Back to Journals » OncoTargets and Therapy » Volume 7
Clinical significance of UGT1A1 gene polymorphisms on irinotecan-based regimens as the treatment in metastatic colorectal cancer
Authors Li M, Wang Z, Guo J, Liu J, Li C, Liu L, Shi H, Liu L, Li H, Xie C, Zhang X, Sun W, Fang S, Bi X
Received 16 May 2014
Accepted for publication 16 July 2014
Published 23 September 2014 Volume 2014:7 Pages 1653—1661
DOI https://doi.org/10.2147/OTT.S67867
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Minmin Li,1 Zhehai Wang,2 Jun Guo,2 Jie Liu,2 Changzheng Li,2 Lin Liu,2 Huan Shi,2 Liyan Liu,2 Huihui Li,2 Chao Xie,2 Xia Zhang,2 Wenwen Sun,1 Shu Fang,1 Xiang Bi1
1School of Medicine and Life Sciences, University of Jinan-Shandong Academy of Medical Sciences, 2Department of Oncology, Shandong Cancer Hospital, Shandong, People's Republic of China
Purpose: The primary aim of this research was to investigate the association between uridine diphosphate glucuronosyltransferase (UGT)1A1 gene polymorphisms and the toxicities of irinotecan-based regimens in Chinese patients with metastatic colorectal cancer.
Methods: The study analyzed the distribution of UGT1A1*28/*6 gene polymorphisms by polymerase chain reaction amplification and pyrosequencing. The adverse reactions and tumor response were evaluated according to National Cancer Institute Common Toxicity Criteria for Adverse Events, Version 3.0, and Response Evaluation Criteria In Solid Tumors, Version 1.0, criteria, respectively. The correlation between UGT1A1 gene polymorphisms and severe delayed diarrhea or neutropenia was analyzed. The influences of UGT1A1*6/*28 polymorphisms on response rate and progression-free survival were also analyzed. Survival analysis was performed by the Kaplan–Meier method, and we used the log-rank test to analyze the effect of genotypes on progression-free survival, the logistic regression model for multivariate analysis, and the Cox regression model for multivariate survival analysis.
Results: A total of 167 patients with metastatic colorectal cancer who were treated with irinotecan-based regimens and with detected UGT1A1 gene polymorphisms were enrolled in this research. The rate of UGT1A1*28 homozygous wild-type TA6/6, heterozygous mutant-type TA6/7, and homozygous mutant-type TA7/7 was 65.3% (109/167), 32.3% (54/167), and 2.4% (4/167), respectively; the incidence of UGT1A1*6 wild-type G/G was 67.1% (112/167), heterozygous mutant-type G/A accounted for 28.7% (48/167), and seven cases were homozygous mutant-type A/A (4.2%; 7/167). The incidence of grade 3 or 4 delayed diarrhea in patients carrying UGT1A1*6 (G/A and A/A) was higher than that in the wild-type (G/G) (P=0.021). The rate was significantly lower in patients with the UGT1A1*28 TA6/6 wide-type genotype than those with TA6/7 and TA7/7 mutant-type genotypes (P=0.027). However, neither UGT1A1*6 (P=0.34) nor UGT1A1*28 (P=0.232) variants were significantly associated with severe neutropenia. Our study found no significant differences of severe neutropenia in patients with different numbers of mutational alleles (P=0.354), but patients with two alleles or single allele variants had more chances to develop severe diarrhea than patients with wild-type (P=0.027). No significant differences of either response rate or progression-free survival were found among different genotypes (P>0.05).
Conclusion: For irinotecan-based regimens in metastatic colorectal cancer, the UGT1A1*28 and UGT1A1*6 locus mutations can be regarded as predictors for irinotecan-associated severe delayed diarrhea, whereas no association between UGT1A1 gene polymorphisms and severe neutropenia was observed. We also found that neither clinical response nor prognosis were significantly associated with UGT1A1 gene polymorphisms.
Keywords: uridine diphosphate glucuronosyltransferase 1A1, gene polymorphism, metastatic colorectal cancer, irinotecan
Introduction
Irinotecan (CPT-11) is a semisynthetic camptothecin; CPT-11 combined with fluorouracil has become one of the main chemotherapies for metastatic colorectal cancer (mCRC). However, its application is limited because of toxicities such as delayed diarrhea and neutropenia. Therefore, CPT-11-based regimens are often in the position of second-line chemotherapy in China.
CPT-11 is one of the S-phase cell-cycle specific drugs. It is transformed into 7-ethyl-10-hydroxycamptothecin (SN-38) by carboxylesterase in vivo. Then SN-38 exerts a cytotoxic effect by inhibiting topoisomerase I, which is required for DNA replication, inducing damage of single-strand DNA and blocking DNA replication.1 The liver is the major site of metabolism of CPT-11. CPT-11 is translated into SN-38, which has stronger cytotoxic activity after injection, and then SN-38 is converted into a water-soluble inactive metabolite, SN-38 glucuronide (SN-38G), by uridine diphosphate glucuronosyltransferases (UGTs). The latter is excreted into the bowel by bile and converted into SN-38 again under the action of β-glucuronidase from the intestinal bacteria. SN-38 leads to intestinal mucosa injury and delayed diarrhea and can also be catalyzed by UGT enzyme into SN-38G in the intestine.2 Therefore, the expression and activity of UGTs enzymes are closely related to the efficacy and adverse reaction of CPT-11, of which UGT1A1 plays a vital role. Studies showed that the activity of UGT1A1 enzyme was closely related to the UGT1A1 gene polymorphisms, especially UGT1A1*6 and UGT1A1*28.3 The change of TA repeats in the TATA box of the UGT1A1 promoter caused UGT1A1*28 polymorphism, resulting in three genotypes: TA6/TA6, TA6/TA7 (heterozygosity), and TA7/TA7 (homozygosity). With the increase in the number of TA repeats, the expression of UGT1A1 will decrease, which leads to the excessive accumulation of SN-38 and then results in CPT-11-related toxicity.2 UGT1A1*6 polymorphism, characterized by a single-nucleotide substitution in exon 1 of UGT1A1 (211G>A; GG, GA, and AA genotypes), occurred at a higher frequency in Asians.3,4 Researchers also found that UGT1A1*6 polymorphism was associated with irinotecan-related diarrhea and neutropenia in Asians, especially neutropenia.5,6
Research has found that UGT1A1 gene polymorphisms can predict the toxicities of CPT-11,7 but it is still controversial. Considering the racial disparities that exist in the UGT1A1 gene polymorphisms, we believe that the prediction of adverse reactions in Chinese patients should comprehensively take into account the effects of UGT1A1*6 and UGT1A1*28 gene polymorphisms. This study analyzed 167 patients who have received irinotecan-based chemotherapy and who have mCRC to explore the relationship between UGT1A1 gene polymorphisms and toxicities of irinotecan-based regimens.
Patients and methods
Patients
A total of 167 patients with mCRC treated in Shandong Cancer Hospital from October 2009 to October 2012 were retrospectively included in this study. Inclusion criteria were that all the cases were confirmed by histopathology or cytology for metastatic colorectal cancer and all patients were older than 18 years; had no prior systemic chemotherapy, or any previous adjuvant chemotherapy or radiotherapy must have been completed at least 4 weeks before study inclusion; had an Eastern Cooperative Oncology Group performance status of 2 or less; had a life expectancy of longer than 3 months; had neutrophil counts of 1.5×109/L or higher and platelets of 80×109/L or higher; had ALT and AST levels 2.5 times the upper limit of normal value or higher (with liver metastasis five times the upper limit of normal value or higher); had a total bilirubin level 1.5 times the upper limit of normal value or higher; had serum creatinine levels at or above the upper limit of normal value; and had a normal electrocardiogram. The study was approved by the Medical Ethics Committee of the Shandong Cancer Hospital, and all patients enrolled in this study gave informed consent.
Treatment
Eighteen patients received irinotecan-based chemotherapy as first-line treatment; it was used as second-line treatment in 141 patients; and it was used as third-line or more treatment in eight patients. The chemotherapy regimens in this study included the following regimens. Each patient received irinotecan-based chemotherapy at least once, and complete blood counts were performed after each administration of irinotecan or before the initiation of the next use.
FOLFIRI regimen
Irinotecan 180 mg/m2, 90 minutes intravenous infusion on day 1; leucovorin (LV) 400 mg/m2 intravenous infusion on day 1; fluorouracil 400 mg/m2 intravenous bolus on day 1; fluorouracil 2,400 mg/m2 intravenous over the course of 46 hours of continuous infusion; repeated every 2 weeks (101 patients).
Irinotecan plus cetuximab/bevacizumab regimen
Cetuximab 400 mg/m2 intravenous infusion on week 1, then 250 mg/m2 intravenous infusion once a week/bevacizumab 5–10 mg/kg intravenous infusion once every 2 weeks; irinotecan 180 mg/m2, 90 minutes intravenous infusion on day 1; repeated every 2 weeks (30 patients).
Irinotecan plus raltitrexed regimen
Irinotecan 180 mg/m2 intravenous infusion on day 1; raltitrexed 2.5 mg/m2 intravenous infusion on day 1; repeated every 2 weeks (18 patients).
Irinotecan plus capecitabine regimen
Irinotecan 180 mg/m2, 90 minutes intravenous infusion on day 1; capecitabine 2,000 mg/(m2 × day) by mouth, twice a day, on days 2–15; repeated every 3 weeks (18 patients).
Toxicity evaluation criteria
Toxicity was evaluated on the basis of National Cancer Institute Common Toxicity Criteria for Adverse Events, Version 3.0, criteria, which showed, as follows, for diarrhea: grade I: increase of more than four stools per day over baseline, mild increase in ostomy output compared with baseline; II: increase of four to six stools per day over baseline with intravenous fluids indicating a mild increase more than 24 hours, a moderate increase in ostomy output compared with baseline, not interfering with activities of daily living (ADL); III: increase of seven or more stools per day over baseline, incontinence, intravenous fluids for 24 hours or longer, hospitalization, severe increase in ostomy output compared with baseline, interfering with ADL; IV: life-threatening consequences (eg, hemodynamic collapse); V: death. For neutropenia, the criteria were I: <LLN (lower limits of normal)–1.5×109/L; II: <1.5–1.0×109/L; III: <1.0–0.5×109/L; and IV: <0.5×109/L; V: death. Grade III or IV delayed diarrhea and neutropenia were defined as severe toxicity.
Tumor response evaluation
The primary endpoint of this study was the severe adverse reactions, and the secondary endpoints included response rate (RR) and progression-free survival (PFS). The adverse reactions were evaluated and graded according to searching for medical history, physical examination, and routine laboratory tests (including biochemical examination, blood tests, and examination of tumor marker) in each cycle of chemotherapy. Tumor response was assessed every 6 weeks by computed tomography (CT). This study used Response Evaluation Criteria In Solid Tumors, Version 1.0, criteria to evaluate the tumor response, which was divided into complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). We used CR plus PR to calculate RR, and CR plus PR and SD to calculate disease control rate. PFS represented the time from initiation of chemotherapy to disease progression or death resulting from any cause.
UGT1A1 gene detection
Sequence analysis of UGT1A1 was identified by pyrosequencing, as described previously.8 Briefly, we extracted genomic DNA from peripheral blood, using QIAamp Blood Kit (Qiagen, Hilden, Germany). Fragments were amplified using TaKaRa polymerase chain reaction (PCR) Amplification Kit (TaKara Bio Inc., Japan). Prosequencing assay was performed with the PyroMark Q24 ID system (Qiagen) following the manufacturer’s instructions.
Statistical analysis
Statistical software Statistical Package for the Social Sciences version 17.0 software (SPSS Inc., Chicago, IL, USA) with chi-squared test, Spearman correlation analysis, and Cochran-Armitage trend test were used to analyze the relationship between genotyping and irinotecan-induced toxicity and clinical response. The chi-squared test to determine genotype frequencies among patients was compatible with the Hardy-Weinberg Equilibrium. Survival analysis was performed by the Kaplan–Meier method with the log-rank test. The logistic regression model was used for multivariate analysis, and the Cox regression model was used for multivariate survival analysis. All statistical analyses were two-sided tests, and P<0.05 was considered a significant difference.
Results
Distribution of UGT1A1 gene polymorphisms
A total of 167 mCRC patients were enrolled in this study: 87 men and 80 women, with a median age of 50 years (range, 27–71 years). Sequencing results of different UGT1A1*6/*28 genotypes are shown in Figure 1A–F. The rate of UGT1A1*28 homozygous wild-type TA6/6, heterozygous mutant-type TA6/7, and homozygous mutant-type TA7/7 was 65.3% (109/167), 32.3% (54/167), and 2.4% (4/167), respectively; UGT1A1*6 wild-type G/G (112 cases, 67.1%), heterozygous mutant-type G/A (48 cases, 28.7%), and homozygous mutant-type A/A (seven cases, 4.2%). In addition, the patients were divided into three categories according to the numbers of mutational alleles: the wild-type (patients with genotype GG and TA6/TA6; n=60 [35.9%]), unit point mutation (patients with genotype GA and TA6/TA6 or GG and TA6/TA7; n=90 [53.9%]), two-point mutation of UGT1A1 (patients with genotype AA and TA6/TA6, GA and TA6/TA7, or GG and TA7/TA7; n=17 [10.2%]; Table 1). We used the chi-squared test to determine that allele and genotype frequencies among our patients were compatible with the Hardy-Weinberg Equilibrium (P>0.05).
  | Figure 1 (A–F) Sequencing results of different UGT1A1*6*28 genotypes. |
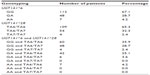  | Table 1 Genotyping of UGT1A1 in 167 metastatic colorectal cancer patients |
The relationship between UGT1A1 genotypes and toxicity
Thirty-four patients (20.4%; 34/167) had severe delayed diarrhea, and 21 patients (12.6%; 21/167) developed severe neutropenia (Table 2). The incidence of severe delayed diarrhea in patients carrying UGT1A1*6 G/A (25.0%; 12/48) and A/A (57.1%; 4/7) mutant-type genotypes was higher than that in the wild-type (G/G, 16.1%; 18/112; P=0.021). However, no significant differences of severe neutropenia were found in patients with GG (10.7%; 12/112), GA (14.6%; 7/48), and AA (28.6%; 2/7) genotypes (P=0.34).
  | Table 2 Distribution of diarrhea and neutropenia in 167 metastatic colorectal cancer patients |
The rates were significantly lower in patients with the UGT1A1*28 TA6/6 (14.7%; 16/109) wide-type genotype than those of TA6/7 (29.6%; 16/54) and TA7/7 (50.0%; 2/4) mutant-type genotypes (P=0.027). In addition, no significant differences of severe neutropenia were seen in patients with TA6/6 (10.1%; 11/109), TA6/7 (18.5%; 10/54), and TA7/7 (0.0%; 0/4; P=0.232). In addition, no significant differences of severe neutropenia were found in patients with different numbers of mutational alleles (P=0.354), but patients with single-allele or two-allele variants were more susceptible to develop severe delayed diarrhea than wild-type patients (24.4% versus 35.3% versus 10.0%; P=0.027), as shown in Table 3. Other adverse reactions such as nausea, vomiting, anemia, and thrombocytopenia were not closely associated with the various genotypes.
  | Table 3 Correlation of UGT1A1*6/*28 polymorphisms with severe toxicities |
Logistic regression model for factors affecting the severe toxicities
We used a multivariate logistic regression model to determine the underlying factors influencing severe toxicities; for instance, UGT1A1*28 and UGT1A1*6, chemotherapy regimen, and chemotherapy line. The result showed that UGT1A1*28 and UGT1A1*6 were significantly associated with severe diarrhea (P=0.01 versus P=0.003); however, our study found no significant differences of severe neutropenia with UGT1A1*28 and UGT1A1*6, and neither chemotherapy regimen nor chemotherapy line were significantly associated with severe toxicities (P>0.05), as shown in Table 4.
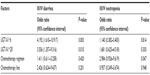  | Table 4 Logistic regression model for factors affecting the severe toxicities |
Correlation of UGT1A1*6/*28 polymorphisms with clinical response
Among the 167 patients, 132 receiving irinotecan-based chemotherapy (79.0%; 132/167) were evaluated for clinical response. Patients with CR, PR, SD, and PD were 0, 24, 62, and 46, respectively. We could not find significant differences between clinical response (RR and disease control rate) and UGT1A1*6 (P=0.290 and P=0.952) or UGT1A1*28 (P=0.286 and P=0.578) polymorphisms (Table 5). In addition, we used a multivariate logistic regression model to determine underlying factors influencing RR, such as combined genotypes, chemotherapy regimen, and chemotherapy line. However, the results showed no significant differences (P>0.05; Table 6).
  | Table 5 Correlation of UGT1A1*6/*28 polymorphisms with clinical response |
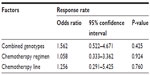  | Table 6 Logistic regression model for factors affecting clinical response |
Correlation of UGT1A1*6/*28 polymorphisms with PFS
The median PFS of UGT1A1*6 G/G, G/A, and A/A genotypes were 5.2, 5.5, and 5.6 months, respectively. There were no significant differences between them (P=0.644; Figure 2). The median PFSs of UGT1A1*28 TA6/6, TA6/7, and TA7/7 genotypes were 5.1, 5.3, and 5.6 months, and no significant differences were seen between them (P=0.557; Figure 3). Moreover, no significant differences of PFS were found in patients with different numbers of mutational alleles (Figure 4). We also used a multivariate Cox regression model to determine underlying factors influencing PFS (eg, combined genotypes, chemotherapy regimen, and chemotherapy line), but there was no statistically significant difference in PFS (P>0.05; Table 7).
  | Figure 2 Correlation of UGT1A1*6 polymorphisms with progression-free survival. |
  | Figure 3 Correlation of UGT1A1*28 polymorphisms with progression-free survival. |
  | Figure 4 Correlation of combined genotypes with progression-free survival. |
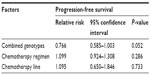  | Table 7 Cox regression model for progression-free survival |
Discussion
The correlation between UGT1A1 gene polymorphisms and CPT-11-related delayed diarrhea and neutropenia received extensive attention, and the US Food and Drug Administration declared that UGT1A1*28 testing be included on the irinotecan label as a risk factor for severe toxicity in 2005. European and American scholars have reported that gene polymorphisms of UGT1A1, UGT1A7, and UGT1A9 were likely to influence the toxicities of CPT-11.9–11 In particular, UGT1A1*28 homozygous mutant type (TA7/7) would increase the incidence of delayed diarrhea and neutropenia.9,10 However, there were significant racial differences in the distribution of UGT1A1*28 gene polymorphisms. For instance, the proportion of UGT1A1*28 homozygous mutant-type TA7/7 in Asians was only 0%–5%,12 which was much lower than the frequency of 12%–27% in Africans and 5%–15% in Caucasians.13 Therefore, whether the polymorphisms of the UGT1A1*28 gene can be the predictors of Chinese patients’ adverse reactions was still controversial. Comparatively, the mutation of UGT1A1*6 gene was more common in Asian populations, which accounted for 13%–23%.3 Studies suggested that UGT1A1*6 and UGT1A1*28 were similar in function, and that both of them can lead to the reduction of metabolism of CPT-11 inactivity in vivo and increase the risk for adverse reactions.14 Similar studies also had reported that UGT1A1*6 can be used to regulate the enzyme activity of UGT1A1 instead of UGT1A1*28; meanwhile the UGT1A1*6 mutation genotype may also increase the risk for toxicities.15
Our study analyzed the relationship between UGT1A1*6 and UGT1A1*28 gene polymorphisms and the toxicities and clinical efficacy of CPT-11. The results showed the correlation between severe delayed diarrhea and UGT1A1*6, UGT1A1*28 clearly, and the combination can also predict the occurrence of delayed diarrhea, yet UGT1A1*28 and UGT1A1*6 had nothing to do with the occurrence of neutropenia. Studies have found that when CPT-11 was administered in low dose (50–180 mg/m2), UGT1A1*28 cannot prompt the increase in hematologic toxicity, and the hematologic toxicity of patients with UGT1A1*28 homozygous mutant type will significantly increase only in the medium or high dose (200–350 mg/m2).16 Other investigators also proposed that the toxicities of low-dose CPT-11 were not affected by the status of the UGT1A1 gene.17 A meta-analysis that included 1,998 patients indicated that when CPT-11 was used in low dose, the UTG1A1*28 genotype was also linked to neutropenia, but the risk for neutropenia was significantly increased after the application of high-dose CPT-11.18 The result of another meta-analysis that contained 1,760 patients and was related to CPT-11-induced diarrhea showed that both UGT1A1*28 homozygous mutant-type and heterozygous mutant-type will increase the risk for delayed diarrhea, but this trend is not significant when the dose of CPT-11 was lower than 125 mg/m2.19 The dose of CPT-11 in this study did not belong to the high dose, and with the limitation of sample size, we did not detect hematologic toxicity being significantly increased in UGT1A1*6 and UGT1A1*28 homozygous mutant-type patients. Although Japanese studies have found that UGT1A1*28 gene polymorphisms were closely related with severe neutropenia,20 and some recent studies also indicated the same point of view.21,22 Many studies also delivered a similar view as ours, reporting that UGT1A1*28 heterozygous mutant type did not increase the risk for severe neutropenia compared with the wild-type.23,24 So the relationship between UGT1A1*28 gene polymorphism and diarrhea was more pronounced in Asia, but the relationship with neutropenia required further study.
Okuyama and colleagues have reported that homozygosity for UGT1A1*28 or UGT1A1*6 and double heterozygosity for both UGT1A1*28 and UGT1A1*6 were significantly associated with severe neutropenia in metastatic colorectal cancer patients.23 The results of our study suggest that the combination can predict the occurrence of delayed diarrhea well; this result may be related to the pharmacokinetics of irinotecan. The enzyme activity of UGT1A1 decreased for UGT1A1*28 or UGT1A1*6 homozygosity, and then AUCSN-38G/AUCSN-38 in the blood was significantly decreased and a high concentration of SN-38 in blood and intestinal tract increased the risk for the hematologic toxicity and diarrhea.25,26
The relationship between UGT1A1 gene polymorphisms and the clinical response of CPT-11-based regimens was one of the hotspots. Clinical studies demonstrated that irinotecan-based chemotherapy can improve the patients’ efficiency, PFS, and overall survival.27,28 The majority of studies found no difference in efficacy between different genotypes of UGT1A1.29,30 Some studies have proposed the clinical response of homozygous mutant-type was better,11 but so far there has been no consistent conclusion. Although this study observed the clinical efficacy of UGT1A1 mutant genotype was better than the wild-type, we found no statistically significant difference.
Whether the polymorphism of UGT1A1 gene can predict the efficacy of CPT-11 has no uniform conclusion. The mutation of UGT1A1 gene can increase the level of SN-38, which is the active metabolite of CPT-11. So the efficacy was likely to increase. However, the majority of clinical trials showed no significant difference between the efficacy of CPT-11 and UGT1A1 genotypes.29,30 Previous study only found that UGT1A1*28 homozygous mutant type was associated with time to progression (TTP) but had nothing to do with overall survival.11 However, another paper reported there was no association between UGT1A1 polymorphism and prognosis.31 Some researchers even reached the opposite conclusion, that UGT1A1*28 homozygous mutant-type patients had the worst prognosis when compared with the wild-type or heterozygous mutant-type patients, and that their TTP and overall survival were the shortest.32 Our study did not find that UGT1A1 gene polymorphism was related to RR, disease control rate, or PFS. This result may be a result of patients with UGT1A1 mutant type accounting for a higher proportion of the occurrence of serious adverse reactions, which often leads to the reduction of drug dose or the termination of chemotherapy treatment and affects the curative effect on patients. Therefore, we should consider not only the UGT1A1 gene polymorphism but also the status of the patients, the intensity of drug dose, the course of treatment, the combination of medication, and the other factors when we predict the clinical effect.
Conclusion
In summary, there was a certain correlation between UGT1A1 gene polymorphism and the occurrence of severe adverse reactions for irinotecan-based regimens in patients with metastatic colorectal cancer. The determination of UGT1A1 gene polymorphism was helpful in preventing the adverse reactions of CPT-11-based regimens. UGT1A1 gene polymorphism cannot, however, predict the clinical efficacy of CPT-11-based chemotherapy. This will require more rigorous experiments and clinical research to verify.
Disclosure
The authors report no conflicts of interest in this work.
References
Zhou CF, Ma T, Su Y, et al. UGT1A1 gene polymorphisms and the toxicities of FOLFIRI in Chinese Han patients with gastrointestinal cancer. Anticancer Agents Med Chem. 2013;13(2):235–241. | |
Hirose K, Kozu C, Yamashita K, et al. Correlation between plasma concentration ratios of SN-38 glucuronide and SN-38 and neutropenia induction in patients with colorectal cancer and wild-type UGT1A1 gene. Oncol Lett. 2012;3(3):694–698. | |
Desai AA, Innocenti F, Ratain MJ. Pharmacogenomics: road to anticancer therapeutics nirvana? Oncogene. 2003;22(42):6621–6628. | |
Park SR, Kong SY, Rhee J, et al. Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Ann Oncol. 2011;22(4):890–896. | |
Takano M, Kato M, Yoshikawa T, et al. Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: a prospective multi-institutional study. Oncology. 2009;76(5):315–321. | |
Minami H, Sai K, Saeki M, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17(7):497–504. | |
Massacesi C, Terrazzino S, Marcucci F, et al. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer. 2006;106(5):1007–1016. | |
Saeki M, Saito Y, Jinno H, et al. Comprehensive UGT1A1 genotyping in a Japanese population by pyrosequencing. Clin Chem. 2003;49(7):1182–1185. | |
Di Paolo A, Bocci G, Polillo M, et al. Pharmacokinetic and pharmacogenetic predictive markers of irinotecan activity and toxicity. Curr Drug Metab. 2011;12(10):932–943. | |
Strassburg CP, Kalthoff S, Ehmer U. Variability and function of family 1 uridine-5′-diphosphate glucuronosyltransferases (UGT1A). Crit Rev Clin Lab Sci. 2008;45(6):485–530. | |
Cecchin E, Innocenti F, D’Andrea M, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27(15):2457–2465. | |
Zhang A, Xing Q, Qin S, et al. Intra-ethnic differences in genetic variants of the UGT-glucuronosyltransferase 1A1 gene in Chinese populations. Pharmacogenomics J. 2007;7(5):333–338. | |
Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33(3):458–465. | |
Sai K, Sawada J, Minami H. [Irinotecan pharmacogenetics in Japanese cancer patients: roles of UGT1A1*6 and *28]. Yakugaku Zasshi. 2008;128(4):575–584. | |
Han JY, Lim HS, Park YH, Lee SY, Lee JS. Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung Cancer. 2009;63(1):115–120. | |
Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99(17):1290–1295. | |
Sugiyama T, Hirose T, Kusumoto S, et al. The UGT1A1*28 genotype and the toxicity of low-dose irinotecan in patients with advanced lung cancer. Oncol Res. 2010;18(7):337–342. | |
Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res. 2010;16(15):3832–3842. | |
Hu ZY, Yu Q, Zhao YS. Dose-dependent association between UGT1A1*28 polymorphism and irinotecan-induced diarrhoea: a meta-analysis. Eur J Cancer. 2010;46(10):1856–1865. | |
Sunakawa Y, Ichikawa W, Fujita K, et al. UGT1A1*1/*28 and *1/*6 genotypes have no effects on the efficacy and toxicity of FOLFIRI in Japanese patients with advanced colorectal cancer. Cancer Chemother Pharmacol. 2011;68(2):279–284. | |
Gao J, Zhou J, Li Y, Lu M, Jia R, Shen L. UGT1A1 6/28 polymorphisms could predict irinotecan-induced severe neutropenia not diarrhea in Chinese colorectal cancer patients. Med Oncol. 2013;30(3):604. | |
Wang Y, Shen L, Xu N, et al. UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J Gastroenterol. 2012;18(45):6635–6644. | |
Okuyama Y, Hazama S, Nozawa H, et al. Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol. 2011;41(4):477–482. | |
Onoue M, Terada T, Kobayashi M, et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int J Clin Oncol. 2009;14(2):136–142. | |
Yamamoto N, Takahashi T, Kunikane H, et al. Phase I/II pharmacokinetic and pharmacogenomic study of UGT1A1 polymorphism in elderly patients with advanced non-small cell lung cancer treated with irinotecan. Clin Pharmacol Ther. 2009;85(2):149–154. | |
Sai K, Saeki M, Saito Y, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther. 2004;75(6):501–515. | |
Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355(9209):1041–1047. | |
Saltz LB, Cox JV, Blanke C, et al; Irinotecan Study Group. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. N Engl J Med. 2000;343(13):905–914. | |
McLeod HL, Sargent DJ, Marsh S, et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol. 2010;28(20):3227–3233. | |
Glimelius B, Garmo H, Berglund A, et al. Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J. 2011;11(1):61–71. | |
Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFIRI chemotherapy. Pharmacogenomics J. 2008;8(4):278–288. | |
Shulman K, Cohen I, Barnett-Griness O, et al. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer. 2011;117(14):3156–3162. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
