Back to Journals » Neuropsychiatric Disease and Treatment » Volume 17
Circular RNA circPHC3 Promotes Cell Death and Apoptosis in Human BMECs After Oxygen Glucose Deprivation via miR-455-5p/TRAF3 Axis in vitro
Authors Xu X, Wu Z, Qiu H, Wu J
Received 25 October 2020
Accepted for publication 29 December 2020
Published 22 January 2021 Volume 2021:17 Pages 147—156
DOI https://doi.org/10.2147/NDT.S288669
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jun Chen
Xiaonan Xu, Zimu Wu, Hongyan Qiu, Jun Wu
Department of Neurology, Peking University Shenzhen Hospital, Shenzhen 518000, Guangdong Province, People’s Republic of China
Correspondence: Jun Wu Email [email protected]
Objective: Brain microvascular endothelial cells (BMECs) are involved in brain vascular dysfunction in ischemic stroke. Abnormal expression of circular RNAs regulate physiological and pathophysiological processes in the central nervous system. The aim of the present study was to investigate profile circRNAs in human BMECs after oxygen glucose deprivation (OGD), which was an in vitro model of ischemic stroke, and find promising biomarkers in ischemic stroke.
Methods: RNA sequencing (RNA-seq) technology was conducted to analyze the differential expression of circRNAs between BMECs after OGD and non-OGD treated BMECs. RT-qPCR, cell proliferation, cell apoptosis and dual-luciferase assay, and so on, were used to investigate the functions and molecular mechanisms of hsa_circ_0001360 (named circPHC3 in this study) in ischemic stroke.
Results: CircPHC3 was highly expressed in human BMECs after OGD. Knockdown of circPHC3 inhibited cell death and apoptosis in human BMECs treated with OGD. Mechanistically, circPHC3 acted as miR-455-5p sponge to activate TRAF3 to promote cell death and apoptosis in human BMECs after OGD.
Conclusion: In short, circPHC3 promotes cell death and apoptosis in ischemic stroke in vitro model, which might be a novel molecular target for acute cerebrovascular protection.
Keywords: ischemic stroke, circPHC3, TRAF3, miR-455-5p
Introduction
Ischemic stroke and cerebral ischemia reperfusion injury can result in serious dysfunction of the brain, which leads to a high rate of morbidity, disability and mortality.1 Various studies show that ischemia leads to a series of pathological events and activates signaling cascades to promote cell death.2 Besides, a combination of many genetic and environmental factors causes the complex genetic disorder, ischemic stroke.3 The detailed pathogenesis mechanisms of ischemic stroke are still not completely understood. Currently, intravenous administration of recombinant tissue plasminogen activator (tPA) is the only clinically effective therapy for acute ischemic stroke, which is useful only within a narrow window of time.4 Thus, further revealling the molecular mechanisms of ischemic stroke, and developing novel and effective therapies for ischemic stroke are really necessary.
Circular RNA (circRNA) is regarded as a novel class of non-coding RNAs, which has closed covalent loops with neither a polyadenylate tail nor 5ʹ to 3ʹ polarity.5 CircRNA is highly stable and conservative and is abundant in mammalian cells.6 Various studies have shown that circRNAs act as miRNA sponges to regulate gene expression in different diseases including neurological disorders.7–9 CircRNAs play critical roles in multiple diseases including neural development and plasticity, Alzheimer’s disease, heart senescence, hypertrophy and failure, and cell growth through interaction analysis between circRNA and disease-associated miRNAs.8,10,11 However, the molecular mechanisms of circRNA in ischemic stroke still remains un clear.
In this study, RNA-seq technology was used to investigate the transcriptome profiling circRNAs in human brain microvascular endothelial cells (BMECs) after exposure to oxygen glucose deprivation (OGD)12 that is an in vitro mimic of stroke conditions. It is the first time to identify various OGD-responsive circRNAs in BMECs. We mainly focus on a circRNA derived from PHC3, hsa_circ_0001360, termed circPHC3, which was screened from RNA-seq. Till now, there are few studies of hsa_circ_0001360. Wang et al demonstrated that hsa_circ_0001360 was abundant and differentially expressed in maternal and neonatal umbilical cord plasma from SGA (small for gestational age).13 In our study, the expression of circPHC3 was increased significantly in BMECs treated with OGD. Besides, circPHC3 promoted cell death and apoptosis in human BMECs after OGD via acting as a ceRNA for miR-455-5p, and activated TRAF3 expression in ischemic stroke. In conclusion, circPHC3 may be a novel potential target for early diagnosis or treatment of ischemic stroke.
Methods
Cell Culture
Human brain microvascular endothelial cells (BMECs) were purchased from Cell Systems Corp. (USA). Cells were cultured according to a previous study.14 Cells were maintained at 37°C in incubator.
In vitro OGD Model
Human BMECs were treated with OGD through cultivating in glucose-free culture medium and an anaerobic chamber (Forma Scientific, Marietta, OH) containing 95% N2 and 5% CO2 for 16h. Control human BMECs were not exposed to OGD.
RNA Sequencing and Bioinformatics Analysis
Three independent repeated human BMECs without OGD and three independent repeated human BMECs after OGD were chosen for RNA sequencing. TRIzol reagent (Invitrogen, Grand Island, NY, USA) was used to extract RNA from human BMECs according to the manufacturer’s protocol. Three U/μg RNase R was utilized to eliminate linear RNA at 37 °C for 25 min. The HiSeq 2000 (Illumina) was employed to sequenced RNA. The reads were aligned to the latest UCSC transcript set utilizing Bowtie2 version 2.1.0.15 RSEM version 1.2.15 was used to estimate gene expression.16 The reads were mapped to genome using STAR.17 The circRNA expression was measured by DCC.18
Reverse Transcription Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
TRIzol reagent was used to isolate total RNA from human BMECs and 3 U/μg RNase R was used to remove linear RNA for circRNA detection. A PrimeScript RT Reagent Kit with gDNA Eraser (Takara., Dalian, China) was utilized to synthesize the first strand of cDNA for detection of circPHC3 and TRAF3. A commercial miRNA reverse transcription PCR kit (RiboBio, Guangzhou, China) was used to synthesize cDNA for miR-455-5p detection. The SYBR Premix Ex TaqTM kit (Takara, Dalian, China) was used for RT-qPCR analysis on the Roche LightCycler 480 Real-Time PCR System. Levels of GAPDH or U6 were used as an endogenous control to normalize the differences of circRNA and miRNA. The primers were: circPHC3-forward: 5ʹ-TCGTCATCGTCATCTTCCTG-3ʹ, circPHC3-reverse: 5ʹ- GGGTAATACTGCCGCTGGTA-3ʹ; TRAF3-forward: 5ʹ- CAGACTAACC CGCCGCTAAAG-3ʹ, TRAF3-reverse: 5ʹ-GATGCTCTCTTGACACGCTGT-3ʹ; GAPDH-forward: 5ʹ-CGCTCTCTGCTCCTCCTGTTC-3ʹ, GAPDH-reverse: 5ʹ-ATCCG TTGACTCCGACCTTCAC-3ʹ. Primers of miR-455-5p was purchased from GeneCopoeia Co. Ltd., Guangzhou, China. The comparative 2−ΔΔCT method was used to analyze the expression levels.
Subcellular Fractionation Location
NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Scientific, Waltham, MA, USA) was utilized to get cytoplasmic and nuclear fraction from human BMECs. RNAs separated from each of the fractions were used for RT–qPCR analysis to measure the expression levels of cytoplasmic control transcript (GAPDH), nuclear control transcript (U6) and circPHC3.
Cell Transfection
Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) was used to transfect siRNAs, miRNA mimics, miRNA inhibitors or plasmids into BMECs according to the manufacturer’s protocols.
Vector Construction
The siRNA targeting circPHC3 was synthesized by GenePharma (Shanghai, China). Sequence of circPHC3 was synthesized by Sangong (Shanghai, China) and cloned into pCD-ciR vector (Geenseed Biotech Co, Guangzhou, China). The wide-type circPHC3/TRAF3 3ʹ untranslated region (UTR) sequence or circPHC3/TRAF3 3ʹ UTR sequence with mutation of miR-455-5p binding site was inserted into psiCHECK2 vector (Promega, Madison, WI). MiR-455-5p mimics or inhibitors were purchased from RiboBio (Guangzhou, China).
Dual-Luciferase Reporter Assay
Human BMECs were transfected with corresponding plasmids and miRNA mimics or inhibitors. After 48 hour transfection, dual-luciferase reporter assay kit (Promega, Madison, WI, USA) was used to detect luciferase and renilla signals after 48 hour transfection. The ratio (renilla luciferase activity/luciferase activity) was analyzed.
RNA Immunoprecipitation (RIP) Assay
RIP assays were carried out in human BMECs after utilizing the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA). Cells (1×107) were lysed in complete RNA lysis buffer, then cell lysates were incubated with anti-Argonaute2 (AGO2) antibody (Millipore, Billerica, Ma, USA), negative control mouse IgG (Millipore, Billerica, Ma, USA) or total RNAs (input controls) in accordance with the manufacturer’s instructions. Co-precipitated RNA was measured by RT-qPCR as previously described.
Cell Viability Assay
Cell Counting Kit-8 (Beyotime Institute of Biotechnology, Shanghai, China) was utilized to detect cell viability. 2500 cells were seeded into a 96-well plate and 10 μL of CCK-8 was added to each well. Cells were cultivated for 120 minutes. Absorbance was detected at a wavelength of 450 nm employing an ELISA microplate reader (Bio-Rad, Hercules, CA, USA) was used to measure the absorbance at the 450 nm wavelength.
Cell Apoptosis Assay
The FITC Annexin V Apoptosis Detection Kit (TransGen, Peking, China) was utilized to double stain cells with FITC-Annexin V and PI according to the manufacturer’s instructions. Right lower quadrant represents the percentage of early apoptosis cells. Besides, in order to detect the effects of circPHC3 and TRAF3 on cell apoptosis, we employed the caspase-3 ELISA assay that was widely used to measure cell apoptosis (Husabio, Wuhan, China) according to the manufacturer’s protocol.
Western Blot Analysis
The protein concentration was measured by the bicinchoninic acid (BCA) quantification assay (Pierce Biotechnology, Rockford, IL, USA). The membranes were incubated overnight with specific primary antibodies against TRAF3 (1:1000; Abcam, USA) and GAPDH (1:1000; Abcam, USA) after blocked with 5% nonfat dry milk. Horseradish peroxidase conjugated secondary antibody (Amersham, Arlington Heights, IL) was used to incubate the blot for one hour at room temperature on a rocking platform. Finally, super signal chemiluminescence reagents (Thermo Fisher Scientific, Inc., Massachusetts, USA) was used to detect signal intensities.
Statistical Analysis
All the above experiments were performed at least three times in this study. SPSS software version 21.0 (SPSS Inc., Chicago, IL, USA) was used to analyze all statistical analysis. All data are presented as the mean ± standard error (SD). Student’s t-test, was used to analyze the group difference. A P-value < 0.05 (two-sided) was chosen for statistical significance.
Results
RNA-Seq Identification of Transcriptome Profiling circRNAs in Human BMECs After OGD
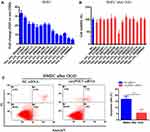
We used an in vitro mimic of stroke conditions, human BMECs after OGD, to determine the transcriptomic profiling circRNAs using RNA-seq. RNA sequencing reads were generated from non-OGD cells and OGD-treated cells. 3978 circRNAs were detected between non-OGD and OGD-treated cells (supplementary material 1). The log2 fold change (> 2) and P < 0.05 of up-regulated or down-regulated circRNAs were considered expressed differentially. 74 circRNAs from OGD Vs Non-OGD, including 14 up-regulated circRNAs and 60 down-regulated circRNAs (Figure 1A), were conformed to this criterion, and heatmap of these 74 differential circRNA expression was shown in Figure 1B.
circPHC3 Promoted Cell Death and Apoptosis in Human BMECs After OGD
Next, we want to know whether these differential circRNAs between non-OGD and OGD-treated BMECs play critical roles in ischemic stroke. The parent genes of 74 circRNAs were involved in different biological process, cellular component and molecular function according to GO analysis (Figure-S1A). Fourteen up-regulated circRNAs were chosen for further study (Table 1). Compared with non-OGD group, the expression levels of 14 circRNAs were highly increased in human BMECs after OGD group through RT-qPCR verification (Figure 2A). The siRNAs were designed for 14 individual circRNAs and the result of CCK-8 assay indicated that hsa_circ_0000566, hsa_circ_0001360 and hsa_circ_0101874 inhibited cell viability significantly (Figure 2B). The most obvious inhibitory effect of hsa_circ_0001360 was presented among these three circRNAs. The hsa_circ_0001360 was picked for in-depth research. The circBase database shows that hsa_circ_0001360, 258 nucleotides in length, was generated from PHC3 gene (Figure-S1B). Thus, hsa_circ_0001360 was named circPHC3 in this study. Flow cytometry assay was used to measure the effect of circPHC3 on the apoptosis of BMECs after OGD. The right lower quadrant of flow chart represents early apoptotic cells. As shown in Figure 2C, cell apoptosis was suppressed significantly after circPHC3 inhibition in BMECs after OGD (P = 0.007406). However, circPHC3 had no effects on angiogenesis (data was not shown). In short, cell death and apoptosis were facilitated markedly by circPHC3 overexpression in human BMECs after OGD.
 |
Table 1 14 Differentially Overexpressed circRNAs in Human BMECs After OGD |
circPHC3 Bound to miR-455-5p in Human BMECs After OGD
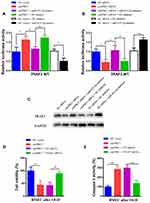
The mechanism of circPHC3 in human BMECs after exposure to OGD was investigated in our further study. CircPHC3 was mainly localized in the cytoplasm of BMECs using subcellular fractionation location assay (Figure 3A). CircRNAs that mainly located in cytoplasm may function as sponges to bind to miRNAs.19 Bioinformatics analysis revealed that circPHC3 may bind to seven miRNAs (Figure 3B). A previous study demonstrates that miR-455-5p suppresses neuronal cell death in cerebral stroke20 and play a protective role in cerebral stroke. Thus, the relationship between circPHC3 and miR-455-5p was verified. Compared with human BMECs with non-OGD, the expression level of miR-455-5p was inhibited significantly in human BMECs after OGD (Figure 3C). It is well known that miRNAs degrade mRNA and suppress translation of their targets in an AGO2-dependent manner.21 Anti-AGO2 precipitation (RIP) was carried out and showed that compared with the control, the expression level of circPHC3 bound to anti-AGO2 antibodies was increased significantly in human BMECs after OGD transfected with miR-455-5p mimics (Figure 3D). Besides, the wide type circPHC3 (WT) with binding sites of miR-455-5p or a mutated circPHC3 (Mut) sequence was inserted into 3ʹUTR of renilla luciferase in the psiCHECK-2 vector (Figure 3E). As shown in Figure 3F, the luciferase activities of WT reporter were decreased markedly after overexpression of miR-455-5p. However, there was no difference in the mutated luciferase reporter. These results suggest that circPHC3 could bind to miR-455-5p in human BMECs.
TRAF3 Was the Functional Target of circPHC3 in Human BMECs After OGD
Yao et al reported that TRAF3, a member of the TRAF families, is an important modulator in neuronal death, and miR-455-5p inhibits neuronal cell death by targeting TRAF3 in cerebral ischemic stroke.20 Thus, we would like to detect whether circPHC3 functioned in human BMECs through TRAF3 regulation. We transfected the luciferase reporter of WT TRAF3 into human BMECs after OGD. As shown in Figure 4A, the luciferase activity was promoted significantly in circPHC3 vector compared to the NC vector group (P < 0.05). The luciferase activity was decreased markedly in circPHC3 vector and miR-455-5p group compared with circPHC3 vector and NC mimics. When cells were transfected with NC vector and miR-455-5p mimics, the luciferase activity was inhibited obviously compared with NC vector and NC mimics group. On the contrary, as shown in Figure 4B and C, compared with NC siRNA group, the luciferase activity and protein expression level of TRAF3 were restrained significantly after inhibition of circPHC3. The luciferase activity and protein expression level of TRAF3 in circPHC3 siRNA plus miR-455-5p inhibitor group were increased obviously compared with circPHC3 siRNA plus NC inhibitor group. Compared with the corresponding control, after co-transfection of NC siRNA and miR-455-5p inhibitor, the luciferase activity and protein expression level of TRAF3 were promoted notably. These results demonstrated that circPHC3 and miR-455-5p have joint influence on the expression of TRAF3. Furthermore, as presented in Figure 4D, overexpression of circPHC3 inhibited cell viability, but this effect could be counteracted significantly by TRAF3 inhibition. ELISA assay was used to detect cell apoptosis in human BMECs. High caspase-3 activity means high cell apoptosis rate.22 The result of ELISA showed that overexpression of circPHC3 caused increased caspase-3 activity significantly in human BMECs after OGD (Figure 4E, P < 0.01). However, the effect of circPHC3 on the caspase-3 activity was offset by suppression of TRAF3 (Figure 4E, P < 0.01). In a word, these results revealed that TRAF3 was the functional target of circPHC3 in human BMECs after OGD.
Discussion
Cerebral ischemia causes irreversible neuronal injury in the ischemic region through various biological mechanisms.23 Many studies have revealed that neuron death is a highlight characteristic after stroke.24 Now, thrombolytic therapy using tissue type plasminogen activator is regarded as the only globally approved treatment for ischemic stroke, but this therapy is limited by a short treatment window of time and low recanalization rates.25 Thus, refraining from ischemic-reperfusion injury is critical for the treatment of patients with stroke.
CircRNAs have been verified to involve in various diseases, including cerebrovascular diseases, nervous system diseases, cancer, etc.26 In this study, the effects of circRNAs in ischemic stroke disease were studied. Human BMECs after OGD was used as an in vitro mimic of stroke conditions and RNA-seq was performed between human BMECs without OGD and after OGD. 14 increased differentially circRNAs were chosen for study and circPHC3 was picked owing to the greatest impact on cell viability inhibition among these 14 circRNAs. In addition, cell apoptosis of BMECs after OGD were promoted significantly. CircPHC3 was mainly distributed in cell cytoplasm and suggested that it may function as a ceRNA to regulate the expression of target genes. Mechanistically, circPHC3 promoted cell death and apoptosis through sponging miR-455-5p to activate the expression of TRAF3.
OGD was the main pathophysiological cause for ischemic stroke. But only a few patients could benefit from timely initiation of reoxygenation/reperfusion.27 Thus, we focus on in vitro OGD model in this study. Besides, reoxygenation after OGD will be investigate in our future work. A recent study has revealed the expression profile of circRNAs in acute ischemic stroke in a South Chinese Han population using ischemic stroke patient circulating blood samples.28 It is the first time to identify various OGD-responsive circRNAs in human BMECs. However, there are still two deficiencies in this work. The effects of circPHC3 on cell death and apoptosis are not verified in vivo ischemic stroke model. Besides, whether circPHC3 could bind to other miRNAs still needs investigation. Therefore, we will further investigate the function and mechanism of circPHC3 in ischemic stroke in the future.
Conclusion
Our study illustrated that circPHC3 promoted cell death and apoptosis of human BMECs after OGD, in an in vitro mimic of stroke conditions, via binding to miR-455-5p to activate the expression of TRAF3. CircPHC3 might be regarded as a promising target for ischemic disease.
Acknowledgments
We thank all of the donors whose names were not included in the author list, but who participated in this program. This research was funded by the Shenzhen Science and Technology Projects (JCYJ20150605103420338) and Shenzhen Municipal Scheme for Basic Research (JCYJ20190808144005614).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang P, Zhang N, Liang J, Li J, Han S, Li J. Micro-RNA-30a regulates ischemia-induced cell death by targeting heat shock protein HSPA5 in primary cultured cortical neurons and mouse brain after stroke. J Neurosci Res. 2015;93:1756–1768. doi:10.1002/jnr.23637
2. Vahidinia Z, Karimian M, Joghataei MT. Neurosteroids and their receptors in ischemic stroke: from molecular mechanisms to therapeutic opportunities. Pharmacol Res. 2020;160:105163. doi:10.1016/j.phrs.2020.105163
3. Cheng YC, Cole JW, Kittner SJ, Mitchell BD. Genetics of ischemic stroke in young adults, Circulation. Cardiovasc Genet. 2014;7:383–392. doi:10.1161/CIRCGENETICS.113.000390
4. Ba K, Casolla B, Caparros F, et al. Early epileptic seizures in ischaemic stroke treated by mechanical thrombectomy: influence of rt-PA. J Neurol. 2020. doi:10.1007/s00415-020-10155-4
5. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science (New York, N Y). 2013;340:440–441. doi:10.1126/science.1238522
6. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO, Moran JV. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi:10.1371/journal.pgen.1003777
7. Cai L, Qi B, Wu X, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. 2019;130:10–22. doi:10.1016/j.yjmcc.2019.03.007
8. Bai Y, Zhang Y, Han B, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50. doi:10.1523/JNEUROSCI.1348-17.2017
9. Deng G, Mou T, He J, et al. Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer Res. 2020;39:1. doi:10.1186/s13046-019-1487-2
10. Xie L, Mao M, Xiong K, Jiang B. Circular RNAs: a novel player in development and disease of the central nervous system. Front Cell Neurosci. 2017;11:354. doi:10.3389/fncel.2017.00354
11. He JH, Han ZP, Luo JG, et al. Hsa_Circ_0007843 acts as a mIR-518c-5p sponge to regulate the migration and invasion of colon cancer SW480 cells. Front Genet. 2020;11:9. doi:10.3389/fgene.2020.00009
12. Zhang J, Yuan L, Zhang X, et al. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol. 2016;277:162–170. doi:10.1016/j.expneurol.2015.12.014
13. Wang Y, Li SF, Dang YJ, et al. Differentially expressed circular RNAs in maternal and neonatal umbilical cord plasma from SGA compared with AGA. J Cell Biochem. 2020;121:713–722. doi:10.1002/jcb.29317
14. Wang F, Zou Z, Gong Y, Yuan D, Chen X, Sun T. Regulation of human brain microvascular endothelial cell adhesion and barrier functions by memantine. J Mol Neurosci. 2017;62:123–129. doi:10.1007/s12031-017-0917-x
15. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi:10.1038/nmeth.1923
16. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi:10.1186/1471-2105-12-323
17. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 2013;29:15–21. doi:10.1093/bioinformatics/bts635
18. Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics (Oxford, England). 2016;32:1094–1096. doi:10.1093/bioinformatics/btv656
19. Bolha L, Ravnik-Glavač M, Glavač D. Circular RNAs: biogenesis. Int J Genomics. 2017;2017:6218353.
20. Yao S, Tang B, Li G, Fan R, Cao F. miR-455 inhibits neuronal cell death by targeting TRAF3 in cerebral ischemic stroke. Neuropsychiatr Dis Treat. 2016;12:3083–3092. doi:10.2147/NDT.S121183
21. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027
22. Liu Y, Zeng Y, Liu L, et al. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat Commun. 2014;5:5393. doi:10.1038/ncomms6393
23. Choi IY, Ju C, Anthony Jalin AM, Lee DI, Prather PL, Kim WK. Activation of cannabinoid CB2 receptor-mediated AMPK/CREB pathway reduces cerebral ischemic injury. Am J Pathol. 2013;182:928–939. doi:10.1016/j.ajpath.2012.11.024
24. Brenna S, Altmeppen HC, Mohammadi B, et al. Characterization of brain-derived extracellular vesicles reveals changes in cellular origin after stroke and enrichment of the prion protein with a potential role in cellular uptake. J Extracell Vesicles. 2020;9:1809065. doi:10.1080/20013078.2020.1809065
25. Kuo PC, Weng WT, Scofield BA, et al. Interferon-β alleviates delayed tPA-induced adverse effects via modulation of MMP3/9 production in ischemic stroke. Blood Adv. 2020;4:4366–4381. doi:10.1182/bloodadvances.2020001443
26. Zhao Y, Li J, Li J, Xu L, Lian W. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100. Eur Rev Med Pharmacol Sci. 2020;24:4420–4429. doi:10.26355/eurrev_202004_21024
27. Benedek A, Cernica D, Mester A, et al. Modern concepts in regenerative therapy for ischemic stroke: from stem cells for promoting angiogenesis to 3D-bioprinted scaffolds customized via carotid shear stress analysis. Int J Mol Sci. 2019;20:2574. doi:10.3390/ijms20102574
28. Li S, Chen L, Xu C, et al. Expression profile and bioinformatics analysis of circular RNAs in acute ischemic stroke in a South Chinese Han population. Sci Rep. 2020;10:10138. doi:10.1038/s41598-020-66990-y
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.