Back to Journals » Infection and Drug Resistance » Volume 16
Clinical and Bacteriological Analyses of Biofilm-Forming Staphylococci Isolated from Diabetic Foot Ulcers
Authors Mamdoh H, Hassanein KM, Eltoony LF, Khalifa WA, Hamed E, Alshammari TO, Abd El-Kareem DM , El-Mokhtar MA
Received 30 November 2022
Accepted for publication 28 January 2023
Published 24 March 2023 Volume 2023:16 Pages 1737—1750
DOI https://doi.org/10.2147/IDR.S393724
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Hend Mamdoh,1 Khaled M Hassanein,2 Lobna Farag Eltoony,3 Walaa A Khalifa,3 Entsar Hamed,2 Taher Obaid Alshammari,4 Doaa M Abd El-Kareem,5 Mohamed A El-Mokhtar2
1Department of Microbiology and Immunology, Faculty of Pharmacy, Sphinx University, New Assiut, Egypt; 2Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, Egypt; 3Department of Internal Medicine, Faculty of Medicine, Assiut University, Assiut, Egypt; 4Department of Ophthalmology, Prince Mohammed Medical City, Al Jouf, Saudi Arabia; 5Department of Clinical Pathology, Faculty of Medicine Assiut University, Assiut, Egypt
Correspondence: Mohamed A El-Mokhtar, Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, Egypt, Email [email protected]
Background: Diabetes mellitus is a chronic disease that is associated with increased morbidity and mortality. Unfortunately, foot ulcers and amputations due to diabetes are very common in developing countries. The purpose of this study was to characterize the clinical presentation of diabetic foot ulcer (DFU) infections, isolate the causative agent, and analyze the biofilm formation and distribution of biofilm-related genes among isolated Staphylococci.
Material and Methods: The study included 100 diabetic patients suffering from DFUs attending Assiut University Hospital. Swabs were collected and antimicrobial susceptibility testing of the isolates was performed. Biofilm formation was tested phenotypically among staphylococcal isolates and the frequency of different biofilm genes was analyzed by PCR. Clinical presentations of diabetic foot ulcers were correlated with bacterial genetic characteristics. Spa types were determined using DNA Gear-a software.
Results: Microbiological analysis showed that 94/100 of the DFUs were positive for bacterial growth. The majority of infections were polymicrobial (54%, n=54/100). Staphylococci were the most commonly detected organisms, of which S. aureus represented 37.5% (n=24/64), S. haemolyticus 23.4% (n=15/64), S. epidermidis 34.3% (n=22/64) and other CNS 4.7% (n=3/64). Interestingly, co-infection with more than one species of Staphylococci was observed in 17.1% (n=11/64) of samples. A high level of antibiotic resistance was observed, where 78.1% (n=50/64) of Staphylococci spp were multidrug-resistant (MDR). Phenotypic detection showed that all isolated Staphylococci were biofilm-formers with different grades. Analysis of biofilm-forming genes among Staphylococci showed that the most predominant genes were icaD, spa, and bap. Isolates with a higher number of biofilm-related genes were associated with strong biofilm formation. Sequencing of the spa gene in S. aureus showed that our isolates represent a collection of 17 different spa types.
Conclusion: The majority of DFUs in our hospital are polymicrobial. Staphylococci other than S.aureus are major contributors to infected DFUs. MDR and biofilm formation are marked among isolates, which is paralleled by the presence of different categories of virulence-related genes. All severely infected wounds were associated with either strong or intermediate biofilm formers. The severity of DFU is directly related to the number of biofilm genes.
Keywords: diabetes mellitus, biofilm formation, Staphylococci, diabetic foot ulcers, ica genes
Introduction
Morbidity and mortality related to diabetes and its complications are rapidly emerging, particularly in developing countries. It is predicted that by 2040, there will be over 642 million people with diabetes in the world. Generally, diabetes is more prevalent in countries of low income.1 Egypt is ranked the eighth country worldwide in the number of diabetic patients, and by 2045 it is expected to be the 6th country.2 Unfortunately, foot ulcers and amputations are very common in developing countries. Infection of foot ulcers appears in more than half of all cases which required hospitalization and 20% of lower extremity infections will lead to amputation.3 Amputation in people with diabetes is greater than those in non-diabetic people.4 Someone loses a leg due to the complications of diabetes every 20 seconds in the world and more than 50% of these people will die within 5 years after such amputations.5
According to the International Diabetes Foundation, 40–60 million persons suffer from diabetic foot ulcers (DFUs), which markedly outnumbers the cases from 2015, that ranged from 9–26 million.6 Several studies have shown that DFUs are classically polymicrobialy-infected, and Staphylococcus aureus (S. aureus) is the most frequently isolated pathogen.7 These infected ulcers are characterized by dysregulated extracellular matrix remodeling, abnormal growth factors for angiogenesis, and excessive inflammatory reactions leading to impeding of the wound’s recovery.8 DFUs caused by antibiotic-resistant bacteria, particularly Methicillin Resistant Staphylococcus aureus (MRSA), are associated with more severe infections. The prevalence of MRSA in DFUs varies among countries, with an increasing rate in the developing countries.9,10 Pathogen- and host-related factors (eg, immunity status, nerve damage, microbial virulence, and degree of antibiotic resistance) influence the prevalence, severity, and pathophysiology of the DFUs.11
A biofilm is a sessile layer generated from microbes and embedded in a matrix of extracellular polymeric substances and exhibits an altered phenotype with regard to growth, gene expression, and protein production.12,13 The polysaccharide intercellular antigen (PIA) codes for biofilm-related genes in Staphylococci and was described as a specific polysaccharide antigen. Intercellular adhesion (ica) locus can control the production of PIA in vitro from UDP-N-acetylglucosamine.14 Importantly, the ica locus, which includes the regulatory icaR and the biosynthetic genes icaADBC, is essential for the synthesis and virulence of the biofilms. However, biofilms can also occur in an ica-independent fashion and are upregulated in response to anaerobic growth which is provided by the biofilm environment.15 In addition, Staphylococcal protein A (spa) production was reported to be essential for biofilm formation in an ica-deletion mutant of S. aureus. In these spa mutants, biofilm development could be recovered by the addition of an exogenous spa, indicating that spa does not need to be covalently anchored to the cell wall.16 Furthermore, fibronectin-binding proteins (FnBPs) can mediate biofilm development via the major autolysin (Atl) and sigB. Additionally, S. aureus’s biofilm-associated protein (Bap) and Bap-related proteins, which are distinguished by their high molecular weight, presence on the bacterial surface, and crucial virulence function, can promote biofilm development independently of PIA production through cell-to-cell aggregation.17
In S. epidermidis, PIA-independent biofilms were mediated through the accumulation-associated protein (Aap).18,19 The agr quorum sensing system indirectly decreases initial biofilm formation by reducing the expression of genes encoding adhesion factors, which lead to less adherence. Also, agr tend to upregulate the expression of detergent-like peptides and nucleases that seem to increase biofilm detachment.20 Induction of agr through an essential component of the quorum sensing system auto-inducing peptides (AIP) leads to the spread of seeding microorganisms in developed biofilms.21
The aim of this work was to isolate and identify bacterial pathogens associated with diabetic foot ulcers, to determine the antimicrobial susceptibility pattern of the isolated bacterial pathogens, to evaluate the ability of the isolated Staphylococcal species to form biofilms quantitatively, screen for genes that are responsible for biofilm formation and molecular typing of S. aureus strains to identify strain relatedness.
Materials and Methods
Ethical Consideration
Informed consent was obtained from all cases. This study complied with the Declaration of Helsinki and the study protocol was approved by the scientific ethics committee of the Faculty of Medicine, Assiut University (IRB no: 17101272).
Bacterial Isolates
This cross-sectional observational study was conducted over the duration of July 2018 to September 2019. A total of 100 diabetic patients attending Diabetes, Endocrine Centre and Vascular Surgery outpatient clinics in Assiut University Hospitals and suffering from DFU were enrolled. Sterile disposable cotton swabs were obtained from diabetic foot ulcers. Standard procedures were used for isolating, processing, and identifying all isolates.22 Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus haemolyticus identification were confirmed using a multiplex PCR method.23
Phenotypic Testing of Biofilm Formation
The microtiter plate (MTP) was used for the quantitative evaluation of biofilm formation. Briefly, a bacterial suspension was prepared in trypticase soy broth containing 1% glucose (Merck, Darmstadt, Germany). Culture was added to a 96-well plate and incubated overnight at 37°C. Then, supernatant was discarded, wells were washed with PBS and incubated at 65°C until dry. Adherent bacteria were stained with 0.1% crystal violet dye (Sigma Chemical Co., St Louis, MO, USA), then washed with deionized water to get rid of the excess dye. For solubilization, 70% ethanol with 10% isopropyl alcohol was added and the optical density was read at 570 nm by an ELISA plate reader (BioTek, Germany). Each test was performed in triplicate.23
Screening for Biofilm related Determinants
The presence of different virulence-related genes was tested by PCR using different primer combinations, as described in Table 1. Uniplex PCR was used for the detection of adhesin genes icaA, icaB, icaC and icaD24 and biofilm-associated protein gene bap, fnbA, aap and altE. Multiplex PCR was used for the detection of accessory regulator genes agrI, agrII, agrIII and agrIV.25
 |
Table 1 Summary of the Target and Sequence of the Oligonucleotide Primers Used in the PCR |
Evaluation of Antibiotic Susceptibility and Detection of Methicillin Resistance
Susceptibility of isolates to different antibiotics was evaluated using the modified Kirby–Bauer method and results were interpreted according to CLSI guidelines.26 The following antimicrobial agents were tested: cefoxitin (30µg), Teicoplanin (30 µg), Vancomycin (30 µg), Gentamycin (10 µg), Azithromycin (15 µg), Erythromycin (15 µg), Ciprofloxacin (5 µg), Levofloxacin (5 µg), Tetracycline (30 µg), Doxycycline (30 µg), Trimethoprim/sulfamethoxazole (1.25/23.75 µg), Clindamycin (2 µg), Linezolid (30 µg), Chloramphenicol (30 µg), Amoxicillin/clavulanate (10/20µg), Cephalexin (30µg), Ceftazidime (30 µg), Ceftriaxone (30 µg), Meropenem (30 µg), Tobramycin (30 µg), Amikacin (30 µg) and Rifampin (30 µg). Methicillin resistance in Staphylococci was defined as the isolates that showed resistance to cefoxitin and a positive mecA PCR test.27
Isolates were regarded to be multidrug-resistant (MDR) if they exhibited resistance to three or more antimicrobials that belonged to distinct antibiotic classes and bacterial targets, and extensively drug resistant (XDR) if strains were resistant to at least one agent in all but two or fewer antimicrobial categories (ie, bacterial isolates remain susceptible to only one or two antimicrobial categories).28
Staphylococcal Protein a (Spa) Typing
Amplification of the spa gene was performed using the primer pairs shown in Table 1, as described previously.29 PCR products were sequenced by Macrogen (Korea) and spa types were determined using DNA Gear-A software and the Ridom SpaServer (http://www.spaserver.ridom.de).30 A clonal complex was defined as a group of at least two related spa types. A spa type that was not part of a clonal complex was considered.31
Results
Demographic and Clinical Characteristics
Among the admitted diabetic patients, 58% (n=58/100) were males, 50% were aged 40 years or above and most of them (63%, n=63/100) were illiterate. About half of the cases (55%) were non-smokers. Based on history records, most of the cases had a previous family history, about 49% were diagnosed incidentally and most of them had had the disease for more than 10 years. Hypertension and obesity were commonly reported co-morbidities (37% and 64%, respectively).
The diabetic patients suffered from complications of diabetis such as peripheral neuropathy (55%), retinopathy (33%), peripheral vascular diseases (24%), cerebrovascular diseases, and presence of coronary artery diseases (13% for each) (Table 1).
The analysis of glycemic control using HbA1C showed that 88% of the cases were uncontrolled diabetic patients. Analysis of random blood glucose levels showed that blood glucose levels were above 200 in most cases, which is consistent with HbA1c results (71% of cases had HbA1c values of >9) (Figure 1 and Table 2). Concerning the treatment, the majority of patients (63%) partially managed the disease with insulin and 24% with oral hypoglycemic agents.
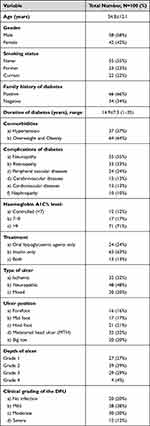 |
Table 2 Demographic and Clinical Characteristics of Enrolled Patients |
 |
Figure 1 Random blood glucose levels of the cases. |
The majority of ulcers were neuropathic (48%) and located under the heads of the metatarsal region of the foot (32%). Most ulcers were deep (grade 2 or 3) according to Wagner–Meggitt classification.
Microbiological Analysis of DFU
Microbiological analysis of the wound swabs showed that 94% of these ulcers were infected, 54% (n=54/100) were polymicrobial, 40% (40/100) were monomicrobial and 6% (n=6/100) were negative. Gram-negative bacteria were detected in 59.49% (n=94/158) of isolates. Gram-positive isolates were detected in 40.5% (n=64/158) of isolates either as mono-infection or coupled with other Gram-negative bacteria. All Gram-positive infected cases contained staphyhlococcal spp.
Among the 64 Staphylococci isolates, 37.5% (n=24/64) were identified as S. aureus, 23.4% (n=15/64) S. haemolyticus, and 34.3% (n=22/64) S. epidermidis. Interestingly, some samples (n=11/64, 17.1%) were co-infected with more than one type of Staphylococci. six samples showed a co-infection of S. aureus and S. epidermidis, 3 samples showed co-infection of S. aureus and S. haemolyticus, and two samples showed mixed infection of S. epidermidis and S. haemolyticus as shown in Figure 2.
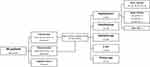 |
Figure 2 Flow chart showing the microbiological analysis of DFUs. Abbreviations: SA, Staphylococcus aureus; SE, Staphylococcus epidermidis; SH, Staphylococcus haemolyticus. |
Most Staphylococci were susceptible to linezolid, chloramphenicol, and rifampicin; however, a high level of resistance was observed to amoxiclav, cefoxitin, and oxacillin, as shown in Figure 3. Among the 64 Staphylococci isolates tested, cefoxitin resistance and mecA gene were observed in 68.7% (n=44/64) of samples, pointing to the high frequency of methicillin resistance among the isolates.
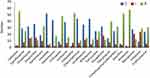 |
Figure 3 Antibiotic sensitivity pattern of isolated Staphylococci. |
Screening for Biofilm-Related Genes
Staphylococci were further analyzed for the presence of different virulence-related genes. Most isolates carried ica-dependent genes of biofilm formation, where 40.6% (n=26/64) of Staphylococci had icaA, 28.1% (n=18/64) had icaB and 84.3% (n=54/64) were positive for icaD. All isolates were negative for icaC. Concerning ica-independent genes, spa and bap were detected in about 70%, aap in 65.6%, altE in 20.3%, and fnbA in 25%.
The quorum-sensing genes agrI and agrII were detected in 15.6% (n=10/64) and 18.7% (n=12/64) of the Staphylococci, respectively. Both agrIII and agrIV were detected in 14% (n=9/64). All isolates infected with S. aureus were positive for spa.
Correlation Between Strength of Biofilm Formation and Number of Bioflim related Genes
Biofilm formation was assessed by microtiter plate which showed that all Staphylococci were biofilm producers. The majority (59.3%, n=38/64) were strong biofilm producers, 32.8% (n=21/64) were moderate and 7% (n=5/64) were weak biofilm producers. The number of biofilm genes was significantly associated with higher MTP values (p value=0.011), as shown in Table 3.
 |
Table 3 Number of Positive Genes According to Strength of Biofilm |
Distribution of Biofilm-Related Genes Among Isolated Staphylococci
Staphylococci isolates tend to form biofilms in different ways and this can be illustrated by the fact that most samples were positive for more than one gene; 93.7% (n=60/64) were positive to multiple biofilm genes and 6.2% (n=4/64) were positive to one gene. Spa, icaD, bap and aap were the most detected genes. Interestingly, the number of biofilm-related genes played an important role in biofilm formation; isolates that were positive to 7 and 9 genes were strong biofilm formers (Figure 4).
 |
Figure 4 Relation between number of biofilm-associated genes and strength of biofilm. |
Strength of Biofilm Formation is Correlated with Severity of Infection and Type of Ulcer
Biofilm formation is highly prevalent among different types of ulcers. Also, it exacerbates the systemic infection associated with diabetic foot. All severely infected wounds were either strong or intermediate biofilm formers. Weak biofilm formers were only in moderate infections. About 50% (n=19/38) of strong biofilm formers were isolated from mildly infected wounds, 19% from moderate infections, 31% from severely infected ulcers. In addition, 41% of intermediate biofilm formers were isolated from mildly infected ulcers, 29% from wounds clinically evaluted as non-infected, 4% from moderate infections and 26% from severely infected wounds.
Correlation Between Strength of Biofilm Formation and Resistance to Antibiotics
As predicted, there was a direct relation between biofilm formation and drug resistance: 78.9% (n=30/38) of strong biofilm forming isolates were MDR and all XDR were strong biofilm formers.
Results of Spa Typing
Sequencing of spa gene in S. aureus isolates showed that our isolates represent a collection of 17 different spa types like t034, t023, t068, t016, t033, t031, t057, t021, t012, t192, t072, t017, t020, t223, t133, t058 and t162. Each of t034, t016, t057, t012 and t021 were detected in 8.3% (n=2/24) while t033 was in 12.5% (n=3/24) of spa type Staphylococci. As shown in Table 4, spa types that were strong biofilm formers showed positive results for agr genes, mostly for agrII, and were resistant virulent isolates. There was no predominant type, probably because samples were from outpatient clinics.
 |
Table 4 Summary of Spa Genotyping, Resistance Profiles and Biofilm Production |
Discussion
Diabetic foot ulcers are an important cause of increased mortality and morbidity, particularly when coupled with pathogenic microorganisms such as S. aureus due to a high load of different virulence factors especially biofilm formation. One of the main problems of bacterial biofilms is the associated resistance to a wide variety of antibiotics.39
In this investigational study, a strong relationship between uncontrolled HA1c, severity of infection and strength of biofilm formation was observed. About 14.7% of strong biofilm forming strains belonged to patients with uncontrolled diabetes as their HA1C was 12–19% and had severely infected diabetic foot. Of strong biofilm isolates, 55% were mildly infected diabetic foot and A1C was 6.5–15. Callahan et al40 highlighted the role of uncontrolled HA1c as a risk factor for increasing risk of infected diabetic foot.
Regarding type of ulcer, we found that neuropathic ulcers accounted for 48%, ischemic ulcers for 32% and neuroischemic (mixed) ulcers for 20%. These percentages were consistent with studies that were done in the MENA region,41 which showed that neuropathic ulcers accounted for 36% and ischemic ulcers for 12%. Another study, by Petropoulos et al,42 showed that Egypt has the second highest percentage of neuropathic ulcers (61.3%) in the MENA region.
Approximately 7% of diabetic patients had heel ulcers. Similar to our findings, a larger cohort of patients with foot ulcers found that 10% of all foot ulcers were located in the heel.43 In clinical practice, heel ulcers are usually regarded as poorly healing ulcers mainly due to reduced blood supply, and researchers have found that these ulcers heal more slowly than those on the metatarsophalangeal joint or the toes.44
Our Staphylococcus isolates were mostly sensitive to linezolid (80%) and this was similar to a study performed by Abubakar et al.45 Limited use of rifampicin, used mostly in cases of tuberculosis, and clindamycin in treatment of cases of diabetic foot infections in Egypt is a factor explaining their relative sensitivity pattern.
In our study we have 15 (23.4%) S. haemolyticus isolates either alone or mixed with other Staphylococcus species like S. aureus and S. epidermidis. Although, S. haemolyticus is a less common species of Staphylococcus, its prevalence was relatively high. Similar findings were reported by other studies.46,47 However, other studies, such as that carried out by Mottola et al,48 showed a lower percentage (1.8%) of S. haemolyticus. Wang et al49 reported that 2.5% of S. haemolyticus were isolated from diabetic foot ulcers. Ge et al50 isolated S. haemolyticus at a percentage of 9% from diabetic foot ulcers. This difference in percentages may be due to the variation in sample types, number of studied population and geographical locations, but it pointed to the importance of atypical microorganisms that contributed to infection of the diabetic foot ulcers. Another important factor for limited isolation of S. haemolyticus is the misidentification of mannitol salt agar because both Staphylococcus aureus and S. haemolyticus cause fermentation of mannitol and produce yellow colonies.50,51
Staphylococcal isolates were mainly strong and moderate biofilm formers (59.3% strong, 32.8% moderate and 7% weak). Our data were in line with Tiwari et al52 whereby isolated Staphylococci were from chronic wounds and formed mainly strong and moderate biofilms. Another study in Egypt, by Gad et al,53 reported that 51.4% of biofilms were strong, 37.1% were moderate, and 11.4% were weak. A lower rate of biofilm formation was demonstrated by Neopane et al54 and their biofilm strength results were 6.97% strong, 27.9% moderate and 34.88% weak. Variation may be due to different types of samples, presence of foreign bodies, different growth conditions and the use of various sugar supplementation for biofilm formation in Staphylococci.
Concerning the distriubtuion of biofilm-related genes among staphylococci, Diemond-Hernández et al33 reported icaA in 10.3% and icaD in 97.5% of S. aureus isolates and did not detect icaB or icaC. Mirzaee et al55 showed higher percentages than our study. Torlak et al56 reported high prevalence of ica genes among S. aureus where all isolates were reported to possess icaA and icaD genes. Arciola et al24 and Gad et al53 detected icaA and icaD genes in all biofilm-forming S. aureus isolates. The discrepancy between the results of different studies may be caused by the diversity of bacterial genetic characterization, source of isolation, and environmental factors.
In our study, 8 isolates (12.5%) were negative to ica (A, B, C and D) genes. However, these isolates were found to be biofilm formers phenotypically by MTP method. One of them was a weak biofilm former, one was an intermediate biofilm former and 6 were strong biofilm formers. This was in line with results of Mirzaee et al55 who reported a S. aureus isolate that is negative for all ica genes but still produced biofilm. Therefore, our study includes both ica genes and genes responsible for biofilm formation using ica-independent method. On the other hand, inability of biofilm formation in some Staphylococcal strains, despite the presence of ica genes, can be caused by insertion of a 1332-bp insertion element causing its inactivation.57 Our results were in agreement with Vindel et al58 that showed agrII is the most prevalent agr gene followed by agrI (14.8%), agrIII (3%) and agrIV (0%). agr deficiency represents an adaptive approach allowing the microorganism to avoid host immune system and to promote colonization.59 In fact, the majority of agr-deficient strains have been linked to biofilm formation.60 In line with our results, 55.8% of agr-negative strains were strong biofilm formers, 38.2% were intermediate and 5.8% were weak.
In an ica-deletion mutant of S. aureus strain protein A (spa) production was found essential for biofilm formation.61 Most strong biofilm formers were positive to 4–8 genes. However, 2 (5.2%) of strong biofilm former isolates were positive only to spa gene and were found to be strong biofilm formers. Moreover, 5 (13.1%) of our samples were positive for only 2 genes and were found to be strong biofilm formers. in these samles spa wasone of these two positive genes, and the other gene was agrI, bap or icaD. Our results showed that spa is predominant in strong and moderate biofilm forming isolates. Exogenous spa was able to restore biofilm formation in spa mutants, indicating that spa does not need to be covalently attached to the cell wall.61 Spa-negative strains were found to form biofilm but were also negative for other tested genes in our study, which could be explained by the role of other biofilm genes not tested in our study. Some studies have linked the absence of spa protein to point mutations.62 Hospital-associated (HA) MRSA are typically multidrug resistant; in our study they mainly belong to agr types I or II and carry mecA gene in many types of spa like t034, t023, t068, t016, t033, t031, t057, t021, t012, t192, t072, t017, t020, t223, t133, t058 and t162. This was similarly reported by Monecke et al63 and Ćirković et al.64
In our study, t034 spa type was detected. This genotype carried the mecA gene, was vancomycin sensitive, and produced strong biofilms, as similarly reported by Köck et al65 t034 is considered to be the most widely distributed spa type in Oman.66 Spa types t012 and t021 have been previously isolated from wound infections13 and were HA MRSAe. Our study declared that isolates which belonged to spa t016 were agrII type and were MRSA, and strong biofilm formers, while the study of Goudarzi et al67 reported that t016 isolates belonged to agrI. Spa t031 has been isolated in MENA region from nasal swabs and reported by Verhoeven et al.68 In our study, it was a strong biofilm former and belonged to agrII group. Von Eiff et al69 detected MRSA strains of spa types t033 and t057 circulating in Germany and other parts of Europe on the basis of several well-characterized S. aureus strain collections. We have isolated both types from our S. aureus samples. t033 is considered the most widely distributed type in the Netherlands.70 Spa type t068 was isolated in the Netherlands71 from bloodstream samples and its presence was associated with bacteraemia and it had TSST.
Other spa types such as t017, t020, t023, t058, t072, t133, t162, t192 and t223 were intermediate biofilm formers and were negative for agr genes. t017 was also detected in the UK.72 t020 was found in Scotland using high-resolution melting analysis.72 t023 was first discovered in Germany73 and t223 is the most predominant type in Turkey.70
In conclusion, infection of diabetic foot ulcers is a serious medical situation. The majority of DFUs is polymicrobial. S. aureus and other coagulase-negative staphylococci are major contributors to infected DFUs. MDR and biofilm formation are marked among isolates. The severity of DFU is directly related to the number of biofilm genes. Therefore, cleaning the wounds, monitoring for signs of infection, and receiving proper antimicrobial therapy are extremely important measures to avoid further complications from MDR and biofilm-forming bacteria.
Abbreviations
DFU, diabetic foot ulcers; S. aureus, Staphylococcus aureus; MRSA, methicillin resistant Staphylococcus aureus.
Funding
This work was partially funded by the grant office, Faculty of Medicine, Assiut university (fund no 2018-01-04-003).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Soldevila-Boixader L, Fernández AP, Laguna JM, et al. Local antibiotics in the treatment of diabetic foot infections: a narrative review. Antibiotics. 2023;12(1):124. doi:10.3390/antibiotics12010124
2. Cho N, Shaw JE, Karuranga S, et al. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–281. doi:10.1016/j.diabres.2018.02.023
3. Armstrong DG, Boulton AJ, Bus SA, Ingelfinger JR. Diabetic foot ulcers and their recurrence. N Engl J Med. 2017;376(24):2367–2375. doi:10.1056/NEJMra1615439
4. Moxey P, Gogalniceanu P, Hinchliffe RJ, et al. Lower extremity amputations—a review of global variability in incidence. Diabetic Med. 2011;28(10):1144–1153. doi:10.1111/j.1464-5491.2011.03279.x
5. Bakker K, Apelqvist J, Lipsky BA, et al. The 2015 IWGDF guidance documents on prevention and management of foot problems in diabetes: development of an evidence‐based global consensus. Diabetes Metab Res Rev. 2016;32:2–6. doi:10.1002/dmrr.2694
6. McDermott K, Fang M, Boulton AJM, et al. Etiology, epidemiology, and disparities in the burden of diabetic foot ulcers. Diabetes Care. 2023;46(1):209–221. doi:10.2337/dci22-0043
7. Richard J-L, Lavigne J-P, Got I, et al. Management of patients hospitalized for diabetic foot infection: results of the French OPIDIA study. Diabetes Metab. 2011;37(3):208–215. doi:10.1016/j.diabet.2010.10.003
8. Chang M, Nguyen TT. Strategy for treatment of infected diabetic foot ulcers. Acc Chem Res. 2021;54(5):1080–1093. doi:10.1021/acs.accounts.0c00864
9. Commons RJ, Robinson CH, Gawler D, et al. High burden of diabetic foot infections in the top end of Australia: an emerging health crisis (DEFINE study). Diabetes Res Clin Pract. 2015;110(2):147–157. doi:10.1016/j.diabres.2015.09.016
10. Markanday A. Diagnosing diabetic foot osteomyelitis: narrative review and a suggested 2-step score-based diagnostic pathway for clinicians. In: Open Forum Infectious Diseases. Oxford University Press; 2014.
11. Jalilian M, Ahmadi Sarbarzeh P, Oubari S. Factors related to severity of diabetic foot ulcer: a systematic review. Diabetes Metab Syndr Obes. 2020;13:1835–1842. doi:10.2147/DMSO.S256243
12. Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi:10.1128/CMR.15.2.167-193.2002
13. Fitzpatrick F, Humphreys H, O’gara J. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infect. 2005;11(12):967–973. doi:10.1111/j.1469-0691.2005.01274.x
14. Cramton SE, Gerke C, Schnell NF, et al. The intercellular adhesion (ica) locus is present in staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67(10):5427–5433. doi:10.1128/IAI.67.10.5427-5433.1999
15. Cramton SE, Ulrich M, Gotz F, et al. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69(6):4079–4085. doi:10.1128/IAI.69.6.4079-4085.2001
16. Dunyach-Remy C, Ngba Essebe C, Sotto A, et al. Staphylococcus aureus toxins and diabetic foot ulcers: role in pathogenesis and interest in diagnosis. Toxins. 2016;8(7):209. doi:10.3390/toxins8070209
17. Lasa I, Penadés JR. Bap: a family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157(2):99–107. doi:10.1016/j.resmic.2005.11.003
18. Hennig S, Wai SN, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297(2):117–122. doi:10.1016/j.ijmm.2006.12.001
19. Rohde H, Burandt EC, Siemssen N, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic Hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720. doi:10.1016/j.biomaterials.2006.11.046
20. Chan WC, Coyle BJ, Williams P. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem. 2004;47(19):4633–4641. doi:10.1021/jm0400754
21. Boles BR, Horswill AR, Cossart P. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. doi:10.1371/journal.ppat.1000052
22. Mendes J, Marques-Costa A, Vilela C, et al. Clinical and bacteriological survey of diabetic foot infections in Lisbon. Diabetes Res Clin Pract. 2012;95(1):153–161. doi:10.1016/j.diabres.2011.10.001
23. Pereira EM, Schuenck RP, Malvar KL, et al. Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus haemolyticus: methicillin-resistant isolates are detected directly in blood cultures by multiplex PCR. Microbiol Res. 2010;165(3):243–249. doi:10.1016/j.micres.2009.03.003
24. Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaDGenes and slime production in a collection of Staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39(6):2151–2156. doi:10.1128/JCM.39.6.2151-2156.2001
25. Gilot P, Lina G, Cochard T, et al. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40(11):4060–4067. doi:10.1128/JCM.40.11.4060-4067.2002
26. Hsueh P-R, Ko W-C, Wu -J-J, et al. Consensus statement on the adherence to Clinical and Laboratory Standards Institute (CLSI) Antimicrobial Susceptibility Testing Guidelines (CLSI-2010 and CLSI-2010-update) for Enterobacteriaceae in clinical microbiology laboratories in Taiwan. J Microbiol Immunol Infect. 2010;43(5):452–455. doi:10.1016/S1684-1182(10)60070-9
27. Institute, C.L.S. Performance standards for antimicrobial susceptibility testing: twenty‐third informational supplement M100‐S23; 2013.
28. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
29. Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37(11):3556–3563. doi:10.1128/JCM.37.11.3556-3563.1999
30. Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi:10.1128/JCM.41.12.5442-5448.2003
31. Faroq A-T, Brunel A-S, Bouzinbi N, et al. DNAGear-A free software for spa type identification in Staphylococcus aureus. BMC Res Notes. 2012;5(1):642. doi:10.1186/1756-0500-5-642
32. Cafiso V, Bertuccio T, Santagati M, et al. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004;10(12):1081–1088. doi:10.1111/j.1469-0691.2004.01024.x
33. Diemond-Hernández B, Solórzano-Santos F, Leaños-Miranda B, et al. Production of icaADBC-encoded polysaccharide intercellular adhesin and therapeutic failure in pediatric patients with staphylococcal device-related infections. BMC Infect Dis. 2010;10(1):68. doi:10.1186/1471-2334-10-68
34. Salgueiro VC, Iorio NLP, Ferreira MC, et al. Methicillin resistance and virulence genes in invasive and nasal Staphylococcus epidermidis isolates from neonates. BMC Microbiol. 2017;17(1):15. doi:10.1186/s12866-017-0930-9
35. Nicoleta M. Phenotypic and genotypic investigation of resistance and virulence features of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients. Rom Biotechnol Lett. 2016;21(3):11591.
36. Potter A, Ceotto H, Giambiagi-deMarval M, et al. The gene bap, involved in biofilm production, is present in Staphylococcus spp. strains from nosocomial infections. J Microbiol. 2009;47(3):319–326. doi:10.1007/s12275-009-0008-y
37. Larsen A, Stegger M, Sørum M. spa typing directly from a mecA, spa and pvl multiplex PCR assay—a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin Microbiol Infect. 2008;14(6):611–614. doi:10.1111/j.1469-0691.2008.01995.x
38. Lina G, Boutite F, Tristan A, et al. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol. 2003;69(1):18–23. doi:10.1128/AEM.69.1.18-23.2003
39. Khan F, Shukla I, Rizvi M, et al. Detection of biofilm formation in Staphylococcus aureus. Does it have a role in treatment of MRSA infections. Trends Med Res. 2011;6(2):116–123. doi:10.3923/tmr.2011.116.123
40. Callahan D, Keeley J, Alipour H, et al. Predictors of severity in diabetic foot infections. Ann Vasc Surg. 2016;33:103–108. doi:10.1016/j.avsg.2016.01.003
41. Majeed A, El-Sayed AA, Khoja T, et al. Diabetes in the Middle-East and North Africa: an update. Diabetes Res Clin Pract. 2014;103(2):218–222. doi:10.1016/j.diabres.2013.11.008
42. Petropoulos IN, Javed S, Azmi S, et al. Diabetic neuropathy and painful diabetic neuropathy in the Middle East and North Africa (MENA) region: much work needs to be done. J Taibah Univ Med Sci. 2016;11(4):284–294. doi:10.1016/j.jtumed.2016.06.002
43. Örneholm H, Apelqvist J, Larsson J, et al. Heel ulcers do heal in patients with diabetes. Int Wound J. 2017;14(4):629–635. doi:10.1111/iwj.12654
44. Pickwell KM, Siersma VD, Kars M, et al. Diabetic foot disease: impact of ulcer location on ulcer healing. Diabetes Metab Res Rev. 2013;29(5):377–383. doi:10.1002/dmrr.2400
45. Abubakar U, Sulaiman SA. Prevalence, trend and antimicrobial susceptibility of Methicillin Resistant Staphylococcus aureus in Nigeria: a systematic review. J Infect Public Health. 2018;11(6):763–770. doi:10.1016/j.jiph.2018.05.013
46. Ghatak S, Muthukumaran RB, Nachimuthu SK. A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. JBT. 2013;24(4):224. doi:10.7171/jbt.13-2404-001
47. Demetriou M, Papanas N, Panagopoulos P, et al. Atypical microbial isolates from infected diabetic foot ulcers: a case series from Greece. RDS. 2017;14(2–3):258. doi:10.1900/RDS.2017.14.258
48. Mottola C, Semedo-Lemsaddek T, Mendes JJ, et al. Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J Biomed Sci. 2016;23(1):33. doi:10.1186/s12929-016-0250-7
49. Wang S-H, Sun Z-L, Guo Y-J, et al. Meticillin-resistant Staphylococcus aureus isolated from foot ulcers in diabetic patients in a Chinese care hospital: risk factors for infection and prevalence. J Med Microbiol. 2010;59(10):1219–1224. doi:10.1099/jmm.0.020537-0
50. Ge Y, MacDonald DL, Holroyd KJ, et al. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob Agents Chemother. 1999;43(4):782–788. doi:10.1128/AAC.43.4.782
51. Shrestha LB, Bhattarai NR, Khanal B. Comparative evaluation of methods for the detection of biofilm formation in coagulase-negative staphylococci and correlation with antibiogram. Infect Drug Resist. 2018;11:607. doi:10.2147/IDR.S159764
52. Tiwari S, Rajak S, Mondal DP, et al. Sodium hypochlorite is more effective than 70% ethanol against biofilms of clinical isolates of Staphylococcus aureus. Am J Infect Control. 2018;46(6):e37–e42. doi:10.1016/j.ajic.2017.12.015
53. Gad GFM, El-Feky MA, El-Rehewy MS, et al. Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. J Infect Dev Ctries. 2009;3(5):342–351. doi:10.3855/jidc.241
54. Neopane P, Nepal HP, Shrestha R, et al. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance</em>. Int J Gen Med. 2018;11:25. doi:10.2147/IJGM.S153268
55. Mirzaee M. Detection of intracellular adhesion (ica) gene and biofilm formation Staphylococcus aureus isolates from clinical blood cultures. J Med Bacteriol. 2015;3(1–2):1–7.
56. Torlak E, Korkut E, Uncu AT, et al. Biofilm formation by Staphylococcus aureus isolates from a dental clinic in Konya, Turkey. J Infect Public Health. 2017;10(6):809–813. doi:10.1016/j.jiph.2017.01.004
57. DuMont AL. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol. 2011;79(3):814–825. doi:10.1111/j.1365-2958.2010.07490.x
58. Vindel A. Methicillin-resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. J Clin Microbiol. 2009;47(6):1620–1627. doi:10.1128/JCM.01579-08
59. Becker K, Friedrich AW, Lubritz G, et al. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol. 2003;41(4):1434–1439. doi:10.1128/JCM.41.4.1434-1439.2003
60. Nhan T-X, Leclercq R, Cattoir V. Prevalence of toxin genes in consecutive clinical isolates of Staphylococcus aureus and clinical impact. Eur J Clin Microbiol Infect Dis. 2011;30(6):719–725. doi:10.1007/s10096-010-1143-4
61. Costerton JW. Microbial biofilms. Annu Rev Microbiol. 1995;49(1):711–745. doi:10.1146/annurev.mi.49.100195.003431
62. Kristian SA, Golda T, Ferracin F, et al. The ability of biofilm formation does not influence virulence of Staphylococcus aureus and host response in a mouse tissue cage infection model. Microb Pathog. 2004;36(5):237–245. doi:10.1016/j.micpath.2003.12.004
63. Monecke S, Coombs G, Shore AC, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(4):e17936. doi:10.1371/journal.pone.0017936
64. Ćirković I, Radenkovic D, Svabic Vlahovic M, et al. National surveillance reveals findings of Panton–Valentine leukocidin positive meticillin-resistant Staphylococcus aureus in Serbia. J Med Microbiol. 2013;62(2):342–344. doi:10.1099/jmm.0.051219-0
65. Köck R. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One. 2013;8(2):e55040. doi:10.1371/journal.pone.0055040
66. Asadollahi P, Farahani NN, Mirzaii M, et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and-susceptible Staphylococcus aureus around the world: a review. Front Microbiol. 2018;9:163. doi:10.3389/fmicb.2018.00163
67. Goudarzi M, Navidinia M, Beiranvand E, et al. Phenotypic and molecular characterization of methicillin-resistant Staphylococcus aureus clones carrying the panton-valentine leukocidin genes disseminating in iranian hospitals. Microb Drug Resist. 2018;24(10):1543–1551. doi:10.1089/mdr.2018.0033
68. Verhoeven P, Gautret P, Haddar CH, et al. Molecular dynamics of Staphylococcus aureus nasal carriage in Hajj pilgrims. Clin Microbiol Infect. 2015;21(7):650. e5–650. e8. doi:10.1016/j.cmi.2015.03.020
69. von Eiff C, Maas D, Sander G, et al. Microbiological evaluation of a new growth-based approach for rapid detection of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2008;61(6):1277–1280. doi:10.1093/jac/dkn122
70. Kluytmans J, Van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–520. doi:10.1128/CMR.10.3.505
71. Ulrich M, Bastian M, Cramton SE, et al. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol. 2007;65(5):1276–1287. doi:10.1111/j.1365-2958.2007.05863.x
72. Kiem S, Oh WS, Peck KR, et al. Phase variation of biofilm formation in Staphylococcus aureus by IS 256 insertion and its impact on the capacity adhering to polyurethane surface. J Korean Med Sci. 2004;19(6):779–782. doi:10.3346/jkms.2004.19.6.779
73. Jefferson KK, Pier DB, Goldmann DA, et al. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186(8):2449–2456. doi:10.1128/JB.186.8.2449-2456.2004
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
