Back to Journals » Neuropsychiatric Disease and Treatment » Volume 18
Combining Multiple Indices of Diffusion Tensor Imaging Can Better Differentiate Patients with Traumatic Brain Injury from Healthy Subjects
Authors Abdelrahman HAF , Ubukata S, Ueda K, Fujimoto G, Oishi N , Aso T , Murai T
Received 26 December 2021
Accepted for publication 1 July 2022
Published 23 August 2022 Volume 2022:18 Pages 1801—1814
DOI https://doi.org/10.2147/NDT.S354265
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Taro Kishi
Hiba Abuelgasim Fadlelmoula Abdelrahman,1 Shiho Ubukata,2 Keita Ueda,1 Gaku Fujimoto,1 Naoya Oishi,2 Toshihiko Aso,3 Toshiya Murai1
1Kyoto University Graduate School of Medicine-Department of Psychiatry, Kyoto, 606-8507, Japan; 2Kyoto University Graduate School of Medicine-Medical Innovation Center, Kyoto, 606-8507, Japan; 3Laboratory for Brain Connectomics Imaging, RIKEN Center for Biosystems Dynamics Research, Kobe, 650-0047, Japan
Correspondence: Hiba Abuelgasim Fadlelmoula Abdelrahman, Email [email protected]
Aim: Diffuse axonal injury (DAI) is one of the most common pathological features of traumatic brain injury (TBI). Diffusion tensor imaging (DTI) indices can be used to identify and quantify white matter microstructural changes following DAI. Recently, many studies have used DTI with various machine learning approaches to predict white matter microstructural changes following TBI. The current study sought to examine whether our classification approach using multiple DTI indices in conjunction with machine learning is a useful tool for diagnosing/classifying TBI patients and healthy controls.
Methods: Participants were adult patients with chronic TBI (n = 26) with DAI pathology, and age- and sex-matched healthy controls (n = 26). DTI images were obtained from all participants. Tract-based spatial statistics analyses were applied to DTI images. Classification models were built using principal component analysis and support vector machines. Receiver operator characteristic curve analysis and area under the curve were used to assess the classification performance of the different classifiers.
Results: Tract-based spatial statistics revealed significantly decreased fractional anisotropy, as well as increased mean diffusivity, axial diffusivity, and radial diffusivity in patients with TBI compared with healthy controls (all p-values < 0.01). The principal component analysis and support vector machine-based machine learning classification using combined DTI indices classified patients with TBI and healthy controls with an accuracy of 90.5% with an area under the curve of 93 ± 0.09.
Conclusion: These results highlight the potential of our approach combining multiple DTI measures to identify patients with TBI.
Keywords: diffuse axonal injury, diffusion tensor imaging, machine learning, screening, traumatic brain injury
Corrigendum for this paper has been published.
Introduction
Traumatic brain injury (TBI) is a common cause of mortality and morbidity worldwide.1 Individuals with TBI have an elevated risk of developing numerous neurocognitive and psychiatric illnesses.2,3 Diffuse axonal injury (DAI) is one of the most common and important pathological features of TBI.4,5 DAI following TBI occurs as a result of acceleration/deceleration trauma to the brain, which can result in the shearing and disruption of white matter axons.6,7 DAI may include extensive microscopic axonal damage, even in the absence of abnormal findings on conventional computed tomography (CT) and magnetic resonance imaging (MRI).8 Unlike conventional imaging techniques, diffusion tensor imaging (DTI) can be used to identify and quantify white matter microstructural changes following DAI.9–12 DTI is therefore considered to be one of the most promising techniques for the study and diagnosis of DAI following TBI.
DTI measures the directional coherence of water diffusion along white matter axons.13,14 A variety of parameters can be obtained from a DTI scan (DTI indices), including fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD). DTI indices reflect the integrity of white matter microstructures. They have been extensively applied as neuroimaging biomarkers in the study of a range of clinical conditions, such as multiple sclerosis, perinatal asphyxial encephalopathy, Alzheimer’s disease, and vascular cognitive impairment.15–17 FA measures anisotropic water diffusion within white matter fibers and can be used to indicate axonal structural integrity. Reduced FA is believed to reflect a loss of axonal integrity, indicating possible damage to myelin or the axon membrane and/or decreased axonal coherence. MD describes the average magnitude of water diffusion, regardless of the diffusion direction. Increased MD is thought to reflect overall structural disintegration. AD is defined as the diffusion of water parallel to white matter fibers, while RD is the diffusion of water perpendicular to white matter fibers.18 Both AD and RD are directionality indices, and can be used to specifically differentiate axonal injury from demyelination in white matter tracts.15,19
Several previous studies have used DTI indices to investigate white matter microstructural changes in patients with TBI at the group level.20–24 A common finding across previous studies is lower FA and higher MD in TBI patients.21,25–30 The majority of these studies have focused on mild TBI (mTBI) with very few studies examining moderate and severe TBI. Although FA and MD changes have been extensively reported, only a small number of studies compared AD and RD between patients with TBI and healthy controls. In one study, Kinnunen et al examined 28 patients with TBI (eight mild TBI and 20 moderate/severe TBI) and 26 healthy controls. The results revealed that patients with TBI exhibited large areas of reduced FA and increased MD, as well as increased AD and RD.11 In another study, Perez et al examined AD and RD in 16 patients with chronic moderate to severe TBI. The results revealed disproportionally high AD and RD in patients with TBI.31 Cubon et al examined 39 patients with chronic TBI (22 mTBI and 17 moderate/severe TBI). In a whole-brain analysis and an analysis using specific regions of interest, it was found that patients with moderate to severe TBI exhibited increased AD and RD.32
Most of the methods used for studying white matter damage following TBI in previous studies have focused on specific large white matter tracts that are known to be most commonly affected by TBI, such as the corpus callosum.8,21–24 These approaches assume that there is a spatial overlap of lesions across affected TBI patients, which is not consistent with the diffuse spatial heterogenous disruption of white matter microstructures reported in human studies and experimental models of TBI.33 In addition, many studies included TBI patients with comorbidities, such as depression and post-traumatic stress disorder (PTSD), which can affect interpretations of DTI findings.30
Despite the ability of DTI indices to reflect white matter microstructural changes following TBI at the group level, it is currently unclear how best to use them in individual patients. In recent years, several studies have investigated the use of machine learning (ML) approaches for studying DTI measures for the prediction of white matter microstructural changes in TBI patients at the individual level to aid in the classification and/or diagnosis of TBI patients. Unlike traditional univariate analysis methods such as voxel-based analyses, supervised ML techniques applying multivariate analyses enable better predictions and inferences at the individual level.34–36
Saha et al recently reviewed 148 papers published between January 2010 and December 2019 related to the human brain, DTI, and ML, and identified five studies that involved the classification of TBI patients versus healthy controls.37–42 Various DTI measures were extracted and used for voxel-based analysis, tractography, and generating connectivity matrices. Support vector machine (SVM) a supervised ML technique were used in all five studies. In one of these studies, Fagerholm et al applied linear SVM analysis to DTI graph metrics of white matter connectivity data from patients with moderate/severe TBI and reported an overall model accuracy of 93.4%.41 In addition to the studies included in Saha et al’s review, some previous studies used different ML techniques other than SVM, such as the study conducted by Mitra et al and Lui et al43,44 Mitra et al applied a decision ensemble ML technique to DTI data (FA and network-based statistics) and reported an overall classification accuracy of 68%.43 Lui et al used other MRI-based imaging parameters in addition to DTI (T1 and rs-fMRI) and tested several different classification approaches on 23 patients with mild TBI and 25 healthy controls.44 The researchers obtained an accuracy of 86% with a multilayer perceptron (neural network) using only relevant variables, and an accuracy of 80% with a Bayesian network using all variables.44
In the current study, we extracted four DTI indices (FA, MD, AD and RD) from all white matter tracts to study the spatial and pathological heterogenous changes in white matter microstructures following TBI. We applied PCA as a feature reduction method followed by SVM to identify the classification model that best differentiated between the TBI and healthy control group. To the best of our knowledge, this is the first study to investigate the use of multiple DTI features extracted throughout the white matter tracts to identify white matter microstructural changes across the whole spectrum of TBI severity.
Materials and Methods
Participants
Participants were recruited through specialist TBI outpatient clinics of the neuropsychology unit at the Department of Psychiatry and the Department of Neurosurgery at Kyoto University Hospital. Participants were recruited in the chronic phase (mean time since injury = 110.3 months, standard deviation [SD] = 90.2 months, range 6–355 months). Inclusion criteria were: 1) age > 18 years, 2) an injury sustained through significant trauma; 3) a brain MRI or CT scan showing possible diffuse pathology (without large focal lesions [> 10 mm3]); 4) the injury occurred ≥ 6 months before the study; 5) ability to give informed consent for participation; and 6) ability to undergo MRI. Exclusion criteria were: 1) history of another TBI with altered consciousness; 2) history of drug or alcohol abuse; 3) history of neurological or psychiatric disorders before TBI onset; and 4) contraindications to MRI (eg, implanted metal, claustrophobia). Neuropsychiatrists (UK, TM) specialized in the neuropsychiatric aspects of TBI assessed patients’ MRI and CT scans and confirmed the information concerning the clinical history and residual symptoms related to the inclusion and exclusion criteria mentioned above. For comparison purposes, age and sex matched healthy control subjects were also recruited to the study.
Imaging Acquisition
For DTI, diffusion-weighted volumes were acquired using a 3.0-T whole body scanner (MAGNETOM Tim Trio; Siemens, Erlangen, Germany) using a 40-mT/m gradient and a receiver-only eight-channel phased-array head coil. The scanning parameters were: TE = 96 ms, repetition time TR = 10,500 ms, matrix = 96 × 96, field of view = 192×192 mm, 70 contiguous axial slices of 2.0-mm thickness, 64 non-collinear axis motion-probing gradients and b = 1500 s/mm2. The b = 0 images were acquired before each set of nine diffusion-weighted images, giving 90 volumes in total. These images were obtained using a 2-mm-section isotropic voxel (2 × 2×2 mm3). The duration of the DTI sequence was 8 minutes and 5 seconds.
Image Processing
DTI data were processed using the FMRIB Software Library (FSL) version 5.2 (FSL; Oxford Centre for Functional MRI of the Brain; www.fmrib.ox.ac.uk/fsl).45 DTI images were registered to the b = 0 image using affine transformations to minimize distortion caused by head motion and eddy currents.46 Images were then brain-extracted using the Brain Extraction Tool. Using the DTIFIT program, diffusion tensors were calculated for whole-brain volumes and fractional anisotropy (FA) maps, axial diffusivity maps (AD [λ1]), radial diffusivity maps (RD [(λ2+λ3)/2]), and mean diffusivity maps (MD [(λ1+ λ2+λ3)/3]) were generated.47–50 MD, AD, and RD are presented in units of mm2/s. TBSS was used for voxel-wise analysis of the DTI indices maps.51 TBSS is a fully automated whole-brain analysis technique for applying voxel-wise statistics to diffusion indices while minimizing the effects of misalignment that can occur using conventional voxel-based analysis methods.51
The TBSS procedure included non-linear registration of all participants’ FA images into the common FMRIB58_FA template space.50 In TBSS, non-linear registration is performed to align each FA image to every other FA image, followed by calculation of the amount of warping needed for the images to be aligned. The most representative image is determined as the one requiring the least warping for all other images to be aligned to it.52 The aligned FA images were then averaged to create a four-dimensional mean FA image, which was then subjected to thinning to create a mean FA skeleton representing the center of all white matter tracts, thereby removing partial-volume confounds. The FA skeleton was then thresholder at an FA value of 0.2 to limit the effects of poor alignment across participants, to exclude areas with extremely low mean FA, and to ensure that gray matter and cerebrospinal fluid voxels were excluded from the skeleton. The same non-linear transformation steps were applied to the MD, AD, and RD maps. For statistical analysis, the randomize tool in FSL was used to conduct non-parametric permutation-based statistics using the threshold-free cluster enhancement (TFCE) method with 10,000 permutations to investigate group differences in FA, MD, AD, and RD. Voxel-wise maps were thresholder at p < 0.05 and corrected for multiple comparisons using family-wise error rate (FWE). The significant white matter tracts were identified with reference to the atlas tool JHU ICBM-DTI-81 white matter labels. To quantitatively assess the structural differences between the patients with TBI and healthy controls, mean DTI indices for each participant were extracted by averaging FA, MD, AD, and RD for the significant white matter clusters using the fslstats tool in FSL. The time required for image processing was 12 minutes per participant, including the TBSS analysis.
Feature Reduction and Feature Extraction
To avoid overfitting and improve prediction accuracy, principal component analysis (PCA) was used as a non-supervised feature reduction technique. PCA reduces redundant features by linearly transforming correlated variables (eg, voxels in a neuroimaging scan) into a lower number of uncorrelated variables known as principal components (PCs). Therefore, high-dimensional neuroimaging data can be transformed into relatively few PCs that maximally explain the variance of the data.53–55 PCA has been successfully used for dimensionality reduction in previous neuroimaging classification studies of schizophrenia, Alzheimer’s disease, and major depressive disorder.56
PCA was applied to the entire FA, MD, AD, and RD skeletonized maps, in which each voxel represented a variable in the cross-validation training dataset, and the same transformation was then applied to the test dataset. A voxel-wise approach was used to combine FA, MD, AD, and RD into one dataset named “ALL”, and PCA was applied to the ALL dataset in the same manner.
Support Vector Machines
A SVM was used to perform ML analysis using the PCs of the DTI indices. The SVM employs a maximum margin classification algorithm designed to derive the optimal separation between two classes of data (TBI patients group and healthy controls group) by identifying a “hyper plane” that crosses the n-dimensional space to separate the training dataset into two pre-defined labels (a TBI patient and a healthy control).40,57
For each DTI index, an SVM classification task was trained to distinguish patients with TBI from healthy controls. PCs of the entire skeletonized maps of each DTI index and the ALL dataset were evaluated. Therefore, five classification tasks were used (FA, MD, AD, RD, and ALL). Five-fold cross validation was used to yield an unbiased assessment of the classification method and prevent overestimation. A linear kernel SVM was chosen as a classifier and the hyperparameter (C) of the linear kernel SVM was fixed to 1.0. To evaluate the different classification tasks, the mean accuracy rate and its SD were calculated for the five-fold cross validation for each classification task (Figure 1). To validate the robustness of the classification results, 1000-times permutation tests were conducted to assess the statistical significance of the classification accuracy scores. To further estimate the performance of the different classification tasks, receiver operating characteristic curves (ROC) were plotted for each classifier and the area under the curve (AUC) values were obtained. The AUC quantifies the overall ability of the classifier to distinguish between the TBI and the healthy participants. The ML analysis including feature extraction was conducted in Python using the Python scikit-learn library. The time required for each classifier was as follows: (ALL) SVM classification: 8.54 s, FA SVM classification: 1.92 s, MD SVM classification: 1.78 s, AD SVM classification: 1.78 s, and RD SVM classification: 1.76 s.
Results
Twenty-six patients with TBI (20 males, mean age: 40.15 years, SD: 14.93) were recruited for the study. The injury mechanism was motor vehicle accident in 22 of the recruited patients with TBI, falling in three patients, and sports-related injury in one patient. We identified “diffuse” injury based on radiological criteria (MRI: fluid-attenuated inversion recovery [FLAIR]; repetition time [TR] = 4200 ms; echo time [TE] = 94 ms; resolution 0.7×0.7 × 3.0 mm3; and susceptibility weighted imaging [SWI]; TR = 28 ms; TE = 20 ms; resolution 0.5×0.5 × 1.2 mm3). Patients with large lesions that included white matter (eg, lobar contusions, hematoma/hemorrhage > 2 mm diameter) were excluded from the study. According to the Glasgow Coma Scale (GCS) or the Japan Coma Scale (JCS; a measure of the severity of impaired consciousness used in Japan), five patients (19.2%) had mild TBI, 2 (7.7%) had moderate TBI and 19 (73.1%) had severe TBI. The relationship between the JCS score and the severity of the injury has been explored previously.58 Regarding comorbidities, two patients had bipolar disorder, which was well-controlled by medication at the time of the study. There were no patients with PTSD.
For comparison purposes, 26 age- and sex-matched healthy controls (20 males, mean age: 40.08 years, SD: 12.82) were recruited for the study. Participants in the healthy control group had no major psychiatric disorders, as assessed using structured clinical interviews (SCID-I). In addition, healthy controls had no past or current neurological or other medical diseases and were not receiving medications.
TBSS Analysis of Multiple DTI Indices
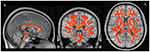
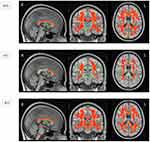
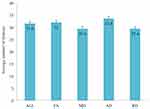
TBSS analysis of the DTI diffusion indices showed a major disruption in the white matter microstructure in the TBI group. Significantly decreased FA and increased MD, AD, and RD were present in the white matter tracts in the TBI group compared with the healthy control group (corrected p-value < 0.01) (Figures 2 and 3). Figure 4 shows boxplots providing a graphical representation for the mean values and standard deviations of FA, MD, AD, and RD for patients with TBI and healthy control groups.
Compared with the healthy control group, the TBI group showed decreased FA in the genu, body, and splenium of the corpus callosum, the middle cerebellar peduncle, the corticospinal tract, the superior cerebellar peduncle, the cerebral peduncle, the internal capsule, the superior and posterior corona radiata, the posterior thalamic radiation, the external capsule, the left cingulum (cingulate gyrus), the fornix, and the superior longitudinal fasciculus (corrected p-value < 0.01). MD was increased in the TBI group in all aforementioned white matter tracts except the superior cerebellar peduncle and the cingulum (cingulate gyrus). In addition, RD was significantly higher in all aforementioned tracts except for the corpus callosum. Unlike FA, MD, and RD, AD was significantly higher in fewer tracts in the genu, body, and splenium of the corpus callosum, the posterior limb of the internal capsule, the superior and posterior corona radiata, the external capsule, the fornix, and the superior longitudinal fasciculus (corrected p-value < 0.01).
Feature Reduction and Feature Extraction
PCA was applied to FA, MD, AD, RD and ALL during cross validation in both training and test datasets. Table 1 shows the average number of PCs that explained 90% of the variance between the TBI and healthy control groups across the five-fold cross-validation process.
 |
Table 1 Mean Accuracy Score for Each Classification Task |
SVM Classification
The ML multivariate analysis using SVM with five-fold cross validation was able to classify the patients with TBI and healthy control groups using PCs of the skeletonized maps of each DTI index (number of voxels = 97,698) and the ALL dataset (number of voxels = 390,792). A high level of accuracy (90.5%) was achieved by the classifier trained on the combined indices dataset (ALL) and (86.5%) by the classifier trained on the FA maps (all p-values < 0.01) (Table 1). Table 1 below shows the average number of PCs and the mean accuracy score for each classifier.
ROC curves for each classifier are shown in Figure 5. The AUCs were 93% and 89% for the ALL and FA classifier tasks, respectively. Figure 6 shows the number of PCs used by each classifier to differentiate patients with TBI from healthy controls.
Discussion
In the current study, we investigated the potential of using multiple DTI indices in conjunction with ML algorithms as a diagnostic measure of TBI with DAI pathology. TBSS analyses revealed a global decrease in FA and increases in MD, AD, and RD in patients with TBI. These results are consistent with those of previous studies.25,28,30,59–61 A recent study by Mohamed et al examined white matter alterations in Vietnam War veterans with moderate-to-severe TBI and PTSD. The findings revealed lower FA, and distinctly higher MD, AD, and RD in the major white matter tracts, including the corpus callosum, external and internal capsules, cingulum, and inferior and superior longitudinal fasciculi.30 Despite the fact that DTI indices are mathematically related to each other, we studied them together as different DTI indices convey different aspects of white matter microstructure, which are potentially complementary for differentiating between the TBI and healthy control groups.18,19
The main purpose of the current study was to investigate whether we could achieve reasonable accuracy in classifying patients with TBI and healthy participants. Thus, DTI indices derived from the diffusion images were used together with PCA and SVMs to classify patients with TBI and healthy controls. Five classification models were examined, and the model using the combined indices (ALL) was found to be the most accurate, with 90.5% accuracy and a high AUC value (93%, SD 0.09). AD and RD had a higher AUC (90%, 91% respectively) in comparison with FA and MD (89%,86% respectively). Previous DTI studies in humans and animal models of human diseases suggest that AD and RD are more specific markers of demyelination and axonal damage than FA and MD.63–66 The RD measure is most affected by the integrity of myelin sheaths while AD may reflect axonal degeneration. Therefore, the direct link with specific neuropathological changes in the case of AD and RD may be the reason behind the increased accuracy of these measures compared to FA and MD.
Previous studies used SVM and other ML statistical techniques with different DTI parameters and obtained variable levels of accuracy.37–44,66,67 Vergara et al applied an SVM approach in mTBI patients, using FA and resting-state functional network connectivity (rsFNC) as features in their models.38 They reported that rsFNC within the default mode network provided the best classification accuracy (84%), followed by FA (75.5%). In the current study, our model achieved higher classification for FA (86.5%). Although combining both rsFNC and FA failed to boost classification accuracy (74.5%) in Vergara et al’s study, the classification accuracy was improved in the current study by combining the four DTI indices.38 Fagerholm et al applied SVM analysis to DTI graph metrics of white matter connectivity data from patients with moderate/severe TBI and reported an overall model accuracy of 93.4%.41 Although Fagerholm et al’s approach obtained higher accuracy than our model (90.5%), a practical advantage of our method is that it uses well-known DTI indices (FA, MD, AD, and RD), rather than graph metrics of white matter connectivity.
A recent similar study conducted by Harrington et al used SVMs and multiple DTI indices to differentiate subjects with blast-related mild traumatic brain injury (bmTBI) from healthy controls.68 The researchers studied 20 subacute/chronic bmTBI and 19 healthy control combat-deployed subjects. Five DTI indices (FA, MD, RD, AD, and AD/RD ratio) were extracted using TBSS throughout white-matter tracts. The results revealed decreased RD, increased FA, and increased axial/radial diffusivity ratio (AD/RD) in the bmTBI group, mostly in anterior white matter tracts. The highest level of accuracy obtained was 89% when combining FA of the anterior/superior corona radiata and the AD/RD ratio of the corpus callosum and anterior limbs of the internal capsule. Although this previous study used a similar approach to that used in the current study, our methods and results differed from theirs in several ways. First, Harrington et al studied only mTBI, whereas we investigated patients with mild, moderate and severe TBI who were recruited to the study on the basis of radiological evidence of the injury. Second, Harrington et al’s findings regarding DTI indices revealed the opposite pattern to that in our results, reporting increased FA and decreased RD with no significant change in MD and AD. This inconsistency is related to the different injury effects caused by blast trauma, and the different post-injury timeframe involved. Finally, our SVM model differed from Harrington et al’s model, because we combined four different DTI indices (FA, MD, AD, and RD) from all white matter tracts and obtained higher accuracy (90.5%).
Regarding using ML techniques other than SVMs, Mitra et al applied a decision ensemble ML technique to FA and network-based statistics and reported an overall classification accuracy of 68%.43 Lui et al obtained an accuracy of 86% using a multilayer perceptron (neural network), and 80% with a Bayesian network.44 Our model achieved higher classification accuracy. Therefore, the high classification accuracy of this combined DTI indices model indicates its potential usefulness as a tool for TBI diagnosis/classification, which we plan to validate in a larger, independent sample of TBI patients and healthy controls in future studies.
Because this study was performed in actual clinical settings, it is sometimes the case that acute-stage clinical information or neuroimaging data are no longer available for some of our TBI subjects, especially those presented several years after the injury onset. Our approach, though it needs further improvement and tuning, might be utilized as an aid for the diagnosis of TBI in various clinical settings. Future studies with a larger number of subjects controlling for clinical variables, especially the injury severity and the time since injury, might bring our method closer to clinical application.
Limitations
The findings of the current study should be considered in the context of several limitations. First, our sample size was relatively small, which may have affected the generalizability of our findings. Subjects with mTBI and those with a short time since injury (acute/Subacute TBI) are less represented in our sample. As for mTBI subjects, it remains to be unanswered to what extent our method is applicable. As for the time since injury, it should be noted that DTI measures change over time after TBI. Thus, future studies, increasing the number of subjects substantially, and covering all ranges of severity as well as time since injury, would address this issue. Second, which is also related to the first limitation, the study did not investigate the influence of clinical and social outcomes because of the limited number of participants. Future studies with larger sample sizes should investigate the impact of the potentially relevant measures, such as acute imaging changes, neurocognitive consequences, and measures of gray matter atrophy. Finally, this study is a cross-sectional study. Future longitudinal studies would increase further utility in clinical settings.
Conclusions
In the present study, we investigated the utility of using combined DTI indices with SVMs for the classification/diagnosis of TBI. Four DTI indices were extracted from all white matter tracts instead of a specific region or tract to elucidate the spatial and pathological heterogeneity of white matter microstructural changes following TBI. Despite the small sample size and other limitations in this study, the high accuracy achieved by our classification model suggests its potential usefulness as a tool to aid in the objective classification/diagnosis of TBI. We plan to validate the robustness of our model in a larger independent sample while incorporating other clinical variables including neuropsychiatric comorbidities in future studies.
Abbreviations
TBI, traumatic brain injury; mTBI, mild traumatic brain injury; DAI, diffusion axonal injury; DTI, diffusion tensor imaging; FA, fractional anisotropy; MD, mean diffusivity; AD, axial diffusivity; RD, radial diffusivity; ALL, voxel-wise combination of FA, MD, AD, and RD in one dataset; TBSS, tract-based spatial statistics; TFCE, threshold-free cluster enhancement; FEW, family-wise error rate; ML, machine learning; PCA, principal component analysis; SVM, support vector machine; PCs, principal components; GCS, Glasgow Coma Scale; JCS, Japan Coma Scale; ROC, receiving operating curve; AUC, area under the curve; bmTBI, blast-related mild traumatic brain injury.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the Committee on Medical Ethics of Kyoto University and all procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all individual participants included in the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by an Industrial Disease Clinical Research Grant (150502-02); a Health Labor Sciences Research Grant; a Grant-in-Aid for Young Scientists (19K17110), B (21H02805), and C (17K10327, 18K07712) from the Japan Society for the Promotion of Science; Innovative Areas (16H06402) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT); a research grant from the National Mutual Insurance Federation of Agricultural Cooperatives; and ISHIZUE 2020 from the Kyoto University Research Development Program. The funding sources had no role in in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Disclosure
The authors declare that they have no competing interests.
References
1. Hyder AA, Wunderlich CA, Puvanachandra P, Gururaj G, Kobusingye OC. The impact of traumatic brain injuries: a global perspective. NeuroRehabilitation. 2007;22(5):341–353. doi:10.3233/nre-2007-22502
2. Koponen S, Taiminen T, Portin R, et al. Axis I and II psychiatric disorders after traumatic brain injury: a 30-year follow-up study. Am J Psychiatry. 2002;159(8):1315–1321. doi:10.1176/appi.ajp.159.8.1315
3. Holsinger T, Steffens DC, Phillips C, et al. Head injury in early adulthood and the lifetime risk of depression. Arch Gen Psychiatry. 2002;59(1):17–22. doi:10.1001/archpsyc.59.1.17
4. Smith DH, Meaney DF. Axonal damage in traumatic brain injury. Neuroscientist. 2000;6(6):483–495. doi:10.1177/107385840000600611
5. Browne KD, Chen X-H, Meaney DF, Smith DH. Mild traumatic brain injury and diffuse axonal injury in swine. J Neurotrauma. 2011;28(9):1747–1755. doi:10.1089/neu.2011.1913
6. Gennarelli TA, Thibault LE, Adams JH, Graham DI, Thompson CJ, Marcincin RP. Diffuse axonal injury and traumatic coma in the primate. Ann Neurol. 1982;12(6):564–574. doi:10.1002/ana.410120611
7. Werner C, Engelhard K. Pathophysiology of traumatic brain injury. Br J Anaesth. 2007;99(1):4–9. doi:10.1093/bja/aem131
8. Hulkower M, Poliak D, Rosenbaum S, Zimmerman M, Lipton ML. A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR Am J Neuroradiol. 2013;34(11):2064–2074. doi:10.3174/ajnr.A3395
9. Klimova A, Korgaonkar MS, Whitford T, Bryant RA. Diffusion tensor imaging analysis of mild traumatic brain injury and posttraumatic stress disorder. Biol Psychiatry Cogn Neurosci Neuroimaging. 2019;4(1):81–90. doi:10.1016/j.bpsc.2018.10.004
10. Assaf Y, Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J Mol Neurosci. 2008;34(1):51–61. doi:10.1007/s12031-007-0029-0
11. Kinnunen KM, Greenwood R, Powell JH, et al. White matter damage and cognitive impairment after traumatic brain injury. Brain. 2010;134(2):449–463. doi:10.1093/brain/awq347
12. Niogi SN, Mukherjee P. Diffusion tensor imaging of mild traumatic brain injury. J Head Trauma Rehabil. 2010;25(4):241–255. doi:10.1097/HTR.0b013e3181e52c2a
13. Alexander AL, Lee JE, Lazar M, Field AS. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4(3):316–329. doi:10.1016/j.nurt.2007.05.011
14. Douglas DB, Iv M, Douglas PK, et al. Diffusion tensor imaging of TBI: potentials and challenges. Top Magn Reson Imaging. 2015;24(5):241. doi:10.3390/medsci7010002
15. Liu Y, Duan Y, He Y, et al. Whole brain white matter changes revealed by multiple diffusion metrics in multiple sclerosis: a TBSS study. Eur J Radiol. 2012;81(10):2826–2832. doi:10.1016/j.ejrad.2011.11.022
16. Porter EJ, Counsell SJ, Edwards AD, Allsop J, Azzopardi D. Tract-based spatial statistics of magnetic resonance images to assess disease and treatment effects in perinatal asphyxial encephalopathy. Pediatr Res. 2010;68(3):205–209. doi:10.1203/PDR.0b013e3181e9f1ba
17. Chen, H.J., Gao, Y.Q., Che, C.H., Lin, H. and Ruan, X.L., Diffusion tensor imaging with tract-based spatial statistics reveals white matter abnormalities in patients with vascular cognitive impairment. Front Neuroanat. 2018;12:53.
18. Aung WY, Mar S, Benzinger TL. Diffusion tensor MRI as a biomarker in axonal and myelin damage. Imaging Med. 2013;5(5):427.
19. Shenton ME, Hamoda H, Schneiderman J, et al. A review of magnetic resonance imaging and diffusion tensor imaging findings in mild traumatic brain injury. Brain Imaging Behav. 2012;6(2):137–192. doi:10.1007/s11682-012-9156-5
20. Inglese M, Makani S, Johnson G, et al. Diffuse axonal injury in mild traumatic brain injury: a diffusion tensor imaging study. J Neurosurg. 2005;103(2):298–303. doi:10.3171/jns.2005.103.2.0298
21. Kraus MF, Susmaras T, Caughlin BP, Walker CJ, Sweeney JA, Little DM. White matter integrity and cognition in chronic traumatic brain injury: a diffusion tensor imaging study. Brain. 2007;130(10):2508–2519. doi:10.1093/brain/awm216
22. Chang MC, Kim SH, Kim OL, Bai DS, Jang SH. The relation between fornix injury and memory impairment in patients with diffuse axonal injury: a diffusion tensor imaging study. NeuroRehabilitation. 2010;26(4):347–353. doi:10.3233/NRE-2010-0572
23. Kasahara K, Hashimoto K, Abo M, Senoo A. Voxel-and atlas-based analysis of diffusion tensor imaging may reveal focal axonal injuries in mild traumatic brain injury—comparison with diffuse axonal injury. Magn Reson Imaging. 2012;30(4):496–505. doi:10.1016/j.mri.2011.12.018
24. Kumar R, Gupta RK, Husain M, et al. Comparative evaluation of corpus callosum DTI metrics in acute mild and moderate traumatic brain injury: its correlation with neuropsychometric tests. Brain Inj. 2009;23(7–8):675–685. doi:10.1080/02699050903014915
25. Aoki Y, Inokuchi R, Gunshin M, Yahagi N, Suwa H. Diffusion tensor imaging studies of mild traumatic brain injury: a meta-analysis. J Neurol Neurosurg Psychiatry. 2012;83(9):870–876. doi:10.1136/jnnp-2012-302742
26. Dodd AB, Epstein K, Ling JM, Mayer AR. Diffusion tensor imaging findings in semi-acute mild traumatic brain injury. J Neurotrauma. 2014;31(14):1235–1248.
27. Gardner A, Kay-Lambkin F, Stanwell P, et al. A systematic review of diffusion tensor imaging findings in sports-related concussion. J Neurotrauma. 2012;29:2521–2538.
28. Arfanakis K, Haughton VM, Carew JD, Rogers BP, Dempsey RJ, Meyerand ME. Diffusion tensor MR imaging in diffuse axonal injury. AJNR Am J Neuroradiol. 2002;23(5):794–802.
29. Kennedy MR, Wozniak JR, Muetzel RL, et al. White matter and neurocognitive changes in adults with chronic traumatic brain injury. J Int Neuropsychol Soc. 2009;15(1):130–136. doi:10.1017/S1355617708090024
30. Mohamed AZ, Cumming P, Nasrallah FA. White matter alterations are associated with cognitive dysfunction decades after moderate-to-severe traumatic brain injury and/or posttraumatic stress disorder. Biol Psychiatry Cogn Neurosci Neuroimaging. 2021;6(11):1100–1109. doi:10.1016/j.bpsc.2021.04.014
31. Perez AM, Adler J, Kulkarni N, et al. Longitudinal white matter changes after traumatic axonal injury. J Neurotrauma. 2014;31(17):1478–1485. doi:10.1089/neu.2013.3216
32. Cubon VA, Putukian M, Boyer C, Dettwiler A. A diffusion tensor imaging study on the white matter skeleton in individuals with sports-related concussion. J Neurotrauma. 2011;28(2):189–201. doi:10.1089/neu.2010.1430
33. Jorge RE, Acion L, White T, et al. White matter abnormalities in veterans with mild traumatic brain injury. AJP. 2012;169(12):1284–1291. doi:10.1176/appi.ajp.2012.12050600
34. Orru G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev. 2012;36(4):1140–1152. doi:10.1016/j.neubiorev.2012.01.004
35. Linden DE. The challenges and promise of neuroimaging in psychiatry. Neuron. 2012;73(1):8–22. doi:10.1016/j.neuron.2011.12.014
36. Zarogianni E, Moorhead TW, Lawrie SM. Towards the identification of imaging biomarkers in schizophrenia, using multivariate pattern classification at a single-subject level. Neuroimage Clin. 2013;3:279–289. doi:10.1016/j.nicl.2013.09.003
37. Saha A, Fadaiefard P, Rabski JE, Sadeghian A, Cusimano MD. Machine learning applications using diffusion tensor imaging of human brain: a PubMed literature review. arXiv preprint arXiv:2012.10517; 2020.
38. Vergara Victor M, MayerAndrew R, KiehlKent A. Detection of mild traumatic brain injury by machine learning classification using resting state functional network connectivity and fractional anisotropy. J Neurotrauma. 2017. doi:10.1089/neu.2016.4526
39. Zheng ZS, Reggente N, Lutkenhoff E, Owen AM, Monti MM. Disentangling disorders of consciousness: insights from diffusion tensor imaging and machine learning. Hum Brain Mapp. 2017;38(1):431–443. doi:10.1002/hbm.23370
40. Hellyer PJ, Leech R, Ham TE, Bonnelle V, Sharp DJ. Individual prediction of white matter injury following traumatic brain injury. Ann Neurol. 2013;73(4):489–499. doi:10.1002/ana.23824
41. Fagerholm ED, Hellyer PJ, Scott G, Leech R, Sharp DJ. Disconnection of network hubs and cognitive impairment after traumatic brain injury. Brain. 2015;138(6):1696–1709. doi:10.1093/brain/awv075
42. Goswami R, Dufort P, Tartaglia MC, et al. Frontotemporal correlates of impulsivity and machine learning in retired professional athletes with a history of multiple concussions. Brain Struct Funct. 2016;221(4):1911–1925. doi:10.1007/s00429-015-1012-0
43. Mitra J, Shen K-K, Ghose S, et al. Statistical machine learning to identify traumatic brain injury (TBI) from structural disconnections of white matter networks. Neuroimage. 2016;129:247–259. doi:10.1016/j.neuroimage.2016.01.056
44. Lui YW, Xue Y, Kenul D, Ge Y, Grossman RI, Wang Y. Classification algorithms using multiple MRI features in mild traumatic brain injury. Neurology. 2014;83(14):1235–1240. doi:10.1212/WNL.0000000000000834
45. Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2014;23:S208–S19. doi:10.1016/j.neuroimage.2004.07.051.
46. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078.
47. Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, Beckmann C, Jenkinson M, Smith SM. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009;45:S173–S86.
48. Ennis DB, Kindlmann G. Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magn Reson Med. 2006;55(1):136–146.
49. Behrens TE, Woolrich MW, Jenkinson M, Johansen–Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM. Characterization and propagation of uncertainty in diffusion–weighted MR imaging. Magn Reson Med. 2003;50:1077–88. doi:10.1002/mrm.10609.
50. Mori S, Wakana S, Van Zijl PC, Nagae-Poetscher L. MRI Atlas of Human White Matter. Elsevier; 2005.
51. Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ Matthews PM. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487 505. doi:10.1016/j.neuroimage.2006.02.024
52. Andersson JL, Jenkinson M, Smith S. Non-linear registration aka spatial normalisation FMRIB technial report TR07JA2. FMRIB Analysis Group of the University of Oxford; 2007.
53. Mwangi B, Tian TS, Soares JC A review of feature reduction techniques in neuroimaging. Neuroinformatics. 2014;12:229–44. doi:10.1007/s12021-013-9204-3
54. Passos IC, Mwangi B, Kapczinski F Big data analytics and machine learning: 2015 and beyond. Lancet Psychiatry. 2016;3:13–5. doi:10.1016/S2215-0366(15)00549-0
55. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–82. doi:10.1162/153244303322753616
56. Joliffe I. Principal Component Analysis. NY: Springer; 2002.
57. O’Dwyer L, Lamberton F, Bokde AL, Ewers M, Faluyi YO, Tanner C, Mazoyer B, O’Neill D, Bartley M, Collins DR. Using support vector machines with multiple indices of diffusion for automated classification of mild cognitive impairment. PloS one. 2012;7:e32441. doi:10.1371/journal.pone.0032441
58. Namiki J, Yamazaki M, Funabiki T, et al. Difficulty and inaccuracy of assessment of the consciousness level by the Glasgow Coma Scale: comparison with the Japan Coma Scale. J Jpn Soc Emerg Med. 2007;10:20–25.
59. Mayer AR, Ling JM, Yang Z, Pena A, Yeo RA, Klimaj S. Diffusion abnormalities in pediatric mild traumatic brain injury. J Neurosci. 2012;32(50):17961–17969. doi:10.1523/JNEUROSCI.3379-12.2012
60. Mannell MALJ, Gasparovic C, Phillips JP, Doezema D, Reichard R, Yeo RA. A prospective diffusion tensor imaging study in mild traumatic brain injury. Neurology. 2010;74(8):643–650. doi:10.1212/WNL.0b013e3181d0ccdd
61. Wallace EJ, Mathias JL, Ward L. Diffusion tensor imaging changes following mild, moderate and severe adult traumatic brain injury: a meta-analysis. Brain Imaging Behav. 2018;12(6):1607–1621. doi:10.1007/s11682-018-9823-2
62. Nakayama N, Okumura A, Shinoda J, et al. Evidence for white matter disruption in traumatic brain injury without macroscopic lesions. J Neurol Neurosurg Psychiatry. 2006;77(7):850–855. doi:10.1136/jnnp.2005.077875
63. Xu J, Rasmussen I-A, Lagopoulos J, Håberg A. Diffuse axonal injury in severe traumatic brain injury visualized using high-resolution diffusion tensor imaging. J Neurotrauma. 2007;24(5):753–765. doi:10.1089/neu.2006.0208
64. Main KL, Soman S, Pestilli F, et al. DTI measures identify mild and moderate TBI cases among patients with complex health problems: a receiver operating characteristic analysis of US veterans. NeuroImage Clin. 2017;16:1–6. doi:10.1016/j.nicl.2017.06.031
65. Hasan KM, Narayana PA. Retrospective measurement of the diffusion tensor eigenvalues from diffusion anisotropy and mean diffusivity in DTI. Magn Reson Med. 2006;56(1):130–137. doi:10.1002/mrm.20935
66. Martinez BI, Stabenfeldt SE. Current trends in biomarker discovery and analysis tools for traumatic brain injury. J Biol Eng. 2019;13(1):16. doi:10.1186/s13036-019-0145-8
67. Mateos-Perez JM, Dadar M, Lacalle-Aurioles M, Iturria-Medina Y, Zeighami Y, Evans AC. Structural neuroimaging as clinical predictor: a review of machine learning applications. Neuroimage Clin. 2018;20:506–522. doi:10.1016/j.nicl.2018.08.019
68. Harrington DL, Hsu PY, Theilmann RJ, et al. Detection of chronic blast-related mild traumatic brain injury with diffusion tensor imaging and support vector machines. Diagnostics. 2022;12(4):987. doi:10.3390/diagnostics12040987
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.