 COMPACT system differed greatly from the BMD for the determination of tigecycline MICs against gram-negative bacilli.9 In this study, most of the methods resulted in MIC50 and MIC90 values similar to those for the BMD method, whereas the Vitek2 detected an MIC90 of 8. Similarly, the resistance rate of 22% measured by the Vitek2 was the highest among all tested methods. Importantly, the ME (9%) rate of the Vitek2, which is the rate at which susceptible strains are reported as being resistant, was the highest among all tested methods. Therefore, this method would misjudge too many strains as being resistant. A recent phenomenon of “false resistance” has been reported. Lat et al reported that the Vitek2 system had a high error rate for the tigecycline susceptibility test against K. pneumoniae, whereas Huang et al reported that the Vitek2 system was consistent when used for tigecycline susceptibility testing against E.coli.22,23 However, in our study, most of the strains were K. pneumoniae, which may explain why the Vitek2 reported an increased resistance rate.
Compared with the other two automated methods, MicroScan was more consistent with the BMD reference method. MicroScan was also identified as an acceptable method according to CLSI standards. The MIC values did not differ significantly between MicroScan and BMD, similar to another study that examined tigecycline susceptibility testing for gram-negative bacilli.9 However, the MicroScan misclassified 6 intermediate strains as susceptible and 1 resistant strain as intermediate, resulting in the highest detection rate of susceptible strains. The primary disadvantage was that the concentrations of tigecycline on the drug susceptibility identification card were limited to 2 and 4 µg/mL; the addition of 1µg/mL or 8 µg/mL concentrations would increase the accuracy of the results.
The BD Phoenix 100 was consistent with the reference BMD method. According to existing research, the BD Phoenix appears to provide the most accurate determination of tigecycline susceptibility in multidrug-resistant Acinetobacter baumannii compared with the Vitek2 and the MicroScan.24 In our study of Enterobacteriales, the BD Phoenix 100 performed well, with a CA of 93%, VME and ME values of 0%, and it qualified as one of the acceptable methods according to CLSI criteria. However, it also committed a lot of mEs, reporting several susceptible strains as intermediate. Although the BD Phoenix 100 had good consistency, the susceptibility rate was much lower than that for the BMD method, which was also observed in our study, which may indicate additional tests would be necessary in clinical practice to improve accuracy.25 One limitation of our study was that the numbers of strains were not evenly distributed among species because CRKP is the most common CRE species in our country. We will continue to collect more CRE species with detailed carbapenemases genotype for future studies.
Conclusion
Compared with the reference method, BMD, ADM was an accurate manual method for performing tigecycline sensitivity tests against CRE. Automated microbial identification and drug susceptibility systems are convenient, and the MicroScan system was acceptable according to CLSI criteria, with better performance than other automated methods. Understanding the advantages and disadvantages of different methods will help us to select appropriate methods to meet the intended purposes of research and provide better and more accurate results.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [Grant/Award Number:81672066].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Doi Y. Treatment options for carbapenem-resistant gram-negative bacterial infections. Clin Infect Dis. 2019;69(Suppl 7):S565–S575. doi:10.1093/cid/ciz830
2. CDC. Diseases and organisms in healthcare settings. Available from: https://www.cdc.gov/hai/organisms/organisms.html.
3. Van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis. 2020;20(6):731–741. doi:10.1016/S1473-3099(19)30755-8
4. Stewardson AJ, Marimuthu K, Sengupta S, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019;19(6):601–610. doi:10.1016/S1473-3099(18)30792-8
5. Gutierrez-Gutierrez B, Salamanca E, de Cueto M, et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Lancet Infect Dis. 2017;17(7):726–734. doi:10.1016/S1473-3099(17)30228-1
6. Ni W, Han Y, Liu J, et al. Tigecycline treatment for carbapenem-resistant Enterobacteriaceae infections: a systematic review and meta-analysis. Medicine. 2016;95:e3126. doi:10.1097/MD.0000000000003126
7. He T, Wang R, Liu D, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4(9):1450–1456. doi:10.1038/s41564-019-0445-2
8. Deng M, Zhu MH, Li JJ, et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother. 2014;58(1):297–303. doi:10.1128/AAC.01727-13
9. Marchaim D, Pogue JM, Tzuman O, et al. Major variation in MICs of tigecycline in Gram-negative bacilli as a function of testing method. J Clin Microbiol. 2014;52(5):1617–1621. doi:10.1128/JCM.00001-14
10. Yin DD, Guo Y, Li M, et al. Performance of VITEK 2, E-test, Kirby–Bauer disk diffusion, and modified Kirby–Bauer disk diffusion compared to reference broth microdilution for testing tigecycline susceptibility of carbapenem-resistant K. pneumoniae and A. baumannii in a multicenter study in China. Eur J Clin Microbiol Infect Dis. 2021:1–6. doi:10.1007/s10096-020-04123-z
11. Clinical and Laboratory Standards Institute (CLSI). The Performance stands for antimicrobial susceptibility testing, M100–S29. 2019.
12. Clinical Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, M07–11th edition, Melvin P. Weinstein, MD: CLSI; 2018.
13. Clinical Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests, M02–13th edition, Melvin P. Weinstein, MD: CLSI; 2018.
14. Clinical Laboratory Standards Institute. Development of in vitro susceptibility testing criteria and quality control parameters, M23–5th edition Matthew A Wikler, MD, MBA, FIDSA. CLSI; 2018.
15. US Food and Drug Administration. FDA-Identified Interpretive Criteria for Tigecycline-Injection products; 2019. Available from: https://www.fda.gov/drugs/development-resources/tigecycline-injection-products.
16. Clinical and Laboratory Standards Institute (CLSI). Verification of commercial microbial identification and antimicrobial susceptibility testing systems M52–1st Edition. 2015.
17. Torrico M, Gonzalez N, Gimenez MJ, et al. Influence of media and testing methodology on susceptibility to tigecycline of Enterobacteriaceae with reported high tigecycline MIC. J Clin Microbiol. 2010;48(6):2243–2246. doi:10.1128/JCM.00119-10
18. Özkök S, Togan T, Yesilkaya A, et al. In vitro susceptibility of tigecycline against multidrug-resistant gram-negative strains: Etest versus agar dilution. Chemotherapy. 2014;60(3):151–156. doi:10.1159/000375440
19. Zhang J, Zhao C, Chen H, et al. Comparative evaluation of tigecycline susceptibility testing methods for Acinetobacter baumannii and Enterobacteriaceae. J Glob Antimicrob Resist. 2015;3(2):75–79. doi:10.1016/j.jgar.2015.02.004
20. Zarkotou O, Pournaras S, Altouvas G, et al. Comparative evaluation of tigecycline susceptibility testing methods for expanded-spectrum cephalosporin- and carbapenem-resistant gram-negative pathogens. J Clin Microbiol. 2012;50(11):3747–3750. doi:10.1128/JCM.02037-12
21. Casal M, Rodriguez F, Johnson B, et al. Influence of testing methodology on the tigecycline activity profile against presumably tigecycline-non-susceptible Acinetobacter spp. J Antimicrob Chemother. 2009;64(1):69–72. doi:10.1093/jac/dkp169
22. Lat A, Clock SA, Wu F, et al. Comparison of polymyxin B, tigecycline, cefepime, and meropenem MICs for KPC-producing Klebsiella pneumoniae by broth microdilution, Vitek 2, and Etest. J Clin Microbiol. 2011;49(5):1795–1798. doi:10.1128/JCM.02534-10
23. Huang TD, Berhin C, Bogaerts P, et al. In vitro susceptibility of multidrug-resistant Enterobacteriaceae clinical isolates to tigecycline. J Antimicrob Chemother. 2012;67(11):2696–2699. doi:10.1093/jac/dks288
24. Idelevich EA, Freeborn DA, Seifert H, et al. Comparison of tigecycline susceptibility testing methods for multidrug-resistant, Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2018;91:360–362.
25. Idelevich EA, Büsing M, Mischnik A, et al. False non-susceptible results of tigecycline susceptibility testing against Enterobacteriaceae by an automated system: a multicentre study. J Med Microbiol. 2016;65(8):877–881. doi:10.1099/jmm.0.000281
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
