Back to Journals » Infection and Drug Resistance » Volume 14
Clinical Comparison of Three Sample-to-Answer Systems for Detecting SARS-CoV-2 in B.1.1.7 Lineage Emergence
Authors Jian MJ, Chung HY, Chang CK, Lin JC, Yeh KM, Chen CW, Li SY, Hsieh SS, Liu MT, Yang JR, Tang SH, Perng CL, Chang FY, Shang HS
Received 7 July 2021
Accepted for publication 4 August 2021
Published 17 August 2021 Volume 2021:14 Pages 3255—3261
DOI https://doi.org/10.2147/IDR.S328327
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Ming-Jr Jian,1,* Hsing-Yi Chung,1,* Chih-Kai Chang,1 Jung-Chung Lin,2 Kuo-Ming Yeh,2 Chien-Wen Chen,3 Shih-Yi Li,1 Shan-Shan Hsieh,1 Ming-Tsan Liu,4 Ji-Rong Yang,4 Sheng-Hui Tang,1 Cherng-Lih Perng,1 Feng-Yee Chang,2 Hung-Sheng Shang1
1Division of Clinical Pathology, Department of Pathology, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan, Republic of China; 2Division of Infectious Diseases and Tropical Medicine, Department of Medicine, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan, Republic of China; 3Division of Pulmonary and Critical Care Medicine, Department of Medicine, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan, Republic of China; 4Centers for Disease Control, Taipei, Taiwan, Republic of China
*These authors contributed equally to this work
Correspondence: Hung-Sheng Shang; Feng-Yee Chang Email [email protected]; [email protected]
Purpose: Accurate molecular diagnostic assays for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, are needed for epidemiology studies and to support infection-control measures. We evaluated the analytical sensitivity and clinical performance of three sample-to-answer molecular-diagnostics systems for detecting SARS-CoV-2 using 325 nasopharyngeal swab clinical samples from symptomatic patients.
Methods: The BioFire Respiratory Panel 2.1 (RP2.1), cobas Liat SARS-CoV-2 and Influenza A/B, and Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV platforms, which have been granted emergency-use authorization by the US FDA, were tested and compared.
Results: The positive percent agreement, negative percent agreement, and overall percent agreement among the three point of care testing systems were 98– 100%, including for the wild-type SARS-CoV-2 (non-B.1.1.7) and a variant of concern (B.1.1.7). Notably, the BioFire RP2.1 may fail to detect the SARS-CoV-2 S gene in the B.1.1.7 lineage because of the spike protein mutation.
Conclusion: All three point of care testing platforms provided highly sensitive, robust, and almost accurate results for rapidly detecting SARS-CoV-2. These automated molecular diagnostic assays can increase the effectiveness of control and prevention measures for infectious diseases.
Keywords: COVID-19, SARS-CoV-2, point of care testing, molecular diagnostics, sample to answer, variant of concern
Introduction
A cluster of pneumonia cases of unknown etiology was reported in December of 2019 and was confirmed to be the novel coronavirus 2019 (2019-nCoV).1–3 By July 2021, more than 200 million reported cases of coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), were reported and were associated with over four million deaths globally (https://covid19.who.int/). Accurate and reliable molecular-diagnostic assays for detecting SARS-CoV-2 may help clinicians better understand the etiology of suspected SARS-CoV-2 infection.4 Nucleic acid-amplification tests are highly sensitive and specific and are considered as the gold standard for SARS-CoV-2 diagnosis.5–7 A correct SARS-CoV-2 diagnosis contributes to disease treatment and regimens. Although many molecular diagnostic platforms have become available to meet the enormous demands during the COVID-19 pandemic,8–10 the clinical performance of these diagnostic methods has not been thoroughly evaluated. Affordable point of care testing (POCT) kits serve as alternative diagnostic methods but are limited by their availability, although they are widely used to test the general public.11,12 Recently, a SARS-CoV-2 test was added to the Liat SARS-CoV-2 and Influenza A/B (Roche Molecular Systems, Inc., Pleasanton, CA, USA), Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV (Cepheid, Sunnyvale, CA, USA), and BioFire Respiratory Panel 2.1 (RP2.1; BioFire Diagnostics, LLC; Salt Lake City, UT, USA) tests, which are commonly used multiplex PCR panels for diagnosing upper respiratory tract infections by multiple co-existing respiratory viruses. Importantly, new SARS-CoV-2 variants of concern (VOCs) may enhance virus transmissibility and/or disease severity, as well as diagnostic and/or treatment failure.13 SARS-CoV-2 lineages carrying the amino acid substitution N501Y spread rapidly in the United Kingdom in late autumn 2020.14 No relevant research exploring the performance and accuracy of POCT platforms for detecting SARS-CoV-2 VOCs has been performed.
Here, we evaluated the analytical and clinical performance of three sample-to-answer POCTs in terms of their abilities to qualitatively detect SARS-CoV-2 RNA from wild-type (non-B.1.1.7) or VOC (B.1.1.7) strains. These POCTs include the BioFire RP2.1 test, Liat SARS-CoV-2 and Influenza A/B, and Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV tests. We independently evaluated the performance characteristics of the three platforms for detecting SARS-CoV-2, particularly with respect to the VOC B.1.1.7 lineage.
Materials and Methods
Study Design and Clinical Specimens
This study was registered on February 8, 2021 and approved by the Tri-Service General Hospital Institutional Review Board (approval number C202005041). We tested 325 de-identified nasopharyngeal swab specimens collected from patients suspected of having COVID-19 using LIBO Specimen Collection and Transport Swab Kits with Universal Transport Medium (New Taipei City, Taiwan). Residual viral transport medium was collected and stored at −80°C. The sample collection periods were between June and September 2020 and May and June 2021. Figure 1 depicts the study design. All SARS-CoV-2 testing results were confirmed by RT-PCR developed by us as described previously.15 Briefly, the SARS-CoV-2 assay simultaneously detected SARS-CoV-2 E and ORF1ab gene along with the human RP gene to monitor the quality of nucleic acid. Results were interpreted as positive or negative based on the detection of E and ORF1ab or lack of detection of those genes, respectively.
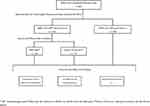 |
Figure 1 Flow chart of study design. |
BioFire Respiratory Panel 2.1
BioFire Respiratory Panel 2.1 (RP2.1; BioFire Diagnostics, LLC; Salt Lake City, UT) tests are commonly used multiplex PCR panels for diagnosing upper respiratory tract infections, including 22 viral and bacterial respiratory pathogens such as influenza A/B, respiratory syncytial virus, and SARS-CoV-2 etc. The BioFire RP2.1 test was performed using the BioFire FilmArray EZ Configuration system according to the manufacturer’s instructions. Briefly, 300 μL of sample was mixed with sample buffer and injected into a test pouch containing all necessary reagents for nucleic extraction, PCR amplification, and detection of the respective targets. The RP2.1 test contains two independent assays targeting the spike (S) and membrane (M) genes in the SARS-CoV-2 genome. The results were interpreted using the BioFire system software, which interprets each assay independently. If either one or both of the assays are positive, the test will show that SARS-CoV-2 was detected. If both assays are negative, the test report result will show that SARS-CoV-2 was not detected.
Liat SARS-CoV-2 and Influenza A/B Test
The Liat SARS-CoV-2 and Influenza A/B test is a multiplex reverse transcriptase (RT)-PCR assay for rapid in vitro detection and discrimination of RNA targets for three viruses, namely, SARS-CoV-2, influenza A virus, and influenza B virus, in nasopharyngeal swabs. The test was performed using the Liat system according to the manufacturer’s instructions. Briefly, 200 μL of sample was added to a Liat tube inserted into the tube slot of the cobas Liat System. An internal control was used for procedural checks. The Liat assay was used to detect two target regions (ORF1a/b and nucleocapsid [N]) of the SARS-CoV-2 genome. The results of Liat SARS-CoV-2 and Influenza A/B are interpreted as follows: SARS-CoV-2 not detected (no SARS-CoV-2 RNA detected), SARS-CoV-2 detected (SARS-CoV-2 RNA present).
Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV Test
The GeneXpert Dx system is an integrated diagnostic device that performs automated specimen processing and real-time RT-PCR analysis. The Xpert Xpress SARS-CoV-2/Flu/RSV test consists of two main components, the Xpert plastic cartridge and GeneXpert instrument. The Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV Test was designed to amplify three viral pathogens: SARS-CoV-2, influenza, and respiratory syncytial virus (RSV). The assay detects the gene sequences of the envelope (E) and nucleocapsid (N2) proteins of SARS-CoV-2. Each cartridge includes a sample processing control and a probe check control to ensure that the sample was processed correctly and the GeneXpert instrument measures the fluorescence signal from the probes to monitor bead rehydration, reaction tube filling, probe integrity, and dye stability. Briefly, 300 µL of each nasal pharyngeal swab specimen was added to a separate Xpert Xpress SARS-CoV-2/Flu/RSV cartridge and loaded into an Xpert instrument. The result was interpreted according to the manufacturer’s instructions. Positive results indicated the presence of RNA of influenza A and B, RSV, or SARS-CoV-2 viruses. The results were interpreted automatically by the GeneXpert System.
Detection of SARS-CoV-2 VOCs
To screen a SARS-CoV-2 VOC (B.1.1.7 lineage), we used VirSNiP SARS-CoV-2 Spike N501Y and Spike del H69/V70 (TIB Molbiol, Berlin, Germany), which use real-time RT-PCR post-melting curve analysis to detect the mutations N501Y and del 69–70 in SARS-CoV-2-positive specimens. Here, we detected the spike gene mutations del 69–70 and N501Y on a LightCycler 480 (Roche Molecular Systems, Inc.) according to the manufacturer’s instructions. To confirm that the results were consistent with those of rapid detection of the SARS-CoV-2 VOC, presumptive cases (denoted as TSGH42 and TSGH43) caused by the VOC were also assessed by whole-genome sequencing as described previously, and the lineages were confirmed as B.1.1.7 (UK) using GISAID software.16
Analytical Tests
The limit of detection (LoD) values for SARS-CoV-2 were determined using AMPLIRUN SARS-CoV-2 RNA controls (Vircell, Granada, Spain), which contained purified genomic RNA from the indicated viruses. These controls were used for absolute quantification. The controls were used to prepare a serial-dilution panel with 1–10 replicates. The analytical sensitivities of the BioFire RP2.1, Liat SARS-CoV-2 and Influenza A/B, and Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV tests were defined as the lowest dilution at which all replicates were identified as positive for SARS-CoV-2.
Comparison of Clinical Test Performance Using Clinical Samples
We included 325 retrospective nasopharyngeal swab specimens from patients hospitalized at the Tri-Service General Hospital (Taipei City, Taiwan). Clinical testing was performed for all 325 clinical specimens, and the results were compared to those of our developed RT-PCR, which was used as the reference method.15
Results
Analytic Sensitivities of Three Sample-to-Answer Platforms for Detecting SARS-CoV-2
We assessed the empirical sensitivity of the BioFire RP2.1, Liat SARS-CoV-2 and Influenza A/B, and Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV for detecting SARS-CoV-2. For consistency, we used the unit copies/mL to compare the results obtained from all three platforms when determining the LoD of SARS-CoV-2. Accordingly, we defined the LoD as the minimum concentration at which a detection rate of 100% could be achieved for 3–10 replicates. The LoD established as per this criterion ranged from 2000 to 25 copies/mL in the three platforms (Table 1).
 |
Table 1 Detection Limit of Three Molecular Diagnostic Assays for SARS-CoV-2 Nucleic Acid |
Clinical Performance of Three Sample-to-Answer Platforms
We analyzed 325 clinical samples in this study; 125 samples tested as positive, which was confirmed by the Taiwan CDC central laboratory, and 200 samples were negative for SARS-CoV-2. The 125 positive samples were further identified as wild-type (non-B.1.1.7) or VOC (B.1.1.7). Concordant and discordant positive results were obtained for all three assays when detecting SARS-CoV-2, when detecting SARS-CoV-2, and when the detection results of any given pair of assays were compared, they showed 98–100% agreement (Table 2 and Supplementary Table 1). The positive percent agreement, negative percent agreement, and overall percent agreement between the Cepheid Xpert Xpress system and cobas Liat System were 100% for wild-type (non B.1.1.7) SARS-CoV-2 specimens, and those between these two assays and BioFire RP2.1 were 98%, 100%, and 99.6% respectively. Among the 50 positive wild-type (non B.1.1.7) SARS-CoV-2 specimens, one sample (sample 7) had cycle threshold (Ct) values of 36.8 and 35.3 for the Cepheid Xpert Xpress system and Cobas Liat System, respectively. However, it was falsely detected as negative with BioFire RP2.1, as BioFire tests do not report Ct values. The Cepheid Xpert Xpress system, BioFire RP2.1, and cobas Liat System showed 100% positive agreement for the other 75 positive VOC (B.1.1.7) SARS-CoV-2 specimens. Unexpectedly, based on further analysis of the results of the BioFire RP2.1 platform, we found one positive result with only M detected, but the test failed to detect S (Supplementary Figure 1). This sample had a Ct value of 26.5 on the cobas Liat System. This result indicates that a novel mutation caused mismatches with the primer for S-gene assays in the VOC of SARS-CoV-2.
 |
Table 2 Comparison of Clinical Performance Among Three Sample-to-Result Molecular Assays for SARS-CoV-2 |
Workflow Evaluation
We next evaluated the total turnaround time (TAT) for each specimen, including the sample preparation, hand-on time, assay detection time, and data interpretation steps. Table 3 shows the overall workflow assessment and a comparison of the three POCT platforms.
 |
Table 3 Characteristics of Three Sample-to-Answer Molecular SARS-CoV-2 Assays |
Analysis of the overall TAT, from sample collection to results, showed that the cobas Liat System had the lowest TAT (approximately 20 min), followed by the Cepheid Xpert Xpress system (42 min) and BioFire RP2.1 (45 min). All three platforms evaluated in this study decreased the hands-on time and reduced the risk of exposure compared to conventional RT-PCR methods, which is particularly important when handling samples from patients suspected of having COVID-19. These POCT platforms are valuable additions to diagnostic laboratories for rapid and accurate diagnostic results in detecting SARS-CoV-2.
Discussion
In vitro diagnostic nucleic acid-amplification tests assays for SARS-CoV-2 employ RT-PCR, which takes less than 1 h or a few hours depending on the number of samples processed or if a specific facility is well-equipped, which facilitates reliable and easy-to-implement in vitro diagnostic assays for the molecular diagnosis of COVID-19.17,18 In this study, we determined the analytic sensitivity of three POCT platforms. Similar LoD values were obtained for the BioFire RP2.1, cobas Liat SARS-CoV-2 and Influenza A/B, and Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV tests. We also examined the clinical performance of the three POCTs. Our findings suggest that the lower diagnostic sensitivity of the BioFire RP2.1 for clinical samples led to false-negative results in samples with very low viral loads, Moreover, this test failed to detect S gene CoV-2 and Influenza A/B and Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV, BioFire RP2.1 showed limitations at lower viral load concentrations in wild-type (non-B.1.1.7) SARS-CoV-2 (eg, sample 7 in our study). Additionally, detection by BioFire RP2.1 was impaired for VOCs (B.1.1.7) of SARS-CoV-2 because of the spike protein mutation.
When relying on fully automated platforms from samples to answers, all of the three evaluated platforms have shortened the time required to obtain results and expanded the number of laboratories capable of testing for SARS-CoV-2, while also testing for co-infecting pathogens (such as other upper respiratory pathogens) as alternative diagnoses, which is key for determining the most effective treatment regimen. Various co-factors influence SARS-CoV-2-detection results, including the input volume, extraction methods, and workflow requirements.19 Our study provides a perspective for deciding which molecular diagnostic test should be implemented in clinical laboratories. To date, several reports of concurrent infections with other pathogens, such as influenza virus and other seasonal coronaviruses, have suggested that coinfection influences the morbidity and mortality of patients with COVID-19.20–22 Therefore, it is crucial for clinicians to rule out SARS-CoV-2 or other upper respiratory viral infections. Notably, BioFire RP2.1 provided more than 20 detection results, including SARS-CoV-2, providing important data for laboratories to rapidly detect and differentiate co-circulating respiratory pathogens and make these tests accessible in remote areas where higher-complexity assays are not feasible.23
Conclusions
The three molecular-diagnostics platforms examined simultaneously test for SARS-CoV-2, influenza A/B, and/or RSV, which can greatly benefit hospitals by enabling the management and control of infectious diseases.
Abbreviations
COVID-19, coronavirus disease 2019; LOD, limit of detection; RT-PCR, reverse transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; POCT, point of care testing; VOC, variant of concern; WHO, World Health Organization.
Institutional Review Board Statement
This study was approved by the Institutional Review Board of Tri-Service General Hospital (TSGHIRB No.: C202005041), registered on Feb 8, 2021.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. This study was conducted in accordance with the Declaration of Helsinki.
Author Contributions
Conceptualization, Jung-Chung Lin, Kuo-Ming Ye, Chien-Wen Chen, and Feng-Yee Chang; Data curation, Ming-Jr Jian and Hsing-Yi Chung; Formal analysis, Shih-Yi Li and Ming-Jr Jian; Investigation, Hsing-Yi Chung, Chih-Kai Chang, Ming-Tsan Liu, Ji-Rong Yang, and Cherng-Lih Perng; Methodology, Chih-Kai Chang, Shan-Shan Hsieh, and Cherng-Lih Perng; Supervision, Hung-Sheng Shang, Jung-Chung Lin, Kuo-Ming Ye, Chien-Wen Chen, Feng-Yee Chang, Ji-Rong Yang, Sheng-Hui Tang, and Cherng-Lih Perng; Writing – original draft, Ming-Jr Jian; Writing – review & editing, Ming-Jr Jian, Feng-Yee Chang and Hung-Sheng Shang. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Tri-Service General Hospital, Taipei, Taiwan, ROC, (grant number TSGH-D-110100). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi:10.1001/jama.2020.2648
2. The Lancet. Emerging understandings of 2019-nCoV. Lancet. 2020;395(10221):311. doi:10.1016/S0140-6736(20)30186-0
3. Xie M, Chen Q. Insight into 2019 novel coronavirus - An updated interim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis. 2020;94:119–124. doi:10.1016/j.ijid.2020.03.071
4. Russo A, Minichini C, Starace M, et al. Current status of laboratory diagnosis for COVID-19: a narrative review. Infect Drug Resist. 2020;13:2657–2665. doi:10.2147/IDR.S264020
5. Mostafa HH, Hardick J, Morehead E, Miller JA, Gaydos CA, Manabe YC. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J Clin Virol. 2020;130:104578. doi:10.1016/j.jcv.2020.104578
6. Caruana G, Croxatto A, Coste AT, et al. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin Microbiol Infect. 2020;26:1178–1182. doi:10.1016/j.cmi.2020.06.019
7. Chung HY, Jian MJ, Chang CK, et al. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg Microbes Infect. 2021;10:161–166. doi:10.1080/22221751.2021.1873073
8. Zhen W, Smith E, Manji R, Schron D, Berry GJ. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol. 2020;58:e00783–20. doi:10.1128/JCM.00783-20
9. Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect. 2020;9:747–756. doi:10.1080/22221751.2020.1745095
10. Smith E, Zhen W, Manji R, Schron D, Duong S, Berry GJ. Analytical and clinical comparison of three nucleic acid amplification tests for SARS-CoV-2 detection. J Clin Microbiol. 2020;58:e01134–20. doi:10.1128/JCM.01134-20
11. Pokhrel P, Hu C, Mao H. Detecting the Coronavirus (COVID-19). ACS Sens. 2020;5:2283–2296. doi:10.1021/acssensors.0c01153
12. Oishee MJ, Ali T, Jahan N, et al. COVID-19 pandemic: review of contemporary and forthcoming detection tools. Infect Drug Resist. 2021;14:1049–1082. doi:10.2147/IDR.S289629
13. Boehm E, Kronig I, Neher RA, et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021. doi:10.1016/j.cmi.2021.05.022
14. Leung K, Shum MHH, Leung GM, Lam TTY, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance. 2021;26:2002106. doi:10.2807/1560-7917.ES.2020.26.1.2002106
15. Jian MJ, Chung HY, Chang CK, et al. Novel automated sample-to-result SARS-CoV-2 laboratory-developed RT-PCR assay for high-throughput testing using LabTurbo AIO 48 system. Clin Chim Acta. 2021;514:54–58. doi:10.1016/j.cca.2020.12.003
16. Jian MJ, Chung HY, Chang CK, et al. Investigation of one familial cluster of COVID-19 in Taiwan: differentiation of genetic variation among isolates and implications for epidemiological investigation and surveillance by genomic assay. Infect Drug Resist. 2021;14:971–977. doi:10.2147/IDR.S298451
17. Cradic K, Lockhart M, Ozbolt P, et al. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020;154:201–207. doi:10.1093/ajcp/aqaa097
18. Vashist SK. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. 2020;10(4):202.
19. Perez-Lopez B, Mir M. Commercialized diagnostic technologies to combat SARS-CoV2: advantages and disadvantages. Talanta. 2021;225:121898. doi:10.1016/j.talanta.2020.121898
20. Ma L, Wang W, Le Grange JM, et al. Coinfection of SARS-CoV-2 and other respiratory pathogens. Infect Drug Resist. 2020;13:3045–3053. doi:10.2147/IDR.S267238
21. Nowak MD, Sordillo EM, Gitman MR, Paniz Mondolfi AE. Co-infection in SARS-CoV-2 infected poatients: where are influenza virus and rhinovirus/enterovirus? J Med Virol. 2020;92:1699–1700. doi:10.1002/jmv.25953
22. Wu X, Cai Y, Huang X, et al. Co-infection with SARS-CoV-2 and influenza A virus in patient with pneumonia, China. Emerg Infect Dis. 2020;26:1324–1326. doi:10.3201/eid2606.200299
23. Creager HM, Cabrera B, Schnaubelt A, et al. Clinical evaluation of the BioFire(R) Respiratory Panel 2.1 and detection of SARS-CoV-2. J Clin Virol. 2020;129:104538. doi:10.1016/j.jcv.2020.104538
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
