Back to Journals » Cancer Management and Research » Volume 11
Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: a systematic and analytical review
Authors Vafaei S , Fattahi F, Ebrahimi M , Janani L, Shariftabrizi A, Madjd Z
Received 17 June 2019
Accepted for publication 15 August 2019
Published 25 September 2019 Volume 2019:11 Pages 8669—8698
DOI https://doi.org/10.2147/CMAR.S219699
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Somayeh Vafaei,1–3 Fahimeh Fattahi,1,2 Marzieh Ebrahimi,3 Leila Janani,4 Ahmad Shariftabrizi,5 Zahra Madjd1,6
1Department of Molecular Medicine, Faculty of Advanced Technologies in Medicine, Iran University of Medical Sciences, Tehran, Iran; 2Student Research Committee, Iran University of Medical Sciences, Tehran, Iran; 3Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran; 4Department of Biostatistics, School of Public Health, Iran University of Medical Sciences, Tehran, Iran; 5Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA; 6Oncopathology Research Center, Iran University of Medical Sciences, Tehran, Iran
Correspondence: Zahra Madjd
Oncopathology Research Center, Department of Molecular Medicine, Iran University of Medical Sciences (IUMS), Hemmat Street (Highway), Next to Milad Tower, Tehran 14496-14530, Iran
Tel +98 218 670 3212
Fax +98 218 862 2608
Email [email protected]
Marzieh Ebrahimi
Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran 16635-148, Iran
Tel +98 2 356 2516
Email [email protected]
Abstract: Nearly half of patients with colorectal cancer (CRC), the third leading cause of cancer deaths worldwide, are diagnosed in the late stages of the disease. Appropriate treatment is not applied in a timely manner and nearly 90% of the patients who experience metastasis ultimately die. Timely detection of CRC can increase the five-year survival rate of patients. Existing histopathological and molecular classifications are insufficient for prediction of metastasis, which limits approaches to treatment. Detection of reliable cancer-related biomarkers can improve early diagnosis, prognosis, and treatment response prediction and recurrence risk. Circulating tumor cells (CTCs) and exosomes in peripheral blood can be used in a liquid biopsy to assess the status of a tumor. Exosomes are abundant and available in all fluids of the body, have a high half-life and are released by most cells. Tumor-derived exosomes are released from primary tumors or CTCs with selective cargo that represents the overall tumor. The current systematic review highlights new trends and approaches in the detection of CRC biomarkers to determine tumor signatures using CTC and exosomes. When these are combined, they could be used to guide molecular pathology and can revolutionize detection tools. Relevant observational studies published until July 24, 2019 which evaluated the expression of tumor markers in CTCs and exosomes were searched in PubMed, Scopus, Embase, and ISI Web of Science databases. The extracted biomarkers were analyzed using String and EnrichR tools.
Keywords: colorectal cancer, circulating tumor cell, CTC, exosomes, diagnosis, prognosis, biomarker, systematic review
Introduction
Colorectal cancer (CRC) is the third highest cause of cancer deaths worldwide.1,2 The time of diagnosis directly influences the overall survival rate of patients. The five-year survival rates are estimated to decrease 12.5% after the occurrence of metastasis vs for localized cancer. Histological examination of tumor tissue is the gold standard for diagnosis, but is invasive, time-consuming, and nonrepeatable over time. There is a need for new methods that are simple, non-invasive, and inexpensive to provide clear clinical evidence and improve early detection or predict a response to treatment.3,4
Serum biomarkers such as carcinoembryonic antigens (CEAs) and carbohydrate antigen 19-9 (CA19-9) along with multi-target stool DNA tests represent the concrete implementation of non-invasive methods for CRC screening5,6 There is urgent need for more reliable molecular markers that demonstrate the heterogeneity of cancer cells during progression. The use of biological fluids as sources of nucleic acid-biomarkers for liquid biopsies in oncology has clinical promise7,8 Molecular characterization of cancer signatures also can provide relevant information for personalized treatment of tumors.9,10 Circulating tumor cells (CTCs) and exosomes are shed from a tumor mass and enter the bloodstream. They can provide a metastatic niche for the invasion and migration of a tumor, so detection of their markers is critical.11
Ashworth et al, first identified CTCs as valuable indicators of cancer progression.12 CTCs detach from the primary tumor, intravasate into the bloodstream, evade immune detection, survive and extravasate into the microvessels of target tissue to establish a micro-metastatic niche.13 They have been identified in many cancers, including colon cancer. CTCs in the bloodstream may exist as single cells with a different EMT phenotypes or as clusters that bind to platelets or macrophages or are reactivated as stromal cells.14,15 The presence and number of CTCs before and during treatment are a strong independent predictor of shorter progression-free survival and overall survival of CRC patients.16 In spite of their advantages, researchers believe that the most challenging obstacles related to research on CTCs are their extremely low numbers, short lifetimes, fragility, and their heterogeneity and plasticity. The investigation of specific and reliable markers for their detection or isolation is an undeniable issue.17
Extracellular vesicles (EVs) generally include microvesicles (100–350 nm), apoptotic bodies (500–1000 nm), and exosomes (30–150 nm).18 Exosomes are nanovesicles with membrane-bound phospholipids which introduced and confirmed by Pan et al,19 and are actively secreted by mammalian cells into body fluids such as urine, plasma, and saliva. Exosomal cargo includes lipids, proteins, DNA, and RNA (mRNA, miRNA, long non-coding RNA) that are selected according to their roles. Exosomes involved in many biological processes, especially intercellular communication, establish a premetastatic niche by carrying oncogenic elements that suppress host immune responses.20
Exosomes are abundant, have high half-lives and are released by most cells. This is in contrast with CTCs, which are tumor specific, rare, fragile, have a short life and are difficult to isolate. It is possible to design a molecular marker common between the exosomes and CTCs for better understanding of the metastasis process. American Society of Clinical Oncology suggests circulating exosomes may provide an alternative platform for monitoring disease progression as opposed to CTCs.21 Several ongoing studies have aimed at quantifying a stress protein or other biomarkers in the blood and urine for monitoring and early diagnosis of malignant solid tumors (https://clinicaltrials.gov). The current analytical review is the first to explore similar molecular mechanisms and pathways between CTCs and Exosomes. In this systematic review, all molecular mechanisms that can potentially apply to the diagnosis and prognosis of CRC using CTCs and exosomes are discussed.
Materials and methods
Search strategy for literature mining
Observational studies evaluating the expression of circulating CRC cells and exosomes markers from 1980 to July 24, 2019 were electronically searched for in the PubMed, Scopus, Embase, and ISI Web of Science databases. The search syntax was modified for each database in accordance with their rules, the Mesh terms and keywords as listed in detail in Table 1.
 |  |  |
Table 1 Search strategy of CTC and exosome in colorectal cancer |
The authors (S. Vafaei and F. Fattahi) searched and identified eligible studies and excluded all irrelevant articles after reviewing the publication titles and abstracts. Duplicate publications were excluded. Discrepancies were resolved between the two reviewers by consensus and by consulting the other authors. Next, the full text of the selected publications was retrieved and fully reviewed. This systematic review has been carried out in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses.22
Publication inclusion criteria
The inclusion criteria for this systematic review followed the criteria of population, intervention, control, and outcomes. Observational studies (case-control) investigating CTC and exosomes mRNA and gene markers for the diagnosis and prognosis of CRC patient samples were included if they met the following criteria:
- The article must be published in English and the full text must be available.
- Studies included those on CRC patient blood samples and human blood for CTC, although tissue or cell lines for exosomes were done because exosomes research is rare and in its initial stages.
- Expression of mRNA and gene markers in patient specimens or cell lines was detected by established molecular methods.
- Studies demonstrated the correlation between mRNA profiling using isolation, detection, or validation methods, included sample type and size and other clinical parameters of diagnosis and prognosis, tumor stage and the frequency of estimated marker expression.
- Study characteristics (first author surname, publication year, and study design) were included.
Publication exclusion criteria
Exclusion criteria included:
- Evidence and article on CTC and exosomes covering review articles, seminars, letters, expert opinions, book chapters, meeting records, commentaries, and clinical guidelines.
- In-vitro or in-vivo experimental studies.
- Articles that were not published in English.
- Full text of the article not available.
Exclusion criteria for CTC articles were:
- Studies performed only on cell lines or tissue samples.
- Studied housekeeping genes, such as glyceraldehyde-3-phosphate dehydrogenase, actin beta, β2-microglobulin, as they are not specific markers for CTC detection and expressed in all cells.
- Bioinformatics analysis or data mining without experimental confirmation of the introduced biomarkers.
- Therapy gaudiness based on the CTC results (perioperative and postoperative) in predicting the clinical outcome, not counting for drug effect on the expression of CTC genes.
- The study only tested the spiked cell lines in human blood donors and not the actual patients.
In exosome studies, because of the limited data, we reviewed all articles on all markers that were introduced using the cell lines, tissue, or blood, even those only introduced through bioinformatics means without experimental confirmation.
Risk of bias (quality) assessment
The quality of each study was assessed using the Newcastle–Ottawa Scale (NOS), a well-known scale for assessing the quality and risk of bias in observational studies.23 NOS gives a score between 0 (minimum) and 9 (maximum). Studies with a NOS score >6 were considered to be of high quality, making them possible for use as potential moderators in meta-regression analysis.
Statistical analysis
Because the studies included were not sufficiently similar in terms of study design, experimental techniques, and heterogeneity of genetic variants, a meta-analysis was not performed.
Bioinformatics approach to systematic search
Molecular pathology is a valuable tool in the development of a cancer signature. The initially extracted markers in this article were subjected to STRING (https://string-db.org/) for better understanding of the significantly related pathway and secondary data were enriched using the EnrichR (amp.pharm.mssm.edu/Enrichr/) web tool. The GO project provided ontologies to describe the attributes of the gene products in the non-overlapping domains of molecular biology. Molecular function describes activities (such as catalytic or binding activities) at the molecular level. Biological processes describe biological goals accomplished by one or more ordered assemblies of molecular functions. Cellular component describes the locations of subcellular structures and macromolecular complexes.24
Results
Literature
The initial search retrieved a total of 607 studies using the search strategy. After primary selection, 497 papers were excluded because they were duplicates, had irrelevant titles or were paper abstracts. Eventually, 110 studies were selected for further evaluation. The schematic of the design and the reasons for exclusions are summarized in Figures 1 and 2 for CTC and exosomes, respectively.
Clinical applications of CTCs and exosomes in CRC as diagnostic markers
CTCs
Antigen expression of circulating cells and their specific phenotypes affects the progression of cancer and patient survival; thus, the focus was on CTC molecular markers that could lead to the detection of CTC rather than isolation in blood samples. CTC detection methods included real-time polymerase chain reaction (RT-PCR), flow cytometry, fluorescence in situ hybridization, and immunocytochemistry. Isolation methods included Cellsearch, OncoQuick, Filration, magnetic-activated cell sorting, fluorescence-activated cell sorting, Adnatest Colon Cancer Select and Detect, CELLection electrophoresis assay, and microfluidic devices.
When attempting to find more reliable markers for CTCs in CRC cases, 6 out of 39 articles described only CK20 mRNA as the target gene, which is not transcribed in normal hematopoietic cells. It has previously been reported through immunohistochemistry by Moll et al,25–30 and has been seen in control blood samples through sensitivity assay and sampling,29 in addition to CK20, CA19-9, and CEA, which is used in clinics routinely for CRC detection, also has been introduced as a marker of CTC in CRC. Six of 39 studied examined CEA alone31–36 or in association with markers such as CK19,37,38 anti-epithelial cell adhesion molecule (EPCAM),39–43 and transmission electron microscopy (TEM)-8.44
Wong et al, used a sensitivity assay for the detection of CTCs and nodal metastases using CD44 splice variants as a tumor marker.45 It has been proven that RT-PCR in combination with positive isolation of epithelial tumor cells (addition of Ber-EP4 immunomagnetic) and negative isolation of non-epithelial cells (CD45 immunomagnetic beads used to deplete leukocytes from MNC) could improve detection.30,36 Guanylyl-cyclase C (GCC) is another marker introduced to detect rare epithelial circulating metastatic cancer cells.46–48
After 2004, researchers focused on multi-marker panels in literature or data mining as listed in Table 2.49–56 Besides these, novel markers such as serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), SERPINB5,57 epidermal growth factor receptor (EGFR),58–60 epithelial cell transforming sequence 2 oncogene (ECT2)61 FAM172A,62 A3 receptor63 have been examined as well as other markers, especially through bioinformatics analysis.64
 |  |  |  |
Table 2 The biomarkers which worked for diagnostic of CRC in circulating tumor cells |
Exosomes
Exosome isolation methods consisted of ultracentrifugation, commercial kits, and a combination of several methods based on their physical, chemical, immunological, and molecular markers. Characterization of exosomes was also achieved based on morphology, such as with scanning electron microscopy and TEM, based on size, such as with dynamic light-scattering and nanoparticle tracking assay or based on molecular profiling through conventional enzyme-linked immunosorbent assay, polymerase chain reaction, and Western blotting.
Exosomes carry molecular markers such as DNA, RNA, and proteins. Many reports indicate that exosomes contain miRNAs;65–68 moreover, blood EVs contain a substantial fraction of intact mRNAs69–72 and a large number of assembling spliced junctions-circRNAs73 and long non-coding RNAs.72,74,75 Exosomal proteins belong to the following functional groups: tetraspanins, including CD63 antigen (CD63), CD9 antigen (CD9), CD81 antigen (CD81), heat shock proteins (HSC70 and HSC90), and endosomal sorting complexes required for transport proteins such as Alix and TSG101, found in a wide range of exosomes.76 The size of the extra vesicles varied and could influence gene expression. Larger vesicles (<100 nm) exhibited the greatest amount of EPCAM in extracted exosomes of HCT116 (CRC cell line) cells.77 The level of glypican-1 was evaluated in exosomes of patients before and after surgical treatment.78
KRTAP5-4 and MAGEA3 mRNA in the serum of patients could be used as diagnostic biomarkers to detect CRC.79 Ct-OATP1B3 mRNA was present in EVs derived from HCT116, HT-29, and SW480 cells that were declared to be serum-based CRC biomarkers,80 Huang et al, introduced UBC, H3F3A, HIST2H2AA3, AKT3, and HSPA1B as hub genes in bioinformatic analysis to serve as diagnostic markers and therapeutic targets of CRC in the future.81 Table 3 shows all of these results.
 |
Table 3 The biomarkers which worked for diagnostic of CRC in Exosome |
Clinical applications of CTCs and exosomes in CRC as prognostic markers
CTCs
Many researchers had discovered prognostic markers related to CRC as a beneficial tool for the detection of CTC. Five papers reported only CK20-positive as a prognostic marker. It caused significantly shorter survival in patients than the CK 20-negative marker.82–86 However, some studies emphasized only on CEA as a marker (five articles)87–91 and several studies also introduced both CK20 and CEA as prognostic markers.92–96 In most articles, CK20 and/or CEA were accompanied by markers such as CK19,97–102 GCC,96,103,104 Prominin 1 (CD133),95,100,105,106 EPCAM,107–109 survivin,110,111 ProtM,112 mucin 1 (MUC 1),105 and mucin 2 (MUC 2),99 and telomerase reverse transcriptase (hTERT).101,113,114
Douard et al, showed that the expression of carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5; formerly CEA)102,115 and CEACAM7 (formerly CGM2)115,116 was more sensitive than use of a single marker in detecting CTCs, in contrast to the other studies, Bessa et al, showed that assessment of CTCs using RT-PCR CEA before surgery does not have prognostic value for CRC patients.117
Some articles examined markers that had been investigated previously, such as EGFR,107,118–121 Plastin3),122,123 anterior gradient-2,102,124,125 leucine-rich repeat-containing-G-protein-coupled receptor 5,102,109,126–128 double cortin-like kinase 1,109,127 twist family bHLH transcription factor 1,110,129 and aldehyde dehydrogenase 1105,129 as prognostic markers in CRC through CTC.
Gradilone et al, assessed CK19 (75%), CK20 (8%), and EGFR (25%) expression in CTCs of some malignant tumors, including CRC samples, by RT-PCR followed by southern blot hybridization. They reported no correlation between prognostic values of CTCs and clinical manifestations of CRC.130
Histone-like protein (HLM),120 tenascin C,121 aquaporin (AQP5),131 plakophilin 3, tyrosinase, prostate-specific antigen),132 universal MAGE-A,133 disheveled segment polarity protein 1 (DVL1),134 CD47,135 and CD44 variant exon 9 (CD44v9)136 were proposed as markers in a smaller number of articles. The heterogeneity of CTC markers led some researchers to focus on multi-marker panels in data mining as listed in Table 4.101,105,109,110,114,125,129,137–139
 |  |  |  |  |  |  |
Table 4 The biomarkers which worked for prognostic of CRC in circulating tumor cells |
Exosomes
Some prognosis markers have nearly the same functional patterns as molecular markers related to CRC. Studies have reported on colorectal exosome prognostic markers such as ALIX (ALG 2-interacting protein X),140,141 Hsp60,142 Hsp70,141 CEA,143 ATP-binding cassette transporter G1 (ABCG1),144 copine III (CPNE3),145 and ΔNp7370 in cancer patients.
Tauro et al, used multiple isolation methods to detect known exosome markers such as ALIX, TSG101, HSP70, and other specific and novel markers listed in Table 5.141 Chen et al, applied bioinformatic analysis for introduction of two panels and validated them.146 Chiba et al, reported that exosomes derived from CRC cell lines contain mRNA, microRNA, and natural antisense RNA as listed in Table 5.71
 |
Table 5 The biomarkers which worked for prognostic of CRC in exosome |
Risk of bias (quality) assessment
All articles related to CTC (39 diagnosis-related and 57 prognosis-related) were assessed by NOS case-control guidelines as reported in Table S1. Of the diagnosis-related articles (40% of the total), 43%, 43%, and 14% scored 7, 6, and 5, respectively. Of the prognosis-related articles (60% of the total), 49%, 31.5%, 14%, and 1.5% scored 7, 6, 5, and 4, respectively; and 4% could not to be scored.
All articles related to exosomes (Five diagnosis-related and nine prognosis-related) were assessed by the NOS case-control guidelines in Table S2. Of the diagnosis-related articles (36% of total), 20%, 40%, and 40% scored 7, 6, and 5, respectively. Of the prognosis-related articles (64% of total), 67%, 22%, and 11% scored 7, 6, and 5, respectively. The 0–3 and 8–9 scores were not given out in these studies, so the NOS number varied from 4 to 7. About 99.3% of systematically imported articles scored over 5, 20% ofthe articless scored 5, and 79.7% scored 6 or 7.
Bioinformatics approach to systematic results
This systematic search identified 66 CTC gene markers for the diagnosis of CRC, 65 CTC gene markers for prognosis with repetition, 10 exosome gene markers for diagnosis of CRC, and 35 exosome gene markers for prognosis as shown in Tables 2–5.
Protein–protein interaction network via STRING analysis
In the gene network, biochemical functions and identified pathways were obtained from gene expression data, and the results are shown in Figures 3 and 4 and supplementary Table S3 (online resources). Surprisingly, the cellular components of exosomes and CTC highlight extracellular space, region and exosome, plasma membrane, and cell junction. Their molecular function highlights cell adhesion molecule binding and protein binding. Biological processes included regulation of cellular component movement, assembly, localization, organization, and response to external stimuli.
Gene ontology
The results of EnrichR web tools in supplementary Table S4 (online resources) can be used to accurately understand the molecular pathways. The common pathways in biomarkers such as proteoglycans in cancer, focal adhesion pathways in cancer, integrin, Rap1, MAPK signaling pathways, angiogenesis, p53 pathways, and viral processes were similar and related to cancer.
Discussion
CRC is a common malignancy that often has a poor prognosis.147 The tumor microenvironment contributes to its progression148 and cross-talk between cancer cells and exosomes play a critical role in this dynamic network.149 Their identification and characterization are important steps to improve understanding of cellular and molecular cancer metastasis. Tracking of tumor-associated molecular markers in the blood can be used to assess the presence of residual disease, recurrence, and resistance.150 This systematic review highlights new trends and approaches in CRC biomarker discovery using CTC and exosomes.
Evidence related to diagnosis of CRC by means of CTC markers was addressed in 38 articles (Table 2) and 54 articles discussed prognosis of CRC using CTC markers (Table 4). Only 14 articles examined exosomes, five about diagnosis and nine about prognosis (Tables 3 and 5). Our results show that the most common markers introduced in CTCs were CEA (35 of 94 studies) and CK20 (33 of 94 studies), especially using quantitative real-time polymerase chain reaction. Most markers investigated for exosomes, in addition to CD9, CD81, ALIX, and TSG101, were including EPCAM and HSP, especially using ultracentrifugation. Comparison of 131 CTC markers and 45 exosomes markers showed only three common markers (CEA, CD9, and EPCAM) on the gene list as diagnostic and prognostic biomarkers.
A half-century-old investigation of CEA in CRC was the first step in the identification of a much larger family of 12 CEACAMs.151,152 Gene encoding CEA is a member of the immunoglobulin supergene family153 that plays a role in cell adhesion and tumor progression,154 even in protecting the colon from microbial infection.155 CEA is involved in the metastatic cascade process through positive regulation of cell migration and invasion;156–158 thus, the monitoring of CEA as a cost-effective and frequent indicator of recurrence of CRC has been investigated for years.159
Integrin on tumor exosomes may play an important role in modulating organ-specific metastasis in cancer progression. CD9 is a member of the tetraspanin superfamily commonly detected in all types of exosomes involved in pathophysiologic processes such as cellular adhesion, growth, motility, cell–cell fusion, signal transduction, and tumor metastasis.160
EPCAM is a membranous glycoprotein that is a CSC marker in tumor cells in the basolateral surface of most normal epithelial tissue and its role is to connect cells by means of calcium. The expression of this marker increases in benign and malignant tumors that arise from epithelial tissue.161 The first step in metastasis is the separation of cancerous cells from primary tumors. CEA, CD9, and EPCAM are closely correlated with tumor progression as a poor prognostic factor and is required for the survival of CTCs in some cancers.162 Taken together, it appears that the signature of the CTC and exosome biomarkers are similar and follow common pathways; thus, exosomes can be applied as alternative tools for guiding better molecular pathology in the fight against cancer.
Precision medicine is changing clinical practice by tailoring treatment based on an individual’s genetic makeup. Recent studies have shown that CTC and circulating tumor DNA provide complementary information and the use of both approaches to study tumor metastasis is warranted.163 CTC and exosomes can pave a path as diagnostic and prognostic procedures using the heterogeneity of tumor sites as they are released into the blood from live origins and can be analyzed at the DNA, RNA, and protein levels. It is undeniable that more investigation is needed to compare them, especially for cancer patients.
Various CTC isolating techniques each have its own advantages and disadvantages as to their CTC capture capacity and subgrouping of CTCs based on various markers. Similar problems also exist for exosomes, with a lack of a proven rapid and high-yield approach for extracting exosomes for downstream analysis.164 Microfluidic devices and bioinformatics analysis might play an important role in solving the current shortcomings of the liquid biopsy concept. Microfluidics, by using inertial focusing/hydrodynamics (laminar flow in microchannels) and applying spiral, acoustic, electrophoretic, and electromagnetic features passively separate CTCs and exosomes from the other background calls.165 Immobilizing specific antibodies either on micro-posts or in a herringbone design against their marker might be useful; it is easy to explore and yields quantitative readouts with high sensitivity, low cost, and minimal sample handling. Finally, although the potential clinical utility of these techniques is clear, more effort is needed to use the full potential of liquid biopsy in clinical settings.166
Future perspectives
Currently, isolation and purification of tumor-derived exosome in a worm bag of EVs is technically cumbersome and also isolation of CTCs has its own limitations. Therefore, combined use of these two biomarkers together as a liquid biopsy requires large-scale clinical trials. Microfluidic devices and bioinformatics analysis might play an important role in solving the current shortcomings of the liquid biopsy. Additionally, cross talking of CTCs and tumor-derived exosomes in a tumor microenvironment should become a heated question in exploring the premetastatic niche. As such, more research is needed on CTCs and exosome’s overlapping molecular pathways to determine more effective biomarker signatures of CRC, especially in the metastatic form.
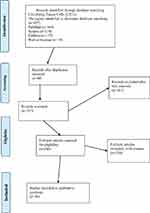 |
Figure 1 Design of PRISMA flow diagram explaining details of our search process was applied during the article selection for circulating tumor cell. |
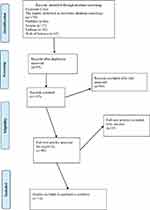 |
Figure 2 Design of PRISMA flow diagram explained details of our search process that applied during the article selection for Exosome. |
Acknowledgments
Systematic Review Network, Vice-Chancellor for Research and Technology, Iran University of Medical Sciences (Grant # 97-4-37-13921), funded this research. We would also like to thank the Royan Stem Cell Technology Company and our colleagues from both centers who provided insight and expertise that greatly assisted the research.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:10.3322/caac.21254
3. Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14(4):235. doi:10.1038/nrclinonc.2016.171
4. Zhai Z, Yu X, Yang B, et al. Colorectal cancer heterogeneity and targeted therapy: clinical implications, challenges and solutions for treatment resistance.
5. Bailey JR, Aggarwal A, Imperiale TF. Colorectal cancer screening: stool DNA and other noninvasive modalities. Gut Liver. 2016;10(2):204–211. doi:10.5009/gnl15420
6. Heichman KA. Blood-based testing for colorectal cancer screening. Mol Diagn Ther. 2014;18(2):127–135. doi:10.1007/s40291-013-0074-z
7. Hench IB, Hench J, Tolnay M. Liquid biopsy in clinical management of breast, lung, and colorectal cancer. Front Med. 2018;5:9. doi:10.3389/fmed.2018.00009
8. Lopez A, Harada K, Mizrak Kaya D, Dong X, Song S, Ajani JA. Liquid biopsies in gastrointestinal malignancies: when is the big day? Expert Rev Anticancer Ther. 2018;18(1):19–38. doi:10.1080/14737140.2018.1403320
9. Ryan E, Creagh E. Emerging methods in colorectal cancer screening. BJS. 2018;105(2):e16–e18. doi:10.1002/bjs.10650
10. Hauptman N, Glavac D. Colorectal cancer blood-based biomarkers. Gastroenterol Res Pract. 2017;2017:2195361. doi:10.1155/2017/2195361
11. Shao H, Chung J, Issadore D. Diagnostic technologies for circulating tumour cells and exosomes. Biosci Rep. 2015;36(1):e00292. doi:10.1042/BSR20150180
12. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75(2):193–208. doi:10.1007/s00018-017-2595-9
13. Thorsteinsson M, Jess P. The clinical significance of circulating tumor cells in non-metastatic colorectal cancer–a review. Eur J Surg Oncol. 2011;37(6):459–465. doi:10.1016/j.ejso.2011.01.025
14. Xu L, Shamash J, Lu Y-J. Circulating tumor cells: a window to understand cancer metastasis, monitor and fight against cancers. J Can Res Updates. 2015;4(1):13–29. doi:10.6000/1929-2279.2015.04.01.2
15. Satelli A, Mitra A, Brownlee Z, et al. Epithelial–mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Cancer Res. 2015;21(4):899–906. doi:10.1158/1078-0432.CCR-14-0894
16. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–3221. doi:10.1200/JCO.2007.15.8923
17. Riggi N, Aguet M, Stamenkovic I. Cancer metastasis: a reappraisal of its underlying mechanisms and their relevance to treatment. Annu Rev Pathol. 2018;13:117–140. doi:10.1146/annurev-pathol-020117-044127
18. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi:10.1016/0092-8674(83)90040-5
19. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789–804. doi:10.7150/thno.18133
20. Soung YH, Nguyen T, Cao H, Lee J, Chung J. Emerging roles of exosomes in cancer invasion and metastasis. BMB Rep. 2016;49(1):18–25. doi:10.5483/BMBRep.2016.49.1.239
21. Traci Pawlowski DS, Tinder T, Kimbrough J, et al. Circulating exosomes may provide an alternate platform to monitor disease progression compared to circulating tumor cells. J Clin Oncol. 2010;28:10580–10580. doi:10.1200/jco.2010.28.15_suppl.10580
22. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi:10.1371/journal.pmed.1000097
23. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi:10.1007/s10654-010-9491-z
24. Harris MA, Clark J, Ireland A, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Databaseissue):D258–D261. doi:10.1093/nar/gkh036
25. Zhang XW, Yang HY, Fan P, Yang L, Chen GY. Detection of micrometastasis in peripheral blood by multi-sampling in patients with colorectal cancer. World J Gastroenterol. 2005;11(3):436–438. doi:10.3748/wjg.v11.i3.436
26. Katsumata K, Sumi T, Mori Y, Hisada M, Tsuchida A, Aoki T. Detection and evaluation of epithelial cells in the blood of colon cancer patients using RT-PCR. Int J Clin Oncol. 2006;11(5):385–389. doi:10.1007/s10147-006-0590-5
27. Soeth E, Vogel I, Roder C, et al. Comparative analysis of bone marrow and venous blood isolates from gastrointestinal cancer patients for the detection of disseminated tumor cells using reverse transcription PCR. Cancer Res. 1997;57(15):3106–3110.
28. Soeth E, Roder C, Juhl H, Kruger U, Kremer B, Kalthoff H. The detection of disseminated tumor cells in bone marrow from colorectal-cancer patients by a cytokeratin-20-specific nested reverse-transcriptase-polymerase-chain reaction is related to the stage of disease. Int J Cancer. 1996;69(4):278–282. doi:10.1002/(SICI)1097-0215(19960822)69:4<278::AID-IJC7>3.0.CO;2-U
29. Vlems FA, Diepstra JH, Cornelissen IM, et al. Limitations of cytokeratin 20 RT-PCR to detect disseminated tumour cells in blood and bone marrow of patients with colorectal cancer: expression in controls and downregulation in tumour tissue. Mol Pathol. 2002;55(3):156–163. doi:10.1136/mp.55.3.156
30. Guo J, Xiao B, Jin Z, et al. Detection of cytokeratin 20 mRNA in the peripheral blood of patients with colorectal cancer by immunomagnetic bead enrichment and real-time reverse transcriptase-polymeras chain reaction. J Gastroenterol Hepatol. 2005;20(8):1279–1284. doi:10.1111/j.1440-1746.2005.03894.x
31. Jonas S, Windeatt S, Fordy C, Allen-Mersh TG. Identification of carcinoembryonic antigen-producing cells circulating in the blood of patients with colorectal carcinoma by reverse transcriptase polymerase chain reaction. Gut. 1996;39(5):717–721. doi:10.1136/gut.39.5.717
32. Castells A, Boix L, Bessa X, Gargallo L, Pique JM. Detection of colonic cells in peripheral blood of colorectal cancer patients by means of reverse transcriptase and polymerase chain reaction. Br J Cancer. 1998;78(10):1368–1372. doi:10.1038/bjc.1998.686
33. Noh YH, Im G, Ku JH, Lee YS, Ahn MJ. Detection of tumor cell contamination in peripheral blood by RT-PCR in gastrointestinal cancer patients. J Korean Med Sci. 1999;14(6):623–628. doi:10.3346/jkms.1999.14.6.623
34. Guadagni F, Kantor J, Aloe S, et al. Detection of blood-borne cells in colorectal cancer patients by nested reverse transcription-polymerase chain reaction for carcinoembryonic antigen messenger RNA: longitudinal analyses and demonstration of its potential importance as an adjunct to multiple serum markers. Cancer Res. 2001;61(6):2523–2532.
35. Hampton R, Walker M, Marshall J, Juhl H. Differential expression of carcinoembryonic antigen (CEA) splice variants in whole blood of colon cancer patients and healthy volunteers: implication for the detection of circulating colon cancer cells. Oncogene. 2002;21(51):7817–7823. doi:10.1038/sj.onc.1205906
36. Guo J, Xiao B, Zhang X, et al. Combined use of positive and negative immunomagnetic isolation followed by real-time RT-PCR for detection of the circulating tumor cells in patients with colorectal cancers. J Mol Med (Berl). 2004;82(11):768–774. doi:10.1007/s00109-004-0590-8
37. Silva JM, Rodriguez R, Garcia JM, et al. Detection of epithelial tumour RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut. 2002;50(4):530–534. doi:10.1136/gut.50.4.530
38. Wong IH, Yeo W, Chan AT, Johnson PJ. Quantitative relationship of the circulating tumor burden assessed by reverse transcription-polymerase chain reaction for cytokeratin 19 mRNA in peripheral blood of colorectal cancer patients with Dukes’ stage, serum carcinoembryonic antigen level and tumor progression. Cancer Lett. 2001;162(1):65–73. doi:10.1016/s0304-3835(00)00630-3
39. Cohen SJ, Alpaugh RK, Gross S, et al. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6(2):125–132. doi:10.3816/CCC.2006.n.029
40. Gogoi P, Sepehri S, Zhou Y, et al. Development of an automated and sensitive microfluidic device for capturing and characterizing Circulating Tumor Cells (CTCs) from clinical blood samples. PLoS One. 2016;11(1):e0147400. doi:10.1371/journal.pone.0147400
41. Mourtzikou A, Kroupis C, Poumpouridou N, et al. Molecular detection of circulating tumor cells in peripheral blood of colon cancer patients. J Gastroenterol Hepatol Res. 2012;1(5):74–79.
42. van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat Res. 2011;728(1–2):23–34. doi:10.1016/j.mrrev.2011.05.002
43. Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi:10.1038/nature06385
44. Raeisossadati R, Farshchian M, Ganji A, et al. Quantitative analysis of TEM-8 and CEA tumor markers indicating free tumor cells in the peripheral blood of colorectal cancer patients. Int J Colorectal Dis. 2011;26(10):1265–1270. doi:10.1007/s00384-011-1230-8
45. Wong LS, Cantrill JE, Odogwu S, Morris AG, Fraser IA. Detection of circulating tumour cells and nodal metastasis by reverse transcriptase-polymerase chain reaction technique. Br J Surg. 1997;84(6):834–839.
46. Fava TA, Desnoyers R, Schulz S, et al. Ectopic expression of guanylyl cyclase C in CD34+ progenitor cells in peripheral blood. J Clin Oncol. 2001;19(19):3951–3959. doi:10.1200/JCO.2001.19.19.3951
47. Tien YW, Chang KJ, Jeng YM, et al. Tumor angiogenesis and its possible role in intravasation of colorectal epithelial cells. Clin Cancer Res. 2001;7(6):1627–1632.
48. Tien YW, Jeng YM, Hu RH, Chang KJ, Hsu SM, Lee PH. Intravasation-related metastatic factors in colorectal cancer. Tumour Biol. 2004;25(1–2):48–55. doi:10.1159/000077723
49. Solmi R, De Sanctis P, Zucchini C, et al. Search for epithelial-specific mRNAs in peripheral blood of patients with colon cancer by RT-PCR. Int J Oncol. 2004;25(4):1049–1056. doi:10.3892/ijo.25.4.1049
50. Yeh CS, Wang JY, Wu CH, et al. Molecular detection of circulating cancer cells in the peripheral blood of patients with colorectal cancer by using membrane array with a multiple mRNA marker panel. Int J Oncol. 2006;28(2):411–420. doi:10.3892/ijo.28.2.411
51. Solmi R, Ugolini G, Rosati G, et al. Microarray-based identification and RT-PCR test screening for epithelial-specific mRNAs in peripheral blood of patients with colon cancer. BMC Cancer. 2006;6:250. doi:10.1186/1471-2407-6-250
52. Lauriola M, Ugolini G, Rosati G, et al. Identification by a Digital Gene Expression Displayer (DGED) and test by RT-PCR analysis of new mRNA candidate markers for colorectal cancer in peripheral blood. Int J Oncol. 2010;37(2):519–525. doi:10.3892/ijo_00000701
53. Barbazan J, Alonso-Alconada L, Muinelo-Romay L, et al. Molecular characterization of circulating tumor cells in human metastatic colorectal cancer. PLoS One. 2012;7(7):e40476. doi:10.1371/journal.pone.0040476
54. Barbazan J, Vieito M, Abalo A, et al. A logistic model for the detection of circulating tumour cells in human metastatic colorectal cancer. J Cell Mol Med. 2012;16(10):2342–2349. doi:10.1111/j.1582-4934.2012.01544.x
55. Alonso-Alconada L, Barbazan J, Candamio S, et al. PrediCTC, liquid biopsy in precision oncology: a technology transfer experience in the Spanish health system. Clin Transl Oncol. 2018;20(5):630–638. doi:10.1007/s12094-017-1760-9
56. Barbazan J, Muinelo-Romay L, Vieito M, et al. A multimarker panel for circulating tumor cells detection predicts patient outcome and therapy response in metastatic colorectal cancer. Int J Cancer. 2014;135(11):2633–2643. doi:10.1002/ijc.28910
57. Findeisen P, Rockel M, Nees M, et al. Systematic identification and validation of candidate genes for detection of circulating tumor cells in peripheral blood specimens of colorectal cancer patients. Int J Oncol. 2008;33(5):1001–1010. doi:10.3892/ijo_00000088
58. Vaiopoulos AG, Kostakis ID, Gkioka E, et al. Detection of circulating tumor cells in colorectal and gastric cancer using a multiplex PCR assay. Anticancer Res. 2014;34(6):3083–3092.
59. Teama SH, Agwa SHA. Detection of circulating tumor cells by nested RT-PCR targeting EGFR/CEA/CK20mRNAs in colorectal carcinoma patients. Egypt J Med Human Genet. 2010;11(2):173–180. doi:10.1016/j.ejmhg.2009.10.001
60. Lankiewicz S, Rother E, Zimmermann S, Hollmann C, Korangy F, Greten TF. Tumour-associated transcripts and EGFR deletion variants in colorectal cancer in primary tumour, metastases and circulating tumour cells. Cell Oncol. 2008;30(6):463–471.
61. Chen CJ, Sung WW, Chen HC, et al. Early assessment of colorectal cancer by quantifying circulating tumor cells in peripheral blood: ECT2 in diagnosis of colorectal cancer. Int J Mol Sci. 2017;18(4):E743. doi:10.3390/ijms18040743
62. Cui CH, Chen RH, Zhai DY, Xie L, Qi J, Yu JL. Detection of FAM172A expressed in circulating tumor cells is a feasible method to predict high-risk subgroups of colorectal cancer. Tumour Biol. 2017;39(6):1010428317699126. doi:10.1177/1010428317699126
63. Gessi S, Cattabriga E, Avitabile A, et al. Elevated expression of A3 adenosine receptors in human colorectal cancer is reflected in peripheral blood cells. Clin Cancer Res. 2004;10(17):5895–5901. doi:10.1158/1078-0432.CCR-1134-03
64. Rodia MT, Ugolini G, Mattei G, et al. Systematic large-scale meta-analysis identifies a panel of two mRNAs as blood biomarkers for colorectal cancer detection. Oncotarget. 2016;7(21):30295–30306. doi:10.18632/oncotarget.8108
65. Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9(4). doi:10.1371/journal.pone.0092921
66. Zhang J, Raju GS, Chang DW, Lin SH, Chen Z, Wu X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer. 2018;124(4):785–796. doi:10.1002/cncr.31062
67. Ostenfeld MS, Jensen SG, Jeppesen DK, et al. miRNA profiling of circulating EpCAM+ extracellular vesicles: promising biomarkers of colorectal cancer. J Extracell Vesicles. 2016;5(1). doi:10.3402/jev.v5.31488
68. Rapado-Gonzalez O, Alvarez-Castro A, Lopez-Lopez R, Iglesias-Canle J, Suarez-Cunqueiro MM, Muinelo-Romay L. Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers. 2019;11(7). doi:10.3390/cancers11070898
69. Hao YX, Li YM, Ye M, et al. KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncol Lett. 2017;13(5):3608–3616. doi:10.3892/ol.2017.5889
70. Soldevilla B, Rodriguez M, Millán CS, et al. Tumor-derived exosomes are enriched in ΔNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014;23(2):467–478. doi:10.1093/hmg/ddt437
71. Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28(5):1551–1558. doi:10.3892/or.2012.1967
72. Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther. 2018;12:229–241. doi:10.1016/j.omtn.2018.05.009
73. Li Y, Zhao J, Yu S, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019;65(6):798–808. doi:10.1373/clinchem.2018.301291
74. Wang L, Duan W, Yan S, Xie Y, Wang C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed Pharmacother. 2019;113. doi:10.1016/j.biopha.2019.108758
75. Liu H, Ye D, Chen A, et al. A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers. Clin Chem Lab Med. 2019;57(7):1073–1083. doi:10.1515/cclm-2019-0052
76. Nakai W, Yoshida T, Diez D, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935. doi:10.1038/srep33935
77. Manri C, Yokoi T, Nishida H. Size-selective harvesting of extracellular vesicles for strategic analyses towards tumor diagnoses. Appl Biochem Biotechnol. 2017;182(2):609–623. doi:10.1007/s12010-016-2348-5
78. Li J, Chen Y, Guo X, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med. 2017;21(5):838–847. doi:10.1111/jcmm.12941
79. Dong L, Lin W, Qi P, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1158–1166. doi:10.1158/1055-9965.EPI-16-0006
80. Morio H, Sun Y, Harada M, et al. Cancer-Type OATP1B3 mRNA in extracellular vesicles as a promising candidate for a serum-based colorectal cancer biomarker. Biol Pharm Bull. 2018;41(3):445–449. doi:10.1248/bpb.b17-00743
81. Huang Q, Shen Z, Zang R, Fan X, Yang L, Xue M. Identification of novel genes and pathways in colorectal cancer exosomes: a bioinformatics study. Transl Cancer Res. 2018;7(3):651–658. doi:10.21037/tcr
82. Wyld DK, Selby P, Perren TJ, et al. Detection of colorectal cancer cells in peripheral blood by reverse-transcriptase polymerase chain reaction for cytokeratin 20. Int J Cancer. 1998;79(3):288–293. doi:10.1002/(sici)1097-0215(19980619)79:3<288::aid-ijc14>3.0.co;2-4
83. Hinz S, Bockhorst J, Roder C, et al. Disseminated tumor cells in the bone marrow negatively influence survival after resection of colorectal liver metastases. Ann Surg Oncol. 2012;19(8):2539–2546. doi:10.1245/s10434-012-2291-9
84. Funaki NO, Tanaka J, Itami A, et al. Detection of colorectal carcinoma cells in circulating peripheral blood by reverse transcription-polymerase chain reaction targeting cytokeratin-20 mRNA. Life Sci. 1997;60(9):643–652. doi:10.1016/s0024-3205(96)00700-x
85. Samija I, Lukac J, Mubrin MK, Kirac I, Kovacevic D, Kusic Z. Detection of cytokeratin-20-positive cells in preoperative and postoperative blood samples from colorectal cancer patients by real-time RT-PCR. Int J Biol Markers. 2013;28(2):174–181. doi:10.5301/jbm.5000003
86. Kust D, Samija I, Kirac I, Radic J, Kovacevic D, Kusic Z. Cytokeratin 20 positive cells in blood of colorectal cancer patients as an unfavorable prognostic marker. Acta Clin Belg. 2016;71(4):235–243. doi:10.1080/17843286.2016.1177264
87. Piva MG, Navaglia F, Basso D, et al. CEA mRNA identification in peripheral blood is feasible for colorectal, but not for gastric or pancreatic cancer staging. Oncology. 2000;59(4):323–328. doi:10.1159/000012190
88. Ito S, Nakanishi H, Hirai T, et al. Quantitative detection of CEA expressing free tumor cells in the peripheral blood of colorectal cancer patients during surgery with real-time RT-PCR on a LightCycler. Cancer Lett. 2002;183(2):195–203. doi:10.1016/s0304-3835(02)00157-x
89. Kanellos I, Zacharakis E, Kanellos D, et al. Prognostic significance of CEA levels and detection of CEA mRNA in draining venous blood in patients with colorectal cancer. J Surg Oncol. 2006;94(1):3–8. doi:10.1002/jso.20549
90. Lu CY, Uen YH, Tsai HL, et al. Molecular detection of persistent postoperative circulating tumour cells in stages II and III colon cancer patients via multiple blood sampling: prognostic significance of detection for early relapse. Br J Cancer. 2011;104(7):1178–1184. doi:10.1038/bjc.2011.40
91. Thorsteinsson M, Soletormos G, Jess P. Low number of detectable circulating tumor cells in non-metastatic colon cancer. Anticancer Res. 2011;31(2):613–617.
92. Yamaguchi K, Takagi Y, Aoki S, Futamura M, Saji S. Significant detection of circulating cancer cells in the blood by reverse transcriptase-polymerase chain reaction during colorectal cancer resection. Ann Surg. 2000;232(1):58–65. doi:10.1097/00000658-200007000-00009
93. Mathur P, Wharton RQ, Jonas SK, Saini S, Allen-Mersh TG. Relationship between tumour vascularity and circulating cancer cells in patients with colorectal carcinoma. Eur J Surg Oncol. 2001;27(4):354–358. doi:10.1053/ejso.2001.1118
94. Guller U, Zajac P, Schnider A, et al. Disseminated single tumor cells as detected by real-time quantitative polymerase chain reaction represent a prognostic factor in patients undergoing surgery for colorectal cancer. Ann Surg. 2002;236(6):
95. Iinuma H, Okinaga K, Egami H, et al. Usefulness and clinical significance of quantitative real-time RT-PCR to detect isolated tumor cells in the peripheral blood and tumor drainage blood of patients with colorectal cancer. Int J Oncol. 2006;28(2):297–306. doi:10.3892/ijo.28.2.297
96. Liu ZP, Li LM, Liu XL, Zhang DX. Comparative analysis of tumor markers and evaluation of their predictive value in patients with colorectal cancer. Onkologie. 2012;35(3):108–113. doi:10.1159/000336816
97. Huang P, Wang J, Guo Y, Xie W. Molecular detection of disseminated tumor cells in the peripheral blood in patients with gastrointestinal cancer. J Cancer Res Clin Oncol. 2003;129(3):192–198. doi:10.1007/s00432-003-0425-y
98. Nakamori S, Kameyama M, Furukawa H, et al. Genetic detection of colorectal cancer cells in circulation and lymph nodes. Dis Colon Rectum. 1997;40(10 Suppl):S29–S36. doi:10.1007/bf02062017
99. Hardingham JE, Hewett PJ, Sage RE, et al. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel disease. Int J Cancer. 2000;89(1):8–13. doi:10.1002/(SICI)1097-0215
100. Shimada R, Iinuma H, Akahane T, Horiuchi A, Watanabe T. Prognostic significance of CTCs and CSCs of tumor drainage vein blood in Dukes’ stage B and C colorectal cancer patients. Oncol Rep. 2012;27(4):947–953. doi:10.3892/or.2012.1649
101. Xi L, Nicastri DG, El-Hefnawy T, Hughes SJ, Luketich JD, Godfrey TE. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007;53(7):1206–1215. doi:10.1373/clinchem.2006.081828
102. Winter M, Cai Z, Winkler K, et al. Circulating tumour cell RNA characterisation from colorectal cancer patient blood after inertial microfluidic enrichment. MethodsX. 2019;6:1512–1520. doi:10.1016/j.mex.2019.06.012
103. Liu Y, Cheng G, Qian J, et al. Expression of guanylyl cyclase C in tissue samples and the circulation of rectal cancer patients. Oncotarget. 2017;8(24):38841–38849. doi:10.18632/oncotarget.16406
104. Liu Y, Qian J, Feng JG, et al. Detection of circulating tumor cells in peripheral blood of colorectal cancer patients without distant organ metastases. Cell Oncol. 2013;36(1):43–53. doi:10.1007/s13402-012-0112-6
105. Chalopin A, Tellez-Gabriel M, Brown HK, et al. Isolation of circulating tumor cells in a preclinical model of osteosarcoma: effect of chemotherapy. J Bone Oncol. 2018;12:83–90. doi:10.1016/j.jbo.2018.07.002
106. Iinuma H, Watanabe T, Mimori K, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol. 2011;29(12):1547–1555. doi:10.1200/JCO.2010.30.5151
107. Zieglschmid V, Hollmann C, Mannel J, et al. Tumor-associated gene expression in disseminated tumor cells correlates with disease progression and tumor stage in colorectal cancer. Anticancer Res. 2007;27(4a):1823–1832.
108. Sato N, Hayashi N, Imamura Y, et al. Usefulness of transcription-reverse transcription concerted reaction method for detecting circulating tumor cells in patients with colorectal cancer. Ann Surg Oncol. 2012;19(6):2060–2065. doi:10.1245/s10434-011-1889-7
109. Mirzaei A, Tavoosidana G, Rad AA, et al. A new insight into cancer stem cell markers: could local and circulating cancer stem cell markers correlate in colorectal cancer? Tumor Biol. 2016;37(2):2405–2414. doi:10.1007/s13277-015-3989-7
110. Wu S, Liu S, Liu Z, et al. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One. 2015;10(4):e0123976. doi:10.1371/journal.pone.0123976
111. Shen C, Hu L, Xia L, Li Y. Quantitative real-time RT-PCR detection for survivin, CK20 and CEA in peripheral blood of colorectal cancer patients. Jpn J Clin Oncol. 2008;38(11):770–776. doi:10.1093/jjco/hyn105
112. Schuster R, Max N, Mann B, et al. Quantitative real-time RT-PCR for detection of disseminated tumor cells in peripheral blood of patients with colorectal cancer using different mRNA markers. Int J Cancer. 2004;108(2):219–227. doi:10.1002/ijc.11547
113. Wang JY, Wu CH, Lu CY, et al. Molecular detection of circulating tumor cells in the peripheral blood of patients with colorectal cancer using RT-PCR: significance of the prediction of postoperative metastasis. World J Surg. 2006;30(6):1007–1013. doi:10.1007/s00268-005-0485-z
114. Wang JY, Lin SR, Wu DC, et al. Multiple molecular markers as predictors of colorectal cancer in patients with normal perioperative serum carcinoembryonic antigen levels. Clin Cancer Res. 2007;13(8):2406–2413. doi:10.1158/1078-0432.CCR-06-2054
115. Douard R, Moutereau S, Serru V, et al. Immunobead multiplex RT-PCR detection of carcinoembryonic genes expressing cells in the blood of colorectal cancer patients. Clin Chem Lab Med. 2005;43(2):127–132. doi:10.1515/CCLM.2005.021
116. Douard R, Le Maire V, Wind P, et al. Carcinoembryonic gene member 2 mRNA expression as a marker to detect circulating enterocytes in the blood of colorectal cancer patients. Surgery. 2001;129(5):587–594. doi:10.1067/msy.2001.112485
117. Bessa X, Elizalde JI, Boix L, et al. Lack of prognostic influence of circulating tumor cells in peripheral blood of patients with colorectal cancer. Gastroenterology. 2001;120(5):1084–1092. doi:10.1053/gast.2001.23245
118. De Luca A, Pignata S, Casamassimi A, et al. Detection of circulating tumor cells in carcinoma patients by a novel epidermal growth factor receptor reverse transcription-PCR assay. Clin Cancer Res. 2000;6(4):1439–1444.
119. Clarke LE, Leitzel K, Smith J, Ali SM, Lipton A. Epidermal growth factor receptor mRNA in peripheral blood of patients with pancreatic, lung, and colon carcinomas detected by RT-PCR. Int J Oncol. 2003;22(2):425–430. doi:10.3892/ijo.22.2.425
120. Fournier MV, Guimaraes Da Costa F, Paschoal ME, Ronco LV, Carvalho MG, Pardee AB. Identification of a gene encoding a human oxysterol-binding protein-homologue: a potential general molecular marker for blood dissemination of solid tumors. Cancer Res. 1999;59(15):3748–3753.
121. Gazzaniga P, Nofroni I, Gandini O, et al. Tenascin C and epidermal growth factor receptor as markers of circulating tumoral cells in bladder and colon cancer. Oncol Rep. 2005;14(5):1199–1202. doi:10.3892/or.14.5.1199
122. Yokobori T, Iinuma H, Shimamura T, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013;73(7):2059–2069. doi:10.1158/0008-5472.CAN-12-0326
123. Sugimachi K, Yokobori T, Iinuma H, et al. Aberrant expression of plastin-3 via copy number gain induces the epithelial-mesenchymal transition in circulating colorectal cancer cells. Ann Surg Oncol. 2014;21(11):3680–3690. doi:10.1245/s10434-013-3366-y
124. Valladares-Ayerbes M, Díaz-Prado S, Reboredo M, et al. Bioinformatics approach to mRNA markers discovery for detection of circulating tumor cells in patients with gastrointestinal cancer. Cancer Detect Prev. 2008;32(3):236–250. doi:10.1016/j.cdp.2008.08.002
125. Mostert B, Sieuwerts AM, Bolt-de Vries J, et al. mRNA expression profiles in circulating tumor cells of metastatic colorectal cancer patients. Mol Oncol. 2015;9(4):920–932. doi:10.1016/j.molonc.2015.01.001
126. Valladares-Ayerbes M, Blanco-Calvo M, Reboredo M, et al. Evaluation of the adenocarcinoma-associated gene AGR2 and the intestinal stem cell marker LGR5 as biomarkers in colorectal cancer. Int J Mol Sci. 2012;13(4):4367–4387. doi:10.3390/ijms13044367
127. Mirzaei A, Tavoosidana G, Modarressi MH, et al. Upregulation of circulating cancer stem cell marker, DCLK1 but not Lgr5, in chemoradiotherapy-treated colorectal cancer patients. Tumour Biol. 2015;36(6):4801–4810. doi:10.1007/s13277-015-3132-9
128. Wang W, Wan L, Wu S, et al. Mesenchymal marker and LGR5 expression levels in circulating tumor cells correlate with colorectal cancer prognosis. Cell Oncol. 2018;45:1–10. doi:10.1007/s13402-018-0386-4
129. Ning Y, Zhang W, Hanna DL, et al. Clinical relevance of EMT and stem-like gene expression in circulating tumor cells of metastatic colorectal cancer patients. Pharmacogenomics J. 2018;18(1):29–34. doi:10.1038/tpj.2016.62
130. Gradilone A, Gazzaniga P, Silvestri I, et al. Detection of CK19, CK20 and EGFR mRNAs in peripheral blood of carcinoma patients: correlation with clinical stage of disease. Oncol Rep. 2003;10(1):217–222. doi:10.3892/or.10.1.217
131. Shan T, Cui X, Li W, Lin W, Li Y. AQP5: a novel biomarker that predicts poor clinical outcome in colorectal cancer. Oncol Rep. 2014;32(4):1564–1570. doi:10.3892/or.2014.3377
132. Burchill SA, Perebolte L, Johnston C, Top B, Selby P. Comparison of the RNA-amplification based methods RT-PCR and NASBA for the detection of circulating tumour cells. Br J Cancer. 2002;86(1):102–109. doi:10.1038/sj.bjc.6600014
133. Miyashiro I, Kuo C, Huynh K, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47(3):505–512.
134. Huang MY, Yen LC, Liu HC, et al. Significant overexpression of DVL1 in Taiwanese colorectal cancer patients with liver metastasis. Int J Mol Sci. 2013;14(10):20492–20507. doi:10.3390/ijms141020492
135. Steinert G, Scholch S, Niemietz T, et al. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 2014;74(6):1694–1704. doi:10.1158/0008-5472.CAN-13-1885
136. Katoh S, Goi T, Naruse T, et al. Cancer stem cell marker in circulating tumor cells: expression of CD44 variant exon 9 is strongly correlated to treatment refractoriness, recurrence and prognosis of human colorectal cancer. Anticancer Res. 2015;35(1):239–244.
137. Bao H, Burke PA, Huang J, et al. Circulating tumor cells: application as a biomarker for molecular characterization and predictor of survival in an all-comer solid tumor phase i clinical study. PLoS One. 2013;8(8):e58557. doi:10.1371/journal.pone.0058557
138. Chang YT, Huang MY, Yeh YS, et al. A prospective study of comparing multi-gene biomarker chip and serum carcinoembryonic antigen in the postoperative surveillance for patients with stage I-III colorectal cancer. PLoS One. 2016;11(10):e0163264. doi:10.1371/journal.pone.0163264
139. Bahnassy AA, Salem SE, Mohanad M, et al. Prognostic significance of circulating tumor cells (CTCs) in Egyptian non-metastatic colorectal cancer patients: A comparative study for four different techniques of detection (Flowcytometry, CellSearch, Quantitative Real-time PCR and Cytomorphology). Exp Mol Pathol. 2019;106:90–101. doi:10.1016/j.yexmp.2018.12.006
140. Valcz G, Galamb O, Krenacs T, et al. Exosomes in colorectal carcinoma formation: ALIX under the magnifying glass. Mod Pathol. 2016;29(8):928–938. doi:10.1038/modpathol.2016.72
141. Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. doi:10.1016/j.ymeth.2012.01.002
142. Silva J, Garcia V, Rodriguez M, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51(4):409–418. doi:10.1002/gcc.21926
143. Campanella C, Rappa F, Sciumè C, et al. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer. 2015;121(18):3230–3239. doi:10.1002/cncr.29499
144. Namba Y, Sogawa C, Okusha Y, et al. Depletion of lipid efflux pump ABCG1 triggers the intracellular accumulation of extracellular vesicles and reduces aggregation and tumorigenesis of metastatic cancer cells. Front Oncol. 2018;8:376. doi:10.3389/fonc.2018.00376
145. Sun B, Li Y, Zhou Y, et al. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234(2):1416–1425. doi:10.1002/jcp.26936
146. Chen MS, Xu R, Ji H, et al. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci Rep. 2016;6:38397. doi:10.1038/srep38397
147. Galon J, Fridman WH, Pages F. The adaptive immunologic microenvironment in colorectal cancer: a novel perspective. Cancer Res. 2007;67(5):1883–1886. doi:10.1158/0008-5472.CAN-06-4806
148. Peddareddigari VG, Wang D, Dubois RN. The tumor microenvironment in colorectal carcinogenesis. Cancer Microenviron. 2010;3(1):149–166. doi:10.1007/s12307-010-0038-3
149. Fu Q, Zhang Q, Lou Y, et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. 2018;37:6105–6118. doi:10.1038/s41388-018-0391-0
150. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi:10.1038/nrclinonc.2013.110
151. Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32(3–4):643–671. doi:10.1007/s10555-013-9444-6
152. Gold P, Freedman SO. Specific carcinoembryonic antigens of the human digestive system. J Exp Med. 1965;122(3):467–481. doi:10.1084/jem.122.3.467
153. Thomas P, Toth CA, Saini KS, Jessup JM, Steele G
154. Oikawa S, Inuzuka C, Kuroki M, Matsuoka Y, Kosaki G, Nakazato H. Cell adhesion activity of non-specific cross-reacting antigen (NCA) and carcinoembryonic antigen (CEA) expressed on CHO cell surface: homophilic and heterophilic adhesion. Biochem Biophys Res Commun. 1989;164(1):39–45. doi:10.1016/0006-291x(89)91679-3
155. Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9(2):67–81. doi:10.1006/scbi.1998.0119
156. Blumenthal RD, Hansen HJ, Goldenberg DM. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen). Cancer Res. 2005;65(19):8809–8817. doi:10.1158/0008-5472.CAN-05-0420
157. Ordonez C, Screaton RA, Ilantzis C, Stanners CP. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 2000;60(13):3419–3424.
158. Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 2004;23(2):465–473. doi:10.1038/sj.onc.1207036
159. Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 2001;47(4):624–630.
160. Kim KJ, Kwon HJ, Kim MC, Bae YK. CD9 expression in colorectal carcinomas and its prognostic significance. J Pathol Transl Med. 2016;50(6):459–468. doi:10.4132/jptm.2016.10.02
161. Mokhtari M, Zakerzade Z. EPCAM expression in colon adenocarcinoma and its relationship with TNM staging. Adv Biomed Res. 2017;6:56. doi:10.4103/2277-9175.205529
162. Han S, Zong S, Shi Q, et al. Is Ep-CAM expression a diagnostic and prognostic biomarker for colorectal cancer? A systematic meta-analysis. EBioMedicine. 2017;20:61–69. doi:10.1016/j.ebiom.2017.05.025
163. Tan CR, Zhou L, El-Deiry WS. Circulating tumor cells versus circulating tumor DNA in colorectal cancer: pros and cons. Curr Colorectal Cancer Rep. 2016;12(3):151–161. doi:10.1007/s11888-016-0320-y
164. Mathai RAVR, Reddy BS, Thomas L, Udupa K, Kolesar J, Rao M. Potential utility of liquid biopsy as a diagnostic and prognostic tool for the assessment of solid tumors: implications in the precision oncology. J Clin Med. 2019;18(8):3. doi:10.3390/jcm8030373
165. Su W, Yu H, Jiang L, Chen W, Li H, Qin J. Integrated microfluidic device for enrichment and identification of circulating tumor cells from the blood of patients with colorectal cancer. Dis Markers. 2019;2019:8945974. doi:10.1155/2019/8945974
166. Willms A, Muller C, Julich H, et al. Tumour-associated circulating microparticles: a novel liquid biopsy tool for screening and therapy monitoring of colorectal carcinoma and other epithelial neoplasia. Oncotarget. 2016;7(21):30867–30875. doi:10.18632/oncotarget.9018
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.