Back to Journals » OncoTargets and Therapy » Volume 11
Clinical characterization of ERBB2 exon 20 insertions and heterogeneity of outcomes responding to afatinib in Chinese lung cancer patients
Authors Liu Z, Wu L, Cao J, Yang Z, Zhou C , Cao L, Wu H, Shen H, Jin M, Zhang Y, Mao X, Xiang J, Ma K, Li B, Zhang T, Hu Y
Received 7 May 2018
Accepted for publication 7 August 2018
Published 23 October 2018 Volume 2018:11 Pages 7323—7331
DOI https://doi.org/10.2147/OTT.S173391
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Zhefeng Liu,1,* Lin Wu,2,* Jun Cao,3 Zhe Yang,4 Chengzhi Zhou,5 Liming Cao,6 Hao Wu,7 Haibo Shen,8 Meiling Jin,9 Yong Zhang,1 Xinru Mao,10 Jianxing Xiang,10 Ke Ma,10 Bing Li,11 Tengfei Zhang,10 Yi Hu1
1Department of Oncology, Chinese PLA General Hospital, Beijing, People’s Republic of China; 2Department of the Second Chest Medicine, Hunan Cancer Hospital, Changsha, Hunan, People’s Republic of China; 3Department of Oncology, Hunan Cancer Hospital, Changsha, Hunan, People’s Republic of China; 4Department of Radiotherapy, Provincial Hospital Affiliated to Shandong University, Jinan, Shandong, People’s Republic of China; 5State Key Laboratory of Respiratory Diseases, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, Guangdong, People’s Republic of China; 6Department of Respiratory Medicine, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 7Department of Thoracic Surgery, Peking University Shenzhen Hospital, Shenzhen, Guangdong, People’s Republic of China; 8Department of Thoracic Surgery, Ningbo No 2 Hospital, Ningbo, Zhejiang, People’s Republic of China; 9Department of Respiratory Medicine, Zhongshan Hospital Affiliated with Fudan University, Shanghai, People’s Republic of China; 10Department of Medicine, Burning Rock Biotech, Guangzhou, Guangdong, People’s Republic of China; 11Department of Bioinformatics, Burning Rock Biotech, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Purpose: ERBB2 exon 20 insertions (20ins) have been identified as oncogenic drivers in lung cancers. Lung cancer patients with 20ins benefit from afatinib. However, response heterogeneity was observed in patients harboring different 20ins subtypes. In this study, we interrogated clinical characteristics in ERBB2-mutated Chinese lung cancer and investigated the clinical outcomes of specific ERBB2 20ins in response to afatinib.
Experimental design: In this study, we retrospectively collected genomic profiling data of 7,520 lung cancer patients sequenced using next-generation sequencing in a Clinical Laboratory Improvement Amendments-certified laboratory. We analyzed the clinical and molecular features of patients harboring ERBB2 20ins and evaluated clinical outcomes of 19 patients with clinical records after afatinib treatment.
Results: ERBB2 20ins were identified in 2.27% (171/7,520) of this lung cancer cohort. It occurred with a high proportion in females with adenocarcinoma histology. ERBB2 20ins was mutually exclusive with other well-established lung cancer oncogenic driver mutations. The most frequently appearing subtype was Y772_A775dup (69.6%) and several novel insertion subtypes were also identified. The correlations of specific 20ins subtypes and survival were investigated. The presence of a glycine at position 778 in ERBB2 was suggested to be a common feature of drug sensitivity mutations. Patients harboring G778_P780dup (G778) subtype achieved longer median progression-free survival and median overall survival than other 20ins (non-G778) subtypes (median progression-free survival, 10 vs 3.3 months, P=0.32; median overall survival, 19.7 vs 7 months, P=0.16). Moreover, we presented the first clinical case of a lung squamous cell carcinoma patient harboring ERBB2 20ins who achieved partial response to afatinib.
Conclusion: This study interrogated the characteristics of ERBB2 20ins in a large cohort from single ethnicity and demonstrated the response heterogeneity to afatinib among different ERBB2 20ins subtypes. Further studies in a larger cohort are needed to investigate the underlying molecular mechanisms and clinical response of different ERBB2 20ins subtypes.
Keywords: ERBB2, lung cancer, afatinib, survival, heterogeneity
Introduction
Genetic alterations of human epidermal growth factor receptor 2 (ERBB2, also known as HER2), occurring in 2%–4% of lung cancers, have been identified as a oncogenic driver.1–4 HER2 protein, encoded by ERBB2 gene, drives cell proliferative and activates PI3K/AKT and MEK/ERK downstream signaling pathways.5 In-frame insertions of ERBB2 exon 20 (20ins), resulting in constitutive activation of the downstream PI3K/AKT signaling, initiate the protumorigenic signaling cascade.2,3,6 ERBB2 20ins were predominantly observed in females, nonsmokers, and adenocarcinoma patients.3,7,8 No clinical case of ERBB2 20ins has been reported in lung squamous cell cancer. The most frequently occurring subtype is Y772_A775dup, accounting for approximately 60% of all 20ins in non-small-cell lung cancer.3,9
With the rapid advancement of genomic profiling technologies and targeted therapy, several ERBB2 inhibitors have been approved for the treatment of ERBB2-positive breast cancer, such as trastuzumab, lapatinib, and pertuzumab.10–15 However, the development of ERBB2-targeted therapy in lung cancer was not very successful in the beginning when the first drug trastuzumab showed negative result in a Phase II trial, which revealed that combination therapy of trastuzumab and chemotherapy failed to produced additive effect in ERBB2-positive lung cancer patients.16 Despite the dismal results from several studies, some case series have reported that lung cancer patients with ERBB2 20ins can benefit from afatinib. It is an ATP-competitive anilinoquinazoline derivative which can covalently bind to and irreversibly block signaling from HER2.17 The response heterogeneity suggests that ERBB2 mutant patients do not represent a homogenous group. Very limited studies have investigated what contributes to such heterogeneity. In this study, we investigated whether different ERBB2 20ins subtypes are responsible for treatment heterogeneity by correlating specific 20ins subtypes to survival in 171 lung cancer patients.
Materials and methods
Patient information
We retrospectively screened 7,520 lung cancer patients (from eight hospitals) whose tissue or plasma samples were sequenced in a Clinical Laboratory Improvement Amendments-certified genomic profiling laboratory (Burning Rock Biotech, Guangzhou, People’s Republic of China), from October 2015 to January 2018. Their sequencing results were reviewed with the intent to study the association between ERBB2 exon 20 insertions distribution and prognosis after afatinib treatment. A total of 171 patients with ERBB2 exon 20 insertions were identified. The study was approved by the institutional review board of Chinese PLA General Hospital.
Preparation of tissue DNA and plasma cfDNA
Tissue DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Circulating cfDNA was recovered from 4 to 5 mL of plasma by using the QIAamp Circulating Nucleic Acid kit (Qiagen). Quantification of DNA was done with the Qubit 2.0 fluorimeter (Thermo Fisher Scientific, Waltham, MA, USA).
Next-generation sequencing library preparation
DNA shearing was performed with a Covaris M220 focused ultrasonicator (Covaris, Inc., Woburn, MA, USA) followed by end repair, phosphorylation, and adaptor ligation. Fragments with a size of 200–400 bp were selected by using Agencourt AMPure beads (Beckman Coulter, Fullerton, CA, USA) followed by hybridization with capture probes baits, hybrid selection with magnetic beads, and PCR amplification. A high-sensitivity DNA assay was performed to assess the quality and size of the fragments, and indexed samples were sequenced on a Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with paired-end reads. Our profiling panels, consisting of 8, 56, 168, or 295 cancer-related genes, were designed and validated for identification of base substitutions, insertions, deletions, copy-number variations, and gene fusion.
Sequencing data analysis
The sequencing data in the FASTQ format were mapped to the human genome (hg19) using BWA aligner 0.7.10.18 Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan,19,20 respectively.
Statistical analysis
Kaplan–Meier analysis was used to estimate the survival functions, and log-rank test to determine the difference of survival curve between groups. A P-value of <0.05 was considered statistically significant. All these survival analyses were conducted in software R (Version 3.3.3, The R Foundation, Vienna, Austria) with survival package (Version 2.41-3; https://cran.r-project.org/web/packages/survival/index.html).
Results
Patient characteristics
Sequencing results of tissue or plasma samples obtained from 7,520 lung cancer patients were analyzed, and 171 patients were identified with ERBB2 20ins, resulting in an incidence of 2.27%. Clinical features of patients harboring ERBB2 20ins mutations are summarized in Table 1. Patients harboring ERBB2 20ins had a median age of 56 years, ranging from 23 to 86 years. Among them, 38% (65/171) were males and 62% (106/171) were females. A vast majority of them were diagnosed with adenocarcinomas, except for one adenosquamous carcinoma and one squamous cell carcinoma (SCC).
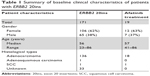  | Table 1 Summary of baseline clinical characteristics of patients with ERBB2 20ins |
Genomic characteristic overview of the ERBB2 20ins patients
To investigate the prevalence and composition of ERBB2 20ins, capture-based targeted sequencing was performed using various panels covering critical exons and introns of lung cancer-related genes. Among the 171 ERBB2 20ins-positive patients, five patients were profiled using 295 cancer-related genes panel. Fifty-three patients were sequenced using a panel consisting of 168 lung cancer-related genes. Thirty-five patients were assessed using a 56-gene panel, and for the remaining 78 patients a panel consisting seven well-known lung cancer driver genes plus KRAS, a well-established prognostic factor, was used.
A total of 432 mutations were identified from the 171 patients harboring ERBB2 20ins, including 139 single-nucleotide variants (SNVs), 172 insertions and deletions (Indels), 83 copy-number amplifications, and 38 frameshift or splice variants. Among them, TP53 ranked as the most frequently mutated gene in this cohort, occurring in 51.6% (48/93) patients who were sequenced using panels covering TP53. All ERBB2 20ins patients did not carry concurrent classic non-small-cell lung carcinoma driver genes (including EGFR, BRAF, MET, ROS1, ALK, RET, and KRAS), which is consistent with previous reports, confirming the strong mutual exclusivity of ERBB2 with other driver oncogenes. Moreover, we found 11.7% (20/171) patients with ERBB2 20ins harboring concomitant ERBB2 amplification.
ERBB2 mutation subtypes overview
Of the 171 patients harboring ERBB2 20ins, 13 insertion subtypes were identified. They included 12-base-pair insertions (Y772_A775dup, A775_G776insSVMA, A775_G776insVVMA, A775_G776insYVMS, Y772_V773insMMAY), 9-bp insertions (G778_P780dup), 3-bp insertions (A775_G776insC), and deletions combined with insertions (G776delinsVC, G776delinsLC, G776delinsVV, G776delinsAVGC, G776delinsIC, G776_V777delinsCVC). The nomenclature of these exon 20 insertions strictly followed the Human Genome Variation Society guidelines. The structural schematic diagrams of those exon 20 insertion subtypes and corresponding insert positions are demonstrated in Figure 1A.
The most frequently appearing subtype was a duplication of 12-bp which resulted in an insertion of YVMA at codon 775 (Y772_A775dup) (Figure 1B). This subtype was detected in 119 patients (69.6%), consistent with previous reports. The second most frequently occurring ERBB2 20ins subtype in our cohort was G778_P780dup, an in-frame 9-bp insertion, which was identified in 18 patients (10.5%). Moreover, several rare or novel ERBB2 20 insertions were identified in this cohort. The Integrative Genomics Viewer screenshots of these rare or novel subtypes are shown in Figure 2.
Correlation of specific 20ins subtypes and survival
To interrogate the clinical outcomes of different ERBB2 20ins in response to afatinib, we selected 19 ERBB2 20ins patients who received afatinib as monotherapy with detailed survival information. Kaplan–Meier survival analysis revealed that patients with ERBB2 G778_P780dup mutations achieved longer median progression-free survival (PFS) (10.0, 3.7, 1.5, 2.5 months, respectively, P=0.69; Figure 3A) and median overall survival (OS) (19.7, 7.0, 1.5, 11.1 months, respectively, P=0.44; Figure 3B) than those harboring Y772_A775dup, G776delinsVC, and other subtypes. However, the differences were not statistically significant owing to the limited patient number of each subgroup.
The glycine at ERBB2 778 site was reported to be a drug-sensitive mutation, which may facilitate inhibitor binding to ERBB2.21 Therefore, we stratified our ERBB2 20ins patients into two subgroups to compare survival between G778 patients (G778_P780dup) and non-G778 patients (other amino acids at 778 due to insertion). The Kaplan–Meier analysis revealed that patients with G778 displayed longer median PFS (10.0 vs 3.3 months, P=0.32; Figure 3C) and median OS (19.7 vs 7.0 months, P=0.16; Figure 3D) than non-G778 patients. But due to the small cohort size of G778 patients, the differences did not reach statistically significant. This result warrants further exploration in large cohorts to better understand this phenomenon.
Overall outcomes of afatinib treatment of the ERBB2 20ins patients
In the cohort of 19 patients receiving afatinib treatment, consisting of 18 lung adenocarcinomas and 1 squamous cell lung cancer, 14 patients reached progressive disease (PD) and the other 5 patients were still benefiting from afatinib treatment to date. The median PFS of the 14 patients was 4.5 months and median OS was 11.5 months after afatinib monotherapy. The overall response rate in this cohort was 15.8% (3/19) and disease control rate was 68.4% (13/19). Among them, three patients achieved partial response (PR) to afatinib treatment, ten patients were in stable disease (SD) after afatinib, whereas six patients only had PFS period records and response details were not available due to rapid disease progression. The overall outcomes of afatinib treatment of the 19 patients harboring ERBB2 20ins are demonstrated in a swimmer’s plot in Figure 4.
For the eleven Y772_A775dup patients, eight patients finally achieved PD, with a median PFS of 3 months. The other three patients were still under afatinib treatment, with two achieving PR and one with SD. Two patients harboring G778_P780dup insertion had SD to afatinib treatment and displayed durable response. One had a PFS of 299 days, while another had not developed PD in the last visit after 214 days of treatment. As for rare subtypes of ERBB2 20ins, the patient with G776delinsVV had a PFS of 75 days; another patient with G776delinsLC achieved SD with a PFS of 333 days. The patient detected with G776delinsIC obtained a PFS of 62 days. Two of the three G776delinsVC patients displayed very different responses, with a PFS of 214 and 46 days, respectively. Another G776delinsVC patient achieved PR to afatinib in the last visit at day 24 and was still under treatment. Taken together, our study demonstrated that patients with different ERBB2 20ins subtypes obtained diverse clinical response to afatinib. Due to the limited sample size, larger cohorts are needed to validate our findings.
A squamous cell lung carcinoma patient harboring ERBB2 20ins achieved PR to afatinib
To the best of our knowledge, no ERBB2 20ins has been reported in lung SCC. In our cohort, we identified a SCC patient harboring ERBB2 20ins. A 47-year-old female patient with no history of smoking presented to the outpatient clinic with intermittent cough and thoracalgia. Chest computed tomography scan revealed a mass located in right lung. Computed tomography-guided biopsy followed by hematoxylin–eosin staining revealed the representative histological appearance of lung SCC (Figure 5A). She was diagnosed with stage IV SCC with bone and brain metastasis. Genomic profiling was performed and revealed the presence of ERBB2 exon 20 insertion G776delinsVC. The patient was administered afatinib (40 mg, QD). Three months later, she achieved PR based on Response Evaluation Criteria In Solid Tumors (RECIST) 1.1, evident by tumor shrinkage (Figure 5B) and alleviation of symptoms, such as coughing, thoracalgia, and dyspnea. Collectively, this case is the first report of an SCC patient harboring ERBB2 20ins and showing benefit from afatinib.
Discussion
In this study, we retrospectively interrogated the clinical and molecular characteristics of ERBB2 20ins in a large Chinese lung cancer cohort. We identified several novel ERBB2 20ins subtypes which were first detected in lung cancer patients. Moreover, we investigated the correlations of ERBB2 20ins subtypes and clinical outcomes to afatinib treatment. In addition, we presented the first case of a lung SCC patient harboring ERBB2 20ins and achieving PR to afatinib.
Our study revealed that ERBB2 20ins demonstrated a female preponderance and adenocarcinoma predominance, in an agreement with published data.3,7,8 In this study, 62% of the patients were female, and nearly all the ERBB2 20ins occurred in adenocarcinoma, except for one in SCC and one in adenosquamous. The insertion YVMA at codon 775 (Y772_A775dup) was the most frequently occurring subtype, accounting for 70% of all 20ins in our cohort, consistent with previous publications, which reported a prevalence of 60%.3,9 As ERBB2 mutations were reported to be mutually exclusive with major mutations in EGFR and KRAS,1,2 ERBB2 20ins in all the 171 patients in our study displayed mutually exclusivity with other well-established oncogenic driver mutations, including EGFR, KRAS, BRAF, MET, ALK, ROS1, and RET. In addition, we found that 11.7% (20/171) ERBB2 20ins patients harbored concurrent ERBB2 amplification. To date, the molecular association of ERBB2 mutation and gene amplification in lung cancers has not been distinctly defined. Li et al8 reported that ERBB2 mutation and gene amplification did not overlap in lung cancer and may present distinct clinical entities. However, their coexistence was reported in other studies.7,22,23 Thus, further efforts are needed to interrogate the underlying association between ERBB2 mutation and amplification.
Previous studies have reported response heterogeneity to afatinib with different ERBB2 20ins subtypes.17,24 G778_P780dup were reported to be sensitive to ERBB2 targeted tyrosine kinase inhibitors than other 20ins subtypes in vitro, and the presence of a glycine at ERBB2 position 778 was supposed to be a common feature among the drug-sensitive insertion subtypes.21 In this study, the survival analysis revealed that patients harboring G778_P780dup (with glycine at site 778) displayed favorable PFS and OS compared with those with other insertion subtypes, which further supports the in vitro finding. Kosaka et al21 reported an adenocarcinoma patient with ERBB2 G778_P780dup achieving significant tumor shrinkage for 7 months when afatinib was given as monotherapy. In our cohort, two patients were identified as harboring G778_P780dup insertion at exon 20. They both responded to afatinib treatment and achieved long durable response. One patient achieved a PFS of 299 days, and another is still benefiting from afatinib in the latest reexamination at day 214 after treatment. This observation further confirmed the notion that glycine778 indicates favorable clinical benefits to afatinib. Both biophysical and structural analyses will be required to further investigate the underlying mechanism, and additional clinical validations in larger cohorts will be necessary to confirm our clinical findings.
There were still limitations in this study. The small population size of cohort for survival correlation studies inevitably induced bias into the analysis and cannot fully reflect the overall correlation. Thus, we were not able to adequately illustrate the correlation of different 20ins subtypes and survival after afatinib treatment. Despite this limitation, our study improved the knowledge of ERBB2 20ins-driven non-small-cell lung cancer in terms of clinical outcomes to afatinib. Our study might be helpful to provide guidance for clinicians to design a more precise clinical strategy for different patients based on specific ERBB2 20ins subtypes.
Acknowledgment
The authors would like to thank all the patients and their families.
Disclosure
Xinru Mao, Jianxing Xiang, Ke Ma, Bing Li and Tengfei Zhang were employed by Burning Rock Biotech, Guangzhou, Guangdong, People’s Republic of China, at the time of the study. The authors report no other conflicts of interest in this work.
References
Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431(7008):525–526. | ||
Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65(5):1642–1646. | ||
Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18(18):4910–4918. | ||
Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. | ||
Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2009;27(34):5838–5847. | ||
Perera SA, Li D, Shimamura T, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci U S A. 2009;106(2):474–479. | ||
Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31(16):1997–2003. | ||
Li BT, Ross DS, Aisner DL, et al. HER2 Amplification and HER2 Mutation Are Distinct Molecular Targets in Lung Cancers. J Thorac Oncol. 2016;11(3):414–419. | ||
Kris MG, Camidge DR, Giaccone G, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol. 2015;26(7):1421–1427. | ||
Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. | ||
Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. | ||
Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355(26):2733–2743. | ||
Blackwell KL, Burstein HJ, Storniolo AM, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 Study. J Clin Oncol. 2012;30(21):2585–2592. | ||
Baselga J, Cortés J, Kim SB; CLEOPATRA Study Group. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366(2):109–119. | ||
Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. | ||
Gatzemeier U, Groth G, Butts C, et al. Randomized phase II trial of gemcitabine-cisplatin with or without trastuzumab in HER2-positive non-small-cell lung cancer. Ann Oncol. 2004;15(1):19–27. | ||
Solca F, Dahl G, Zoephel A, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther. 2012;343(2):342–350. | ||
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. | ||
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. | ||
Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. | ||
Kosaka T, Tanizaki J, Paranal RM, et al. Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res. 2017;77(10):2712–2721. | ||
Li C, Sun Y, Fang R, et al. Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol. 2012;7(1):85–89. | ||
Yoshizawa A, Sumiyoshi S, Sonobe M, et al. HER2 status in lung adenocarcinoma: a comparison of immunohistochemistry, fluorescence in situ hybridization (FISH), dual-ISH, and gene mutations. Lung Cancer. 2014;85(3):373–378. | ||
Li BT, Lee A, O’Toole S, et al. HER2 insertion YVMA mutant lung cancer: Long natural history and response to afatinib. Lung Cancer. 2015;90(3):617–619. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.