Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
Clinical Characteristics and Prognosis of Acute Myeloid Leukemia Patients with Protein Tyrosine Phosphatase Non-Receptor Type 11 Gene Mutation
Authors Huang R, Zhang YT, Lin Y, Pang RL, Yang Z, Zhao WH
Received 18 May 2023
Accepted for publication 27 October 2023
Published 11 November 2023 Volume 2023:16 Pages 1011—1026
DOI https://doi.org/10.2147/PGPM.S420254
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Rui Huang,* Yi-Ting Zhang,* Yu Lin, Ru-Li Pang, Zhi Yang, Wei-Hua Zhao
Department of Hematology, the First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei-Hua Zhao, Department of Hematology, the First Affiliated Hospital of Guangxi Medical University, Shuang-Yong Road 6, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China, Tel +86-771-5356746, Fax +86-771-5352681, Email [email protected]
Objective: The purpose of our study was to investigate the clinical characteristics, molecular biological characteristics and prognosis of acute myeloid leukemia (AML) patients with protein tyrosine phosphatase non-receptor type 11 (PTPN11) gene mutation.
Methods: The clinical data of 30 newly diagnosed adult AML patients with PTPN11 gene mutation were analyzed retrospectively. Kaplan-Meier and Cox proportional risk regression model were examined for prognostic analysis and prognostic factor screening.
Results: High-frequency mutation sites of PTPN11 gene are located in exon 3 of chromosome 12, which are D61 and A72 (16.7%), followed by E76 (13.3%). The median variant allele frequency (VAF) of PTPN11 mutant gene is 18.4%. The patients were divided into two groups according to PTPN11 VAF 35.3% (upper quartile). We observed that the peripheral blood leukocyte count in patients with VAF ≥ 35.3% was significantly higher than patients with VAF < 35.3% (p = 0.019) and also closely related to M5 (p = 0.016) and internal tandem duplication (ITD) of FMS-like tyrosine kinase 3 (FLT3) (FLT3-ITD) mutation (p = 0.048). Taking PTPN11 VAF 20% and 35.3% as the cutoff value, the patients were divided into two groups, and the overall survival and event-free survival (EFS) of the two groups were not significant. Multivariate analysis of Cox risk ratio model showed that white blood cell count and Eastern Cooperative Oncology Group (ECOG) physical status score were independent risk factors affecting the EFS.
Conclusion: Our study observed that PTPN11 VAF may not be a prognostic factor in patients with PTPN11mut AML. Newly diagnosed high white blood cell count and poor performance status were independent risk factors for EFS in PTPN11mut AML.
Keywords: acute myeloid leukemia, PTPN11, mutation, clinical characteristics, prognosis
Graphical Abstract:
Introduction
Acute myelogenous leukemia (AML) is a malignant clonal disease originating from hematopoietic stem cells, which can occur in various age groups and is highly heterogeneous. In recent years, with the development of immunology, cytogenetics and molecular biology, the biological characteristics of AML tumor cells have been more deeply understood and laid a foundation for the precise classification, diagnosis, prognosis and selection of the best treatment for AML. Further research on abnormal molecular biology can provide the basis for stratified diagnosis, precise treatment and evaluation of prognosis of leukemia patients.1,2
SHP2 encoded by protein tyrosine phosphatase non-receptor type 11 (PTPN11) is the first carcinogenic non-receptor tyrosine phosphatase found in the cytoplasm, which plays an important role in the occurrence of a variety of solid tumors and hematological tumors. As the first proto oncogene identified to encode tyrosine phosphatase,3 PTPN11 has encoded the SHP2 protein (SRC homology-2 domain containing protein tyrosine phosphatase) which can be recruited by various receptor tyrosine kinases to induce cell signal transduction and participate in a variety of intracellular oncogenic signal cascades, such as Ras/Raf/MAPK, JAK/stat, PI3K/Akt, PD-1/PD-L1 and other pathways,4–7 playing an important role in cell cycle maintenance, proliferation, differentiation and migration.8 The reversible process of phosphorylation of protein tyrosine kinases (PTKs) and dephosphorylation of non-receptor protein tyrosine phosphatases (PTPs) achieves a dynamic balance and maintains the normal physiological function of cells.9 Abnormal regulation of these enzymes often leads to the pathogenesis of various diseases, including diabetes, autoimmune diseases and cancer.10
Germline mutations in PTPN11 can cause Noonan syndrome (NS), a congenital disorder involving multisystem developmental disorders.11 Somatic mutations are associated with the occurrence of a variety of cancers, including lung cancer, colon cancer, breast cancer and glioblastoma in solid tumors.12 Many experimental studies have shown that SHP2 playing a crucial role in hematopoietic development is a positive regulator of hematopoiesis, and the loss or reduced catalytic activity of SHP2 is related to the decrease in the number and function of hematopoietic stem cells.13,14 As SHP2 is a key regulator of RAS/MAPK in the proliferation and survival signaling pathway, its abnormality leads to the increase of RAS-MAPK activity through the increase of phosphatase activity, thereby promoting the occurrence of leukemia.15,16
PTPN11 gene has been rarely studied in acute myeloid leukemia, and the prognosis of this gene for AML has not been clear. The somatic mutation of PTPN11 occurs in about 5% of acute myeloid leukemia. Previous studies on the PTPN11 gene in acute myeloid leukemia were few. In recent years, clinical studies have begun to suggest that the PTPN11 gene is closely related to the prognosis of AML patients. Studies suggest that PTPN11 is associated with poor prognosis in AML.17–19 In addition, PTPN11 gene mutation frequency (variant allele frequency, VAF) may have a certain impact on prognosis.19 However, several studies have shown that PTPN11 mutation has no significant effect on the prognosis of the total AML population.20,21 Whether this gene is an adverse factor in AML patients needs to be confirmed by more large-sample clinical cohort studies.
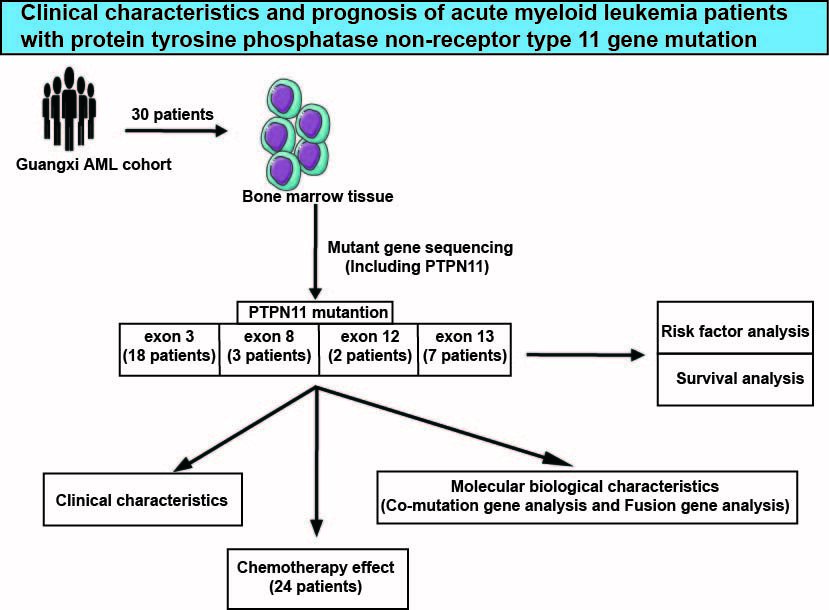
In order to further explore and analyze the clinical characteristics, molecular biological characteristics and prognosis of PTPN11 gene mutation-positive AML patients. Our current study summarized the clinical dataset of 30 patients with PTPN11 mutation-positive acute myeloid leukemia from the First Affiliated Hospital of Guangxi Medical University for comprehensive analysis and exploration.
Materials and Methods
Demographic Data
Through retrospective analysis, the clinical data of 30 patients with acute myeloid leukemia with PTPN11 gene mutation who were treated in the Department of Hematology, the First Affiliated Hospital of Guangxi Medical University from January 2018 to September 2021 were collected. Twenty-seven were primary AML and 3 were MDS-transformed secondary leukemia. There were 12 males (40%) and 18 females (60%), with a median age of 44.5 (14–72) years. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Approval Number: 2023-E232-01). The signed informed consent was obtained from all participants.
Fusion Gene and Genetic Mutations
Fusion Genes
Four milliliters bone marrow fluid samples of newly diagnosed patients were collected and anticoagulated in EDTA tubes, and the detection method was qualitatively determined by multiple nested RT-PCR, which was used to screen the expression of 38 fusion genes that may occur in AML. The specific detection items are shown in Table S1.
Genetic Mutations
Genetic mutations were detection with high throughput sequencing technology, through the extraction of bone marrow in gDNA, extended with multiple PCR technology, one-time grab all of the genes to be detected, and will build a good library with sequencing machine for testing, testing samples related gene detection area inside and outside show and splice site near the transcription and hot spot mutations, CBL, PHF6, CEBPA, PIGA, CSF3R, PTPN11, DNMT3A, RUNX1, ETV6, SETBP1, SF3B1, EZH2, SRSF2, FLT3, STAG2, IDH1 and TET2 were detected, IDH2, TP53, JAK2, U2AF1, KIT, WT1, KRAS, ZRSR2, ASXL1, MPL, BCOR, NF1, CALR, NRAS, BCORL1 and NPM1.
Treatment of Patient
In this study, there were 26 patients aged <60 years old who had at least one course of induction, 23 patients received IA for the first induction chemotherapy regimen, 2 patients were induced with DA regimen, and 1 patient was induced with azacitidine + CAG regimen. This patient was MDS Transformed leukemia, and allogeneic hematopoietic stem cell transplantation was performed at the time of diagnosis of MDS. There are 4 patients over 60 years old, one of them died without treatment after diagnosis, 1 was treated with azacitidine + venetoclax targeted therapy, and 1 was treated with decitabine + CA regimen, 1 list was treated with azacitidine demethylation. For those who did not relieve after 1 course of treatment, re-induction was given with the medium-dose cytarabine + homoharringtonine regimen, MA, azacitidine combined with veneclax, azacitidine alone, and citabine + CAG. Recurrent cases were induced with decitabine + clarithromycin + cytarabine or medium-dose cytarabine + daunorubicin.
Consolidation chemotherapy is generally based on medium- and high-dose cytarabine (Ara-C)-based regimen or standard-dose chemotherapy for consolidation. The total post-remission chemotherapy cycle is ≥6 courses or standard-dose chemotherapy for 3–4 courses of consolidation followed by hematopoiesis stem cell transplantation. Hematopoietic stem cell transplantation was performed for those with transplant indications and successful matching.
Follow‑Up
The follow-up time starts from the time of diagnosis, the dead cases are followed up to the day of death, and the follow-up time ends on February 28, 2022. The follow-up method is to contact by phone and check the inpatient medical records and outpatient medical records. For patients who died during the follow-up period, follow the medical records or check with the patient’s family by phone. The median follow-up time was 26 months, and 2 patients were lost to follow-up.
Statistical Analysis
SPSS 22.0 statistical analysis software was used, and the data were expressed as median or percentage. Appropriate statistical analysis methods were selected for different types of patient data. The comparison of measurement data was performed by t test or nonparametric test, and the comparison of count data was by χ2 test and Kaplan–Meier method was used for OS and EFS survival analysis, and the Cox risk model was used for univariate and multivariate analysis of related variables. After univariate analysis, factors with P < 0.1 were included in the Cox regression risk model for multivariate analysis, and P < 0.1 was set for multivariate analysis. P < 0.05 was considered a statistically significant difference.
Results
Clinical Characteristics
Of the 30 newly diagnosed patients with PTPN11 mutated positive acute myeloid leukemia (PTPN11mut AML) who met the conditions and were included in this study, 27 were primary AML and 3 were secondary leukemia transformed from MDS. There were 12 males (40%) and 18 females (60%), with a median age of 44.5 years.
In 30 patients, 19 (64.3%) showed anemia such as dizziness and fatigue, 9 (30%) showed infection such as fever and abdominal pain, and 2 (6.7%) showed bleeding such as ecchymosis and ecchymosis. Combined with the results of physical examination and abdominal color ultrasound, 2 patients (6.7%) had hepatomegaly, 5 patients (16.7%) had splenomegaly, and 6 patients (20%) had superficial lymph node (supraclavicular, axillary, inguinal, etc.) enlargement. No patient had skin infiltration, green tumor, or central nervous system invasion. According to the ECOG (Eastern Cooperative Oncology Group) score of physical condition, 8 (26.7%) patients scored 0 at the initial diagnosis, 13 (43.3%) scored 1, 7 (23.3%) scored 2, and 2 (6.7%) scored 3.
Median white blood cell count of peripheral blood at initial diagnosis 11.9 × 109/L, median hemoglobin 72.8 g/L, median platelet 49.7× 109/L. The median percentage of bone marrow blast cell was 61.8% (Table 1).
 |
Table 1 Clinical Characteristics of Thirty AML Patients with PTPN11 Mutation |
Clinical characteristics of 30 AML patients with PTPN11 mutation are shown in Table 1. The bone marrow morphology of 30 patients with PTPN11mut AML was more common in M5 type (43.3%), M4 type (33.3%), and M2 type (23.3%). In flow cytometry, CD13 was expressed in 30 cases (100%), CD33 in 30 cases (100%), HLA-DR in 25 cases (83.3%), CD34 in 21 cases (70.0%), MPO in 9 cases (30.0%), CD117 in 24 cases (80.0%), CD15 in 8 cases (26.7%), CD14 in 5 cases (16.7%), CD11b in 5 cases (16.7%), CD64 in 5 cases (16.7%), CD19 in 2 cases (6.7%), CD2 in 1 case (3.3%), CD7 in 6 cases (20.0%), and CD56 in 5 cases (16.7%). In 30 patients with PTPN11mut AML, 29 patients had bone marrow chromosome karyotype analysis at the time of initial diagnosis, 1 patient had no chromosome culture, 19 of the remaining 28 patients (67.9%) had normal karyotype chromosomes, 4 patients (14.3%) had −7 chromosomes and 3 patients (10.0%) had complex karyotypes (Table 2). PTPN11 usually shows normal karyotype, but when the karyotype is abnormal, the incidence of poor prognosis karyotype such as −7 chromosomes is higher.
 |
Table 2 Karyotype Analysis Results of AML Patients with Abnormal Karyotype |
Molecular Biological Characteristics
Distribution of PTPN11 gene mutation sequence: Among all patients in this study, 29 patients detected 1 PTPN11 mutation and 1 patient detected 2 PTPN11 mutations. Eighteen (60.0%) PTPN11 gene mutations were located in exon 3, 7 (23.3%) in exon 13, 3 (10.0%) in exon 8, and 2 (6.7%) in exon 12. There are 20 types of amino acid (AA) mutations, the most common of which are D61G, A72V and N308D. The other mutations are T468M, E76A, A72T, T73I, D61Y, E76K, E76Q, G60A, G60V, G503A, G503E, G503R, M504V, N58Y, Q510H, S502L and S502P. The most common amino acid mutation sites are D61 and A72 (16.7%), followed by E76 (13.3%). All mutations occur in N-SH2 and PTP domains. See Figure 1A for specific mutation types and domains.
Mutation ratio of PTPN11 gene: the VAF value of PTPN11 mutation gene has a large difference, the lowest value is 1.1%, the highest value is 48.3%, the median is 18.4%, and the upper and lower quartiles are 7.2% and 35.3%, respectively. The mutation frequency of PTPN11 gene in each patient is shown in Figure 1B. With the median value of PTPN11 VAF (18.4%) as the cut-off value, the patients were divided into two groups to analyze the characteristics of age, sex, bone marrow immature cells, peripheral white blood cells, hemoglobin, platelets, and whether NPM1 mutation was combined. The results showed that the patients with lower PTPN11 VAF value (<18.4%) had no significant difference in the above clinical characteristics compared with patients with higher VAF value (≥18.4%). With the upper quartile (35.3%) of PTPN11 VAF value as the boundary, the patients were again divided into two groups: low and high VAF. It can be seen that the white blood cell count in the group with high PTPN11 VAF (≥35.3%) was significantly higher than that in the group with low PTPN11 VAF (<35.3%) (p = 0.019, Table 3), which was more likely to be manifested as M5 type (p = 0.016, Table 3) and more likely to be combined with FLT3-ITD mutation (p = 0.048, Table 3). However, there was no significant difference between the two groups in sex, age, proportion of bone marrow blasts, whether NPM1 mutation was combined and complete remission rate (p < 0.05).
 |
Table 3 The Influence of PTPN11 VAF on the Clinical Characteristics of AML with PTPN11 Mutation |
Co-mutation gene: 29 patients (96.7%) were detected with other gene mutations, and 1 patient (3.3%) only had PTPN11 gene mutation, but no associated mutation gene was detected (Table 4). Among patients with co-mutation genes, 3 (10.0%) patients detected 1 co-mutation gene, 12 (40%) patients detected 2 co-mutation genes, 7 (23.3%) patients detected 3 co-mutation genes, 3 (10.0%) patients detected 4 co-mutation genes, and the number of co-mutation genes 5, 6, 7 and 9 was 1 (3.3%) (Table 4). NPM1, NRAS, DNMT3A, TET2, ASXL1, IDH2, FLT3-ITD, KRAS, FLT3-TKD, RUNX1, PHF6, CEBPA, BCORL1, SETBP1, IDH1, WT1, BCOR, NF1, ZRSR2, TP53, SF3B1, ETV6, KIT were detected among 33 myeloid high-frequency mutation genes. Multiple mutant genes can be detected in the same patient at the same time, and the mutation sites of the same mutant gene in different patients are different. Among the 30 patients, NPM1 was the most positive (11 cases (36.7%)), followed by NRAS positive (9 cases (30.0%)), DNMT3A positive (7 cases (26.7%)), ASXL1, KRAS and TET2 positive (6 cases (20%)), IDH2 positive (5 cases (16.7%)), FLT3-ITD and FLT3-TKD positive (4 cases (13.3%)), RUNX1, PHF6, BCORL1, CEBPA and BCOR positive (3 cases (10.0%)), SETBP1, WT1, IDH1 and TP53 positive (2 cases). SF3B1, ETV6, NF1, ZRSR2 and KIT were positive in 1 case, respectively (Figure 1C).
 |
Table 4 The Number of Co-Occurrence of Mutations |
Fusion gene: 30 patients completed the detection of fusion gene, 23 patients (76.7%) were negative for fusion gene, 1 patient had 2 positive fusion genes, and the remaining 6 patients had only 1 positive fusion gene. Among the 38 fusion genes detected, EVI1 rearrangement was most positive in 3 cases (10.0%), MLL-AF6, MLL-AF9, DEK-CAN, AML1-ETO, CBF β/One case was positive for MYH11.
A total of 28 of the 30 PTPN11 positive patients improved MICM typing. According to the 2017 ELN genetic risk stratification, there were 12 in low-risk group, 6 in medium-risk group, and 10 in high-risk group. In the other two patients, one patient’s chromosome was not cultured, but the RUNX1 gene mutation was positive. One patient’s karyotype was not checked, and the ASXL1 gene mutation was positive. The two patients were classified as high-risk group. Therefore, 40.0% (12 cases) of patients in this study are in low-risk group, 20% (6 cases) are in medium-risk group, and 40% (12 cases) are in high-risk group.
Therapeutic Effect
Among 30 PTPN11 positive patients, 22 were treated regularly, 7 were returned to hospital for chemotherapy irregularly, and one died if he did not receive chemotherapy after diagnosis. Among 22 patients with regular treatment, 9 patients received hematopoietic stem cell transplantation, including 3 patients receiving autologous peripheral blood hematopoietic stem cell transplantation and 6 patients receiving allogeneic hematopoietic stem cell transplantation. Among them, 2 patients died of severe pneumonia after recurrence after transplantation. Of the remaining 13 patients, 6 died due to severe infection and uncontrolled leukemia during the treatment process. In the current consolidation and maintenance treatment process of 5 patients, 2 patients completed chemotherapy, and the out-patient took Vinaikara for regular return visits. Among the 7 patients with irregular chemotherapy, 2 patients gave up treatment after one course of induction chemotherapy, and all of them lost the follow-up; Two patients were induced to give up treatment after two courses of treatment and died; One patient did not return to hospital on time for consolidation treatment and died of intracranial hemorrhage after leukemia relapse; One patient did not return to the hospital for consolidation treatment on time, and now is preparing for allogeneic hematopoietic stem cell transplantation after leukemia relapse; One patient did not return to the hospital for chemotherapy as instructed after 4 times of chemotherapy. At present, he is taking vinaikara for follow-up observation in the outpatient department.
After one course of induction chemotherapy and re-examination of bone marrow morphology, the patients achieved a morphological remission rate of 50% (12/24). A total of 19 patients completed one course of induction chemotherapy and flow type minimal residual disease (MRD) detection. Nine patients (47.4%) were MRD positive, and 10 patients (53.6%) were MRD negative. The complete remission rate of bone marrow morphology after 2 courses of chemotherapy was 75.0% (18/24).
By comparing the effects of patient’s sex, age, peripheral blood leukocyte count at the initial diagnosis, proportion of bone marrow protoplasts, chromosome karyotype, NPM1 gene mutation, and ELN2017 risk stratification on CR rate after one course of induced chemotherapy, it was found that patients with bone marrow protoplasts ≥60% and white blood cells ≥17×109/L significantly reduced CR rate after one course of chemotherapy (p = 0.041, p = 0.003, Table 5). There was no significant difference between abnormal karyotype chromosome and −7 chromosomes in CR rate after one course of induction chemotherapy (p = 0.371, p = 0.217). The CR rate of patients with positive NPM1 mutation was lower than that of patients with negative NPM1 mutation after one course of chemotherapy, but there was no statistical.
 |
Table 5 Correlation Analysis of Clinical Parameters Affecting CR in Patients with PTPN11 Mutation AML After One Course of Treatment |
Clinical Outcomes of Patients
As of February 28, 2022, 30 patients with positive PTPN11 mutation had survived in 16 cases, died in 12 cases, and lost follow-up in 2 cases. The median follow-up time was 26 months. As of the follow-up time, the median survival time of patients was not observed. During the follow-up time, the 1-year OS rate was 71.6%, the 2-year OS rate was 52.3%, and the median EFS was 12 months. The survival curve and event-free survival curve are shown in Figure 2A and B. Compared NPM1 positive patients with negative patients in 30 patients, there was no statistical difference between total OS (p = 0.686) and EFS (p = 0.171) (Figure 2C and D). The PTPN11 positive patients were divided into high and low groups according to the cut-off value of VAF 20%. The results showed that the OS (p = 0.579) and EFS (p = 0.325) of the two groups were not statistically significant (Figure 2E and F). The patients were divided into two groups according to the cutoff value of 35.3% VAF. The results showed that the OS (p = 0.796) and EFS (p = 0.236) of the two groups were not statistically significant (Figure 2G and H).
Factors Influencing the Prognosis of PTPN11mut AML
COX risk proportion model was used to conduct EFS single-factor analysis on patients’ age, gender, ECOG score, percentage of myeloblasts, peripheral blood picture at initial diagnosis, PTPN11 VAF value, and NPM1 mutation. The results showed that the influence of ECOG score on EFS was statistically significant (p = 0.047) (Table 6). Factors with P < 0.1 were included in the multifactor analysis, and multivariate analysis showed that peripheral blood leukocyte count and ECOG score were independent risk factors affecting PTPN11mutAML EFS (Table 7).
 |
Table 6 Univariate Cox Proportional Hazard Regression Analysis of Clinical Parameters Affecting EFS in Patients with PTPN11 Mutated AML |
 |
Table 7 Multivariate Cox Proportional Hazard Regression Analysis of Clinical Parameters Affecting EFS in Patients with PTPN11 Mutated AML |
Discussion
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by blocked differentiation and clonal proliferation of hematopoietic stem and progenitor cells, which hinders normal hematopoietic function.22 In recent years, advances in molecular and cellular biology have improved our understanding of the pathophysiology of AML. With the application of next-generation sequencing technology, 40 ~ 50 genes with recurrent somatic mutations have been identified in different AML subtypes.23–26 Cytogenetic results and molecular abnormalities are considered as important prognostic factors, which have high predictive power for complete response rate, disease-free survival, recurrence risk and overall survival.1
In this study, we collected the data of 30 AML patients with PTPN11 mutations, 27 of whom were primary AML and 3 were MDS transformed secondary leukemia. According to foreign reports, the incidence of primary AML, sAML and tAML in PTPN11mut (mutant PTPN11) AML patients is almost similar to that in PTPN11wt (wild-type PTPN11) AML patients.17,18 The number of cases in this study is small, and the conclusion of this study cannot be confirmed. The female incidence rate of this study is slightly higher than that of male, which is consistent with foreign reports.17–19 The median age of the patients was 44.5 (14–67) years, which was slightly smaller than the previous foreign studies (52.5–70 years).19–21 Considering that the sample size included in this study is relatively small, it needs to be further confirmed by expanding the sample size later.
In this study, the patients showed leukocytosis, anemia and thrombocytopenia at the initial diagnosis, which was consistent with the characteristics of general AML. It was reported in the literature that PTPN11mutAML had a higher leukocyte count than PTPN11wtAML (P < 0.05), but there was no significant difference in hemoglobin, platelet count and the proportion of bone marrow blasts.17,18 In this study, the remission rate of patients with peripheral blood leukocyte count ≥17×109/L and bone marrow blasts ≥60% after 1 course of induction treatment was lower. In the EFS multifactor analysis, leukocyte as an independent factor affecting the prognosis of the disease, suggesting that patients with high tumor load at the initial diagnosis had poor response to treatment, low complete remission rate, and had a negative impact on the prognosis of the disease.
Since MICM classification was proposed, it has been widely used in the diagnosis and treatment of leukemia. Bone marrow morphology of 30 patients with PTPN11mut AML: M5: 43.3%, M4: 33.3%, M2: 23.3% (n = 6). No other types were found, which is consistent with previous reports,18,20 M4 or M5 is the most common FAB classification of PTPN11mut AML. In this study, 30 patients were tested for immunophenotype. More than 70% of the patients’ bone marrow leukemia cells expressed CD33, CD13, CD34, HLA-DR, CD117 and other cell surface antigens. A few patients expressed lymphoid markers, of which the positive expression rates of CD19 and CD56 were 6.7% and 16.7%, respectively. PTPN11 mutation is closely related to CD14 expression,20 CD14 is a marker of monocyte line expression, which is consistent with that of patients in FAB M4 and M5. However, in this study, the expression of CD14 is 16.7%, slightly different from the above report, and larger samples are needed to confirm the immunophenotypic characteristics of PTPN11mut AML. Among the 28 patients with cultured chromosomes, 19 (63.3%) were normal karyotype chromosomes, and 4 (13.3%) of the patients with abnormal karyotype were primary leukemia patients, 2 were refractory leukemia patients, one died within 30 days after diagnosis, and one was recurrent after allogeneic hematopoietic stem cell transplantation. −7/del(7q) is a common high-risk cytogenetic abnormality, which often occurs in MDS or high-risk AML.27 It has been reported that the incidence of chromosome −7 in PTPN11mut AML is 23%, and −5/-7 in relapsed/refractory PTPN11mut AML is significantly higher than that in PTPN11wt AML.18,19 It is not difficult to see that PTPN11 mutation may be associated with relapse or refractory treatment, suggesting the possibility of poor prognosis. In addition, 3 cases (10.7%) of complex karyotypes in this study showed no significant increase, similar to those reported abroad (7.1%).17
According to research reports, most of PTPN11mut AML carry PTPN11 single mutation, and a few carry PTPN11 double mutation.17 In this study, PTPN11 gene mutation was multiple single mutations, and only one case was double mutation (3.33%), which was consistent with previous reports. The current literature shows that PTPN11 mutations in AML are most often located in exon 3 (N-SHP domain), and the rest are mostly located in exon 13 (PTP domain). The most common amino acid mutation sites are D61, A72, G60, E76, etc.17,18 In this study, the mutations of PTPN11 gene in exon 3 accounted for 18 (60.0%), exon 13 accounted for 7 (23.3%), exon 8 (10.0%) and exon 12 (6.7%), all located in N-SHP domain or PTP domain. The most common amino acid mutation sites were D61, A72 (16.7%), followed by E76 (13.3%), which was basically consistent with the previous study. Previous studies have shown that mutations in these amino acid sites destroy the self inhibition between the PTP catalytic domain and N-SH2 domain, enhance SHP2 activity, and thus become functional acquired mutations.28 Researchers have observed that germline PTPN11 mutations in NS patients do not overlap substantially with somatic PTPN11 mutations in leukemia patients.3 Mutants in leukemia (such as D61Y and E76K) have stronger phosphatase catalytic activity than NS-related mutants (such as N308D).29 Therefore, it is suggested that higher levels of phosphatase activity will lead to a more severe phenotype, and if mutations with high phosphatase activity occur in the germline, it may lead to embryonic death in utero, and predicted in aborted fetuses with NS, a higher prevalence of leukemia-associated mutations.30
The median VAF of PTPN11 was between 13.4% and 24%.17–19 The median VAF of PTPN11 gene in this study was 18.4%, the lowest value was 1.1%, and the highest value was 48.3%. In addition, the frequency of PTPN11 gene mutation may have a certain impact on the prognosis of AML. Swoboda concluded that VAF >20% was associated with poorer prognosis in PTPN11mut AML (OS 8.9 vs 20.5 months, p = 0.04).19 Stasik17 first used an algorithm to compare the VAF of PTPN11 mutation and the main co-mutated gene to dichotomize PTPN11 mutation into dominant and subclonal mutations (the difference between the two is ≥10% for dominant mutation, <10% for subclonal mutation), found in the majority of patients at the subclonal level (64%), and observed that compared with AML with dominant PTPN11 mutation, subclonal PTPN11 mutation had worse OS (10.38 vs 29.88 months, P = 0.0018), RFS (8.62 vs 39.48 months, P = 0.0061) and lower CR rate (61.8% vs 76.3%, P = 0.046), and higher ED30 (14.7% vs 5.3%, P = 0.003). The results suggest that subclonal mutation is a late event that occurs together with a variety of other adverse features and may lead to poor outcomes, but this classification is more dependent on the effect of co-mutated genes on AML, and the accuracy of its results needs to be further explored. In this study, patients were divided into two groups based on the median value of PTPN11 VAF. Compared with patients with PTPN11 VAF <18.4% and VAF ≥18.4%, there was no significant statistical difference in age, sex, bone marrow immature cells, peripheral blood leukocyte count, hemoglobin, platelet count, complete remission rate and other clinical characteristics and treatment effects. According to previous research conclusions, patients with high VAF value have worse prognosis.19 In this study, the cut-off value was increased, and the patients were again divided into low and high VAF groups with the upper quartile of PTPN11 VAF value (VAF 35.3%) as the boundary. It was found that the white blood cell count in the group with VAF ≥ 35.3% was significantly higher than that in the group with VAF <35.3% (58.1 × 109/L vs 9.7 × 109/L, p = 0.019), which is more likely to be M5 (p = 0.016) and more likely to be combined with FLT3 ITD mutation (p = 0.048). There is no significant difference in sex, age, platelet, and myeloblast. Therefore, patients with PTPN11 VAF ≥ 35.3% should be alert to whether they have hyperleukaemia, which has a heavy tumor load, leading to systemic bone pain, invasion and destruction of blood vessels, leukocyte stasis syndrome, tumor lysis syndrome and other complications. Patients with PTPN11 VAF ≥ 35.3% are more likely to be associated with FLT3-ITD mutation, and FLT3-ITD VAF ≥ 50% is an influential factor for poor prognosis of AML in the 2017 ELN risk stratification.2 Therefore, it may affect the survival prognosis of PTPN11mut AML with FLT3-ITD mutation. However, in this study, the VAF values of 4 patients with FLT3-ITD mutation are less than 50%, which does not affect the risk stratification of AML. In addition, AML with PTPN11 mutations observed in previous studies are less likely to have FLT3-ITD mutations.17,20 The activation of signal pathways between FLT3-ITD and RAS pathway family NRAS, KRAS and PTPN11 overlaps, and there is mutation rejection. Both of them have functional redundancy or independence in driving leukemia formation. The relationship between this gene mutation and PTPN11mut AML and its prognostic impact have needed to be further studied.
In terms of merging fusion genes, 30 patients in this study completed the detection of fusion genes, of which 23 patients (76.7%) were negative for fusion genes and 3 patients (10%) were positive for EVI1 fusion genes. The viral integration site-1 (EVI1) gene is a transcription factor with DNA binding activity zinc finger structure. It can induce the pathogenesis of acute leukemia in multiple links such as cell cycle, apoptosis and stem cell regulation by recognizing and binding the common sequence of DNA.31,32 Previous clinical studies showed that the CR rate of patients with EVI1 positive expression in AML was low, and the long-term disease-free survival rate and relapse-free survival rate were significantly lower than those of patients with EVI1 negative expression.33 At present, there is no report on the influence of EVI1 positive in PTPN11mut AML. If EVI1 is found in PTPN11mut AML, it can predict its poor prognosis and guide intensive treatment.
Studies have shown that in patients with PTPN11mut AML, there is a significant increase in the combination of NPM1 mutations.17–21 Mutations in PTPN11 and NPM1 frequently co-occur, possibly suggesting that these mutations cooperate to promote leukemia. In a study of 78 PTPN11mut AML patients, Swoboda DM’s team found that patients with PTPN11mut/NPM1wt (PTPN11 mutant/NPM1 wild type) had a shorter OS than PTPN11mut/NPM1mut (PTPN11 mutant/NPM1 mutant) patients (10.3 vs 24.4 months, p =0.007).19 Hou’s team reported that among 266 NPM1-positive AML patients, patients with PTPN11 mutations had worse OS than non-mutated patients (p=0.001).20 Among NPM1wt patients analyzed by Fobarer, compared with PTPN11wt patients, the CR rate of PTPN11mut patients was lower (36% vs 61%, p=0.001), and the 3-year EFS was shorter (9% vs 19%, p=0.003).21 It can be concluded from the above studies that in AML with PTPN11 mutation, NPM1 positive patients have a better prognosis; For the patients with negative NPM1 mutation, PTPN11 mutation is a poor risk factor, and its mechanism has not been well explained at present. It may suggest that PTPN11 gene mutation is a bad prognostic factor for AML. Another argument is that other associated gene mutations may affect the overall prognosis of PTPN11mut AML patients with negative NPM1 mutations.20 However, this conjecture has not been confirmed, and more comprehensive studies are needed to explain the molecular mechanism between PTPN11 and NPM1 genes. The number of NPM1 mutations in the co mutation genes of 30 patients in this paper is the largest, reaching 11 (36.7%). Whether there are NPM1 mutations or not is divided into two groups to analyze the impact on CR, OS and EFS after a course of induced chemotherapy. The results show that patients with negative NPM1 mutations have a low CR rate after the first induction (p=0.095), OS (p=0.686) and EFS (p=0.171) are shorter than patients with NPM1 mutations, but there is no significant difference. Considering the small number of cases in this study, It is necessary to further expand the sample analysis. For decades, the standard intensive treatment of AML has been the combination of 7 days of cytarabine+3 days of anthracycline drugs (nordaunorubicin and daunorubicin). The complete remission and cure rates vary with age. The complete remission rate of AML in adults ≤ 60 years old is 60–85%, the cure rate is 35–40%, and the complete remission rate of AML in adults over 60 years old is 40–60%, and the cure rate is 5–15%.34 For PTPN11mut AML patients of all ages, complete remission rates ranged from 33.3% to 67%.17,18 In this study, patients aged ≤60 years were treated with an IA-based induction regimen, and a medium- and high-dose Ara-C-based regimen was generally used for consolidation chemotherapy. Demethylation combined with veneclax should be used in patients over 60 years of age. After one course of induction chemotherapy, the morphological remission rate was 50% (12/24), and the complete remission rate after two courses of induction was 78.6% (18/24). A total of 19 patients completed 1 course of induction chemotherapy and minimal residual disease (MRD) testing, of which 9 patients (47.4%) were MRD negative, Similar to Alfayez’s study (MRD-negative rate of 59%).18 According to literature reports, the median OS of PTPN11mut AML was 9–13.44 months,17,20 and the median EFS was 7.53 months.17 The median follow-up time in this study was 26 months, and 53% of patients were still alive at the end of the follow-up time, so the median OS could not be analyzed. During the follow-up period, the 1-year OS rate was 71.6%, the 2-year OS rate was 52.3%, and the median EFS was 12 months. The median EFS in this study was longer than previous studies. However, the result bias caused by the small number of cases and the large differences in treatment plans is not excluded. Foreign literature reported that the OS of PTPN11mut AML patients who received AHSCT was 24.4 months, while the OS of PTPN11mut AML patients who did not receive AHSCT was 5.6 months.19 In our study, 9 patients underwent hematopoietic stem cell transplantation, and only 2 died so far, suggesting that hematopoietic stem cell transplantation is still the only reliable way to cure AML.
PTPN11 mutation was reported to be an independent prognostic factor for worse OS (HR 1.75).17 Alfayez found that patients with PTPN11-positive AML had significantly shorter median OS than those with negative AML (8.4 vs 13.6 months, p = 0.008) and also in high-intensity chemotherapy (OS 7.6 vs 35.4 months, p < 0.0001).18 These studies suggest that PTPN11 may serve as an independent predictor of poor prognosis in AML. However, studies have shown that PTPN11 mutations have no significant effect on the prognosis of the total AML population. Swoboda reported that there was no significant difference in the overall response rate (ORR) and CR rate of 58 PTPN11-positive patients after treatment among newly diagnosed AML patients compared with 380 PTPN11-negative patients.19 Hou reported that there was no significant difference in the complete response rate (75% vs 62%), median OS (13 vs 25.5 months), and relapse-free survival (RFS) in 14 PTPN11 mutation-positive and 258 PTPN11-negative AML patients (19 vs 25 months).20 The explanation for the difference of the above research results may include the difference of population treatment schemes and the contingency of gene mutation characteristics. To confirm the prognosis of PTPN11 in AML, more multicenter and diversified clinical studies are needed to confirm it. Due to the small number of this case in our center and the short time of research, we cannot compare the prognosis between the positive and negative control groups of PTPN11 gene mutation, and we need to continue to collect relevant data for analysis and verification. In this study, univariate and multivariate analysis by Cox proportional hazards model showed that peripheral blood leukocyte differential count and ECOG score were independent risk factors for PTPN11 mutAML EFS. Patients with high leukocyte have a heavy tumor burden, which can seriously affect the prognosis of patients, and a good physical status can ensure the smooth completion of the chemotherapy regimen and help improve the prognosis of patients.
However, our study found some positive results, but there are still some limitations to be stated. Due to the small sample size and short follow-up time in this study, some results may be biased. Due to the failure to include more detailed clinical parameters, no significant difference of PTPN11 mutation in the prognosis of AML was observed in this study cohort, which still needs to be further verified in a large cohort.
Conclusion
PTPN11mut AML is a heterogeneous disease, and most FAB genotypes of PTPN11mut AML were M5 or M4. Most of them are normal karyotype chromosomes, and −7 chromosome was the most common chromosome abnormality, which was easily associated with NPM1 mutation. Patients with high PTPN11 VAF tend to have high white blood cell counts at the initial diagnosis and are often associated with FLT3-ITD mutations. Patients with high tumor burden at initial diagnosis have low CR rate and poor treatment response after one course of chemotherapy. Newly diagnosed high white blood cell count and poor performance status were independent risk factors for EFS in PTPN11mut AML.
Data Sharing Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Approval Number: 2023-E232-01). The signed informed consent was obtained from all participants. This study complies with the Declaration of Helsinki.
Acknowledgments
We would also acknowledge the support by Key Laboratory of Hematology, Guangxi Medical University, Education Department of Guangxi Zhuang Autonomous Region, Nanning, Guangxi, China.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Rui Huang and Yi-Ting Zhang are co-first authors for this study.
Funding
The present study was supported in part by the development and application of medical and health appropriate technology in Guangxi (grant no. S2022092) and the Self‑raised Scientific Research Fund of the Health and Family Planning Commission of the Guangxi Zhuang Autonomous Region (grant no. Z-A20230475).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31(1):63–76. doi:10.1016/j.blre.2016.08.005
2. De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J. 2016;6(7):e441–e441. doi:10.1038/bcj.2016.50
3. Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109(3):862–867. doi:10.1182/blood-2006-07-028829
4. Quintana E, Schulze CJ, Myers DR, et al. Allosteric Inhibition of SHP2 Stimulates Antitumor Immunity by Transforming the Immunosuppressive Environment. Cancer Res. 2020;80(13):2889–2902. doi:10.1158/0008-5472.CAN-19-3038
5. Xu D, Qu CK. Protein tyrosine phosphatases in the JAK/STAT pathway. Front Biosci. 2008;13(13):4925–4932. doi:10.2741/3051
6. Wu CJ, O’Rourke DM, Feng GS, Johnson GR, Wang Q, Greene MI. The tyrosine phosphatase SHP-2 is required for mediating phosphatidylinositol 3-kinase/Akt activation by growth factors. Oncogene. 2001;20(42):6018–6025. doi:10.1038/sj.onc.1204699
7. Valero V 3rd, Pawlik TM, Anders RA. Emerging role of Hpo signaling and YAP in hepatocellular carcinoma. J Hepatocellular Carcinoma. 2015;2:69–78. doi:10.2147/JHC.S48505
8. Zhang J, Zhang F, Niu R. Functions of Shp2 in cancer. J Cell Mol Med. 2015;19(9):2075–2083. doi:10.1111/jcmm.12618
9. Zhang ZY. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu Rev Pharmacol Toxicol. 2002;42:209–234. doi:10.1146/annurev.pharmtox.42.083001.144616
10. Yuan X, Bu H, Zhou J, Yang CY, Zhang H. Recent Advances of SHP2 Inhibitors in Cancer Therapy: current Development and Clinical Application. J Med Chem. 2020;63(20):11368–11396. doi:10.1021/acs.jmedchem.0c00249
11. Jongmans MC, Van der Burgt I, Hoogerbrugge PM, et al. Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur J Hum Genet. 2011;19(8):870–874. doi:10.1038/ejhg.2011.37
12. Song Y, Zhao M, Zhang H, Yu B. Double-edged roles of protein tyrosine phosphatase SHP2 in cancer and its inhibitors in clinical trials. Pharmacol Ther. 2022;230:107966. doi:10.1016/j.pharmthera.2021.107966
13. Nabinger SC, Chan RJ. Shp2 function in hematopoietic stem cell biology and leukemogenesis. Curr Opin Hematol. 2012;19(4):273–279. doi:10.1097/MOH.0b013e328353c6bf
14. Chan G, Cheung LS, Yang W, et al. Essential role for Ptpn11 in survival of hematopoietic stem and progenitor cells. Blood. 2011;117(16):4253–4261. doi:10.1182/blood-2010-11-319517
15. Alhumaid MS, Dasouki MJ, Ahmed SO, et al. Comprehensive Genomic Analysis of Noonan Syndrome and Acute Myeloid Leukemia in Adults: a Review and Future Directions. Acta Haematol. 2020;143(6):583–593. doi:10.1159/000505715
16. Murakami N, Okuno Y, Yoshida K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131(14):1576–1586. doi:10.1182/blood-2017-07-798157
17. Stasik S, Eckardt JN, Kramer M, et al. Impact of PTPN11 mutations on clinical outcome analyzed in 1529 patients with acute myeloid leukemia. Blood Adv. 2021;5(17):3279–3289. doi:10.1182/bloodadvances.2021004631
18. Alfayez M, Issa GC, Patel KP, et al. The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia. 2021;35(3):691–700. doi:10.1038/s41375-020-0920-z
19. Swoboda DM, Ali NA, Chan O, et al. PTPN11 mutations are associated with poor outcomes across myeloid malignancies. Leukemia. 2021;35(1):286–288. doi:10.1038/s41375-020-01083-3
20. Hou HA, Chou WC, Lin LI, et al. Characterization of acute myeloid leukemia with PTPN11 mutation: the mutation is closely associated with NPM1 mutation but inversely related to FLT3/ITD. Leukemia. 2008;22(5):1075–1078. doi:10.1038/sj.leu.2405005
21. Fobare S, Kohlschmidt J, Ozer HG, et al. Molecular, clinical, and prognostic implications of PTPN11 mutations in acute myeloid leukemia. Blood Adv. 2022;6(5):1371–1380. doi:10.1182/bloodadvances.2021006242
22. Kishtagari A, Levine RL, Viny AD. Driver mutations in acute myeloid leukemia. Curr Opin Hematol. 2020;27(2):49–57. doi:10.1097/MOH.0000000000000567
23. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. doi:10.1056/NEJMoa1516192
24. Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi:10.1056/NEJMoa1112304
25. Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi:10.1056/NEJMoa0903840
26. Ley TJ, Miller C, et al.; Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074.
27. McNerney ME, Brown CD, Peterson AL, et al. The spectrum of somatic mutations in high-risk acute myeloid leukaemia with −7/del(7q). Br J Haematol. 2014;166(4):550–556. doi:10.1111/bjh.12964
28. Shen D, Chen W, Zhu J, et al. Therapeutic potential of targeting SHP2 in human developmental disorders and cancers. Eur J Med Chem. 2020;190:112117. doi:10.1016/j.ejmech.2020.112117
29. Tartaglia M, Gelb BD. Germ-line and somatic PTPN11 mutations in human disease. Eur J Med Genet. 2005;48(2):81–96. doi:10.1016/j.ejmg.2005.03.001
30. Tartaglia M, Martinelli S, Stella L, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78(2):279–290. doi:10.1086/499925
31. Nucifora G. The EVI1 gene in myeloid leukemia. Leukemia. 1997;11(12):2022–2031. doi:10.1038/sj.leu.2400880
32. Aytekin M, Vinatzer U, Musteanu M, Raynaud S, Wieser R. Regulation of the expression of the oncogene EVI1 through the use of alternative mRNA 5’-ends. Gene. 2005;356:160–168. doi:10.1016/j.gene.2005.04.032
33. Groschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28(12):2101–2107. doi:10.1200/JCO.2009.26.0646
34. Newell LF, Cook RJ. Advances in acute myeloid leukemia. BMJ. 2021;375:n2026. doi:10.1136/bmj.n2026
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


