Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA circ_0007142 Facilitates Colorectal Cancer Progression by Modulating CDC25A Expression via miR-122-5p
Authors Yin W, Xu J, Li C, Dai X, Wu T, Wen J
Received 12 November 2019
Accepted for publication 3 March 2020
Published 1 May 2020 Volume 2020:13 Pages 3689—3701
DOI https://doi.org/10.2147/OTT.S238338
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr William C. Cho
Wenzhe Yin,1 Jun Xu,2 Chao Li,1 Xiankui Dai,1 Tong Wu,1 Jifeng Wen2
1Department of Orthopaedics, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150086, People’s Republic of China; 2Department of Gastroenterology, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150086, People’s Republic of China
Correspondence: Jifeng Wen
Department of Gastroenterology, The Second Affiliated Hospital of Harbin Medical University, No. 246 Xuefu Road, Nangang District, Harbin City, Heilongjiang Province, People’s Republic of China
Tel +86 451-86605134
Email [email protected]
Background: Colorectal cancer (CRC) is a common malignant tumor in digestive system. Circular RNA (circRNA) circ_0007142 has been identified as an oncogene in CRC. However, the mechanism of circ_0007142 in CRC was rarely reported.
Materials and Methods: The levels of circ_0007142, dedicator of cytokinesis 1 (DOCK1), microRNA-122-5p (miR-122-5p), and cell division cycle 25A (CDC25A) in CRC tissues (n=31) and cells were examined by quantitative real-time polymerase chain reaction (qRT-PCR). The cell viability and colony-forming ability were evaluated via 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay and colony-formation assay, respectively. The migrated and invaded abilities were monitored by Transwell assay. The dual-luciferase reporter assay was performed to validate the interactions between miR-122-5p and circ_0007142 or CDC25A. The protein level of CDC25A was detected via Western blot assay. The biological role of circ_0007142 was examined by xenograft tumor model in vivo.
Results: The levels of circ_0007142 and CDC25A were enhanced and the level of miR-122-5p was declined in CRC tissues and cells, while the level of DOCK1 had no fluctuation. Circ_0007142 sponged miR-122-5p and CDC25A was a target of miR-122-5p. Circ_0007142 knockdown impeded cell proliferation, colony formation, migration, and invasion in CRC cells by regulating miR-122-5p. Besides, miR-122-5p inhibitor promoted cell proliferation, colony formation, migration, and invasion in CRC cells by modulating CDC25A. Circ_0007142 regulated CDC25A expression in CRC cells by sponging miR-122-5p. Moreover, circ_0007142 knockdown blocked CRC tumor growth in vivo.
Conclusion: Circ_0007142 modulated CDC25A expression to promote CRC progression by sponging miR-122-5p.
Keywords: circ_0007142, miR-122-5p, CDC25A, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common cancer in the world and accompanied by high mortality.1 The 5-year survival rate of CRC patients diagnosed at early stage is as much as 90%; however, only 12% of 5-year survival rate for patients at advanced stage.2 Thus, it is crucial to search for potential therapeutic targets for early-stage diagnosis in CRC. Combined with the improved conventional treatments for CRC in recent decades, it is possible to prolong the life-span of CRC patients.
Circle RNAs (circRNAs), a novel type of stable covalently closed RNAs generated by single pre-mRNAs, play vital roles in modulating gene expression and functions.3,4 Convincing evidence indicated that circRNAs had been reported to implicate in the processes of many types of tumors, including CRC. For example, a study in breast cancer implied that hsa_circ_0052112 was elevated in breast cancer and its knockdown-repressed metastasis via miR-125a-5p.5 Another study in bladder cancer elucidated that circ_0000144 silencing impeded cell viability and metastasis in vitro, and also confined tumor growth in vivo.6 Chen et al documented that circ_101555 was elevated in CRC, and its depletion blocked cell proliferation and facilitated apoptosis.7 Also, circ_0007142, a novel circRNA originated from linear RNA DOCK1, has been documented to be dysregulated in CRC. However, the mechanism of circ_0007142 is still unclear.
MicroRNAs (miRNAs) are a form of small RNA with no translation capacity, and impact gene expression at post-transcriptional stage.8 Increasing data indicated that many miRNAs were aberrantly expressed in CRC and further affected the biological processes in CRC. For instance, Ma et al reported that miR-150 was dramatically declined in CRC.9 Another exploration in CRC revealed that miR-144 was strikingly decreased in CRC, and its overexpression reduced cell viability and mobility by targeting GSPT1 in vitro.10 Accumulating evidence showed that miR-122-5p was also related to tumor progression, including gastric cancer,11 hepatocellular carcinoma,12 and nasopharyngeal carcinoma.13 Interestingly, we found that miR-122 was expressed at the low expression in metastatic CRC,14 implying the involvement of miR-122 in CRC progression.
Cell division cycle 25A (CDC25A) was a gene located on human chromosome 20 and also associated with tumor development. For example, a research in breast cancer uncovered that CDC25A was remarkably elevated in breast cancer, and its knockdown-regulated cell behaviors mediated by miR-99a-5p.15 Nevertheless, the biological mechanism of CDC25A remained unclear in CRC.
In this exploration, our data showed that circ_0007142 was increased in CRC tissues and cells. Functionally, circ_0007142 downregulation restrained cell viability, colony formation, migration, and invasion in CRC cells. Moreover, bioinformatics analysis discovered that there was the underlying binding relationship between miR-122-5p and circ_0007142 or CDC25A. Therefore, this study aimed to explore whether circ_0007142 could regulate CRC progression by the miR-122-5p/CDC25A axis.
Materials and Methods
Tissues Collection
The project was permitted by the Ethics Committee of The Second Affiliated Hospital of Harbin Medical University and performed according to the Declaration of Helsinki Principles. Thirty-one CRC tissue samples (8 patients with clinical stage I–II and 23 patients with stage III–IV) were obtained from The Second Affiliated Hospital of Harbin Medical University, as well as corresponding adjacent normal tissues (non-cancerous paracancer tissues). All tissues were stored at −80°C until further used. The clinical information of the patients included age, gender, tumor size, lymphatic metastasis, distal metastasis, and TNM stage was summarized in Table 1. Written informed consents were provided by all CRC patients.
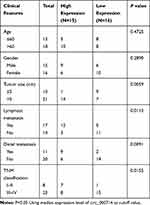 |
Table 1 Correlation Between Circ_0007142 Expression and Clinicopathological Features of Colorectal Cancer |
Cell Culture and Transfection
Two CRC cell lines (HT-29 and HCT-116) and human normal colorectal epithelial cell line (NCM460) were purchased from BioVector (Beijing, China). The CRC cells HT-29 and HCT-116 were cultured in McCoy’s 5a medium (Procell, Wuhan, China) containing with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Rockville, MD, USA), while the NCM460 cells were cultivated in RPMI-1640 medium (Solarbio, Beijing, China) supplemented with 10% FBS (Solarbio, Beijing, China) and maintained in an incubator at 37°C with 5% CO2.
Small interfering RNA (siRNA) against circ_0007142 (si-circ_0007142, 5ʹ-GGAAACAGCTTTTTATAAC-3ʹ) and its mock (si-NC), miR-122-5p mimic (miR-122-5p) and its scramble (miR-NC), miR-122-5p inhibitor (anti-miR-122-5p) and its negative control (anti-miR-NC), siRNA targeting CDC25A (si-CDC25A) and its control (si-NC) were purchased from GenePharma (Shanghai, China). The fragment of circ_0007142 was cloned and inserted into pcDNA3.1 (vector; Invitrogen, Carlsbad, CA, USA) to construct overexpression plasmid (circ_0007142). The transfection was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). This assay was carried out based on the previous description.16
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The RNA in CRC tissues and cells was extracted using TriQuick Reagent (Solarbio, Beijing, China), and AMV Reverse Transcriptase (Solarbio, Beijing, China) was used to perform reverse transcription. Then, the qPCR was carried out using SYBR Green PCR Master Mix (Ambion, Carlsbad, CA, USA). The levels of circ_0007142, DOCK1, and CDC25A were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) while miR-122-5p was standardized by small nuclear RNA U6, then processed by the method of.−ΔΔCt17 The sequences of primers were displayed as follows: circ_0007142: (F, 5ʹ-GAACTCTGCCTCAGGATGAA-3ʹ, and R, 5ʹ-AACGTGTAACCTCGGTACCA-3ʹ); DOCK1: (F, 5ʹ-CCGCCGCAAACTTTTTCCTC-3ʹ, and R, 5ʹ-AGATGTGCACAGTGTCTCCG-3ʹ); miR-122-5p: (F, 5ʹ-GGGGTGGAGTGTGACAATG-3ʹ, and 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ); CDC25A: (F, 5ʹ-TGACATCTTTCAGCTCATCG-3ʹ, and R, 5ʹ-CAGACAAAGTGGCTGTCACAG-3ʹ); GAPDH: (F, 5ʹ-TGTTCGTCATGGGTGTGAAC-3ʹ, and R, 5ʹ-ATGGCATGGACTGTGGTCAT-3ʹ), and U6: (F, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ, and R, 5ʹ-GGAACGCTTCACGAATTTG-3ʹ).
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
This assay was performed following the previous description.18 The MTT (Solarbio, Beijing, China) was used to measure the cell viability of HT-29 and HCT-116 cells. The cells (3×103 per well) were firstly maintained in 96-well plate for 24 h and underwent the transfection. Then, the cells were incubated for another 0 h, 24 h, 48 h, and 72 h, and the MTT was injected into each well and incubated with 4 h at 37°C. Subsequently, dimethyl sulfoxide (DMSO) was added to dissolve the formazan. Absorbance at 490 nm was measured with a Multiscan Spectrum (Potenov, Beijing, China) to represent cell viability.
Colony Formation Assay
Following transfection and cultivation, the HT-29 and HCT-116 cells were digested with trypsin and plated into 6-well plate. Then, the cell colonies were cultured for 2 weeks and fixed with methanol and stained with 0.5% crystal violet for 30 min. Colonies with more than 50 cells were counted under a microscope. This assay was carried out based on the previous description.19
Transwell Assay
The Transwell chambers (Corning, Tewksbury, MA, USA) were used to evaluate the migrated and invaded abilities of HT-29 and HCT-116 cells. For cell migration, the HT-29 and HCT-116 cells (6×103) were plated in upper chamber supplemented with McCoy’s 5a medium with no FBS; however, the lower one was added with medium containing 10% FBS. Following 24-h cultivation, the cells on the backside of polycarbonate film were fixed with 4% methanol and then stained with 0.1% crystal violet for 30 min. The cells in 10 randomly selected fields were counted under a microscope. For cell invasion, the difference is that a Matrigel matrix (Corning, Tewksbury, MA, USA) was covered on the upper chamber. This assay was conducted in line with the previous description.20
Dual-Luciferase Reporter Assay
The potential target of circ_0007142 was searched by starBase v3.0 (http://starbase.sysu.edu.cn/). Also, the putative candidate target of miR-122-5p was predicted by starBase v3.0. The wild type (containing the complementary binding sites) and mutant sequences of circ_0007142 and 3ʹ-untranslated regions (3ʹUTR) of CDC25A were inserted into pmirGLO vector (Promega, Madison, WI, USA) to construct luciferase reporter. The luciferase reporter and miR-122-5p or miR-NC was co-transfected into HT-29 and HCT-116 cells. The luciferase activity was detected using Dual-Lucy Assay Kit (Solarbio, Beijing, China). This test was performed according to the previous description.21
Western Blot Assay
The assay was implemented according to the previous description.22 The protein in CRC was extracted using a protein extraction kit (Beyotime, Shanghai, China). Then, the protein samples were separated by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Subsequently, the membrane was first blocked in skim milk at 37°C for 4 h and incubated with primary antibodies anti-CDC25A (1:2000, ab989; Abcam, Cambridge, MA, USA), anti-GAPDH (1:2500; ab9485, Abcam, Cambridge, MA, USA) overnight at 4°C. Then, the membrane was incubated with secondary goat anti-rabbit antibody (1:10,000; ab175781, Abcam, Cambridge, MA, USA) for 3 h at 37°C. The chemiluminescence intensity was evaluated using an ECL kit (Beyotime, Shanghai, China).
Mice Xenografts
This mouse experiment was approved by the Animal Ethics Committee of the Second Affiliated Hospital of Harbin Medical University. GeneChem (Shanghai, China) provided the Lentiviral-based the short hairpin RNAs (shRNAs) specific to circ_0007142 (sh-circ_0007142) and the corresponding control (sh-NC). Moreover, BALB/c nude mice (5–6 weeks old) were obtained from the National Laboratory Animal Center (Beijing, China), and divided randomly into two groups (n=4). Whereafter, a total of 1× 107 HT-29 cells with sh-circ_0007142 or sh-NC were injected into the right flanks of nude mice, as described previously.23 Tumor volume was measured every 5 days beginning at day 10 after injection. At 30 days after injection, the mice were euthanized, and tumors were excised and weighed, followed by analysis with qRT‐PCR or Western blot assays.
Statistical Analysis
GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA) was used to perform the experiment data. All quantitative data from three independent experiments were presented as the mean ± standard deviation (SD). The differences between two groups were processed by Student’s t-test, while the differences among multiple groups were evaluated by one-way analysis of variance (ANOVA). Statistical significance was defined as P value less than 0.05.
Results
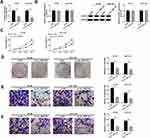
circ_0007142 Was Strikingly Increased in CRC Tissues and Cells
In order to determine the effect of circ_0007142 in CRC, the level of circ_0007142 was firstly measured in 31 paired CRC and adjacent normal tissue samples. As exhibited in Figure 1A, the level of circ_0007142 was conspicuously up-regulated in CRC tissues related to that in adjacent normal tissues. Also, the level of circ_0007142 was apparently increased in CRC cell lines HT-29 and HCT-116 compared with that in human normal colorectal epithelial cell line NCM460 (Figure 1B). Then, we divided the patients into two groups (High and Low) using the median value of circ_0007142 expression levels in CRC patients. Whereafter, we analyzed the potential clinical significance circ_0007142 by Chi-square test. As displayed in Table 1, the high expression of circ_0007142 was significantly associated with Tumor size, lymphatic metastasis (p=0.0113), distal metastasis (p=0.0091) and advanced TNM stage (p=0.0155), suggesting that circ_0007142 might affect the clinical prognosis of CRC patients. These data implied that circ_0007142 may play an important role in CRC.
The Depletion of circ_0007142 Impeded Cell Proliferation, Colony Formation, Migration, and Invasion in HT-29 and HCT-116 Cells
To investigate the functions of circ_0007142 in CRC, si-circ_0007142 was transfected into HT-29 and HCT-116 cells. The level of circ_0007142 was distinctly declined in HT-29 and HCT-116 cells transfected with si-circ_0007142, confirming the knockdown efficiency (Figure 2A). To avoid the production of circ_0007142 by DOCK1 during the transcription, we monitored the level of DOCK1 in HT-29 and HCT-116 cells transfected with si-circ_0007142 or si-NC. As presented in Figure 2B, both mRNA level and protein level of DOCK1 had no obvious change in any group. Following MTT assay indicated that the transfection of circ_0007142 resulted in the remarkable reduction of cell viability in HT-29 and HCT-116 cells (Figure 2C). Furthermore, the colony-forming ability was also down-regulated in HT-29 and HCT-116 cells transfected with si-circ_0007142 in contrast to that in si-NC group (Figure 2D). In addition, the Transwell assay showed that the introduction of si-circ_0007142 markedly reduced the migrated and invaded abilities in HT-29 and HCT-116 cells (Figure 2E–F). Taken together, these results demonstrated that circ_0007142 silencing inhibited CRC progression in vitro.
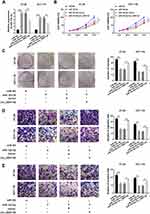
circ_0007142 Sponged miR-122-5p in HT-29 and HCT-116 Cells
To explore the mechanism of circ_0007142 in CRC, starBase v3.0 was used to search the potential targets of circ_0007142. As shown in Figure 3A, miR-122-5p had complementary sequences with circ_0007142. Following dual-luciferase reporter assay indicated that the transfection of miR-122-5p contributed to the notable decline of luciferase activity of WT-circ_0007142 reporter in HT-29 and HCT-116 cells in comparison with that in miR-NC group, while the luciferase activity of MUT-circ_0007142 had no obvious fluctuation in any group (Figure 3B). Besides, the level of miR-122-5p was evidently decreased in CRC tissues and cells (Figure 3C–D). Interestingly, we found that the expression level of circ_0007142 was negatively correlated with miR-122-5p in CRC tissues (Figure 3E). The transfection efficiency was affirmed by the dramatical up-regulation of circ_0007142 in HT-29 and HCT-116 cells transfected with circ_0007142 (Figure 3F). Meanwhile, the level of miR-122-5p was prominently reduced in HT-29 and HCT-116 cells transfected with circ_0007142, while drastically enhanced in si-circ_0007142-transfected HT-29 and HCT-116 cells (Figure 3F). These data indicated that the miR-122-5p was negatively interacted with circ_0007142 in HT-29 and HCT-116 cells.
circ_0007142 Promoted Cell Proliferation, Colony Formation, Migration, and Invasion in HT-29 and HCT-116 Cells by Regulating miR-122-5p
To elucidate the functions of circ_0007142 and miR-122-5p in CRC, miR-122-5p and circ_0007142 were co-transfected into HT-29 and HCT-116 cells. As shown in Figure 4A, the level of miR-122-5p was dramatically elevated in HT-29 and HCT-116 cells transfected with miR-122-5p, while mitigated in HT-29 and HCT-116 cells co-transfected with miR-122-5p and circ_0007142. The MTT assay indicated that the introduction of circ_0007142 regained cell viability in HT-29 and HCT-116 cells repressed by miR-122-5p mimics (Figure 4B). Also, the introduction of circ_0007142 alleviated the colony-forming ability HT-29 and HCT-116 cells refrained by miR-122-5p (Figure 4C). Besides, the migrated and invaded abilities were significantly reduced in HT-29 and HCT-116 cells transfected with miR-122-5p, while partly restored by the re-introduction of circ_0007142 (Figure 4D–E). These results implicated that circ_0007142 accelerated CRC progression by targeting miR-122-5p.
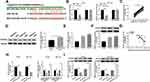
CDC25A Was Negatively Interacted with miR-122-5p in HT-29 and HCT-116 Cells
To explore the mechanism of miR-122-5p in CRC, the putative candidates were searched by starBase v3.0 online database. As displayed in Figure 5A, the CDC25A 3ʹUTR had complementary binding sites with miR-122-5p. The luciferase activity of WT-CDC25A reporter was conspicuously reduced in HT-29 and HCT-116 cells transfected with miR-122-5p mimics, while the luciferase activity of MUT-CDC25A had no apparent change in any group (Figure 5B). Meanwhile, the mRNA and protein levels of CDC25A were both distinctly up-regulated in CRC tissues and cells (Figure 5C-F). Also, our data suggested that the expression level of CDC25A was inversely related to miR-122-5p level in CRC tissues (Figure 5G). Moreover, the level of miR-122-5p was dramatically declined in HT-29 and HCT-116 cells transfected with anti-miR-122-5p compared to that in miR-NC group. In addition, the mRNA level of CDC25A was significantly down-regulated in HT-29 and HCT-116 cells transfected with miR-122-5p, while were strikingly elevated in anti-miR-122-5p-transfected HT-29 and HCT-116 cells (Figure 5H). Apart from that, the protein level of CDC25A also displayed a similar trend in transfected CRC cells (Figure 5I). These data uncovered that CDC25A was a direct target of miR-122-5p in HT-29 and HCT-116 cells.
miR-122-5p Inhibitor Relieved the Restraint Effects on Cell Proliferation, Colony Formation, Migration, and Invasion in HT-29 and HCT-116 Cells Induced by CDC25A Knockdown
To further investigate the mechanism and functions of miR-122-5p and CDC25A in CRC, si-CDC25A and anti-miR-122-5p were transfected into HT-29 and HCT-116 cells. The mRNA and protein levels of CDC25A were effectively decreased in HT-29 and HCT-116 cells transfected with si-CDC25A, while restored in HT-29 and HCT-116 cells co-transfected with si-CDC25A and anti-miR-122-5p (Figure 6A and B). Furthermore, the cell viability was notably declined in si-CDC25A-transfected HT-29 and HCT-116 cells, but partly ameliorated by the re-introduction of anti-miR-122-5p (Figure 6C). Also, the transfection of anti-miR-122-5p reversed the suppressive impact on the colony-forming ability in HT-29 and HCT-116 cells inhibited by si-CDC25A (Figure 6D). Besides, the emergence of anti-miR-122-5p recovered the mitigated and invaded abilities in HT-29 and HCT-116 cells confined by CDC25A depletion (Figure 6E and F). To sum up, CDC25A silencing refrained CRC progression mediated by miR-122-5p.
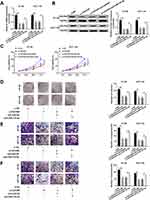
circ_0007142 Positively Regulated CDC25A Expression by Sponging miR-122-5p in HT-29 and HCT-116 Cells
To elucidate the relationship among circ_0007142, miR-122-5p, and CDC25A in CRC, miR-122-5p and circ_0007142 were transfected into HT-29 and HCT-116 cells. As showed in Figure 7A and B, the mRNA and protein levels of CDC25A were notably decreased in HT-29 and HCT-116 cells transfected with miR-122-5p, while recovered by the introduction of circ_0007142. In addition, our results proved that CDC25A upregulation relieved the suppression action of circ_0007142 knockdown on cell proliferation, colony formation, migration, and invasion in HT-29 and HCT-116 cells (Figure S1). These results demonstrated that circ_0007142 modulated CDC25A in HT-29 and HCT-116 cells via miR-122-5p.
circ_0007142 Deficiency Suppressed CRC Cell Growth in vivo
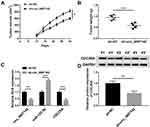
At last, in order to further evaluate the effect of circ_0007142 on tumor growth in vivo, a mouse xenograft model of CRC was established. As indicated in Figure 8A and B, tumor volume and weight were reduced in the presence of circ_0007142 knockdown, supporting that circ_0007142 silencing could inhibit CRC tumor growth in vivo. In addition, our data suggested that the levels of circ_0007142 and CDC25A were remarkably declined in tumor tissues from sh-circ_0007142 group compared with sh-NC group, while miR-122-5p level was enhanced in the tumor tissues (Figure 8C). Meanwhile, our result also verified that CDC25A protein level was decreased in the xenograft (Figure 8D). Hence, it is concluded that circ_0007142 knockdown could hinder CRC tumor growth partly by the miR-122-5p/CDC25A axis in vivo.
Discussion
It is well known that tumor progression is a complex process. Emerging data indicated that circRNAs played crucial roles in cancer.24 The main purpose of the research was to investigate the mechanism and functions of circ_0007142 in CRC. These data unraveled that circ_0007142 knockdown suppressed CRC progression via miR-122-5p/CDC25A axis.
Accumulating evidence disclosed that circRNAs were involved in tumor progression of CRC. For example, a study implied that circHIPK3 was enhanced in CRC, and its silencing confined cell growth and metastasis, while induced apoptosis.25 Zhu et al reported that circRNA BANP was significantly increased in CRC, while the depletion of BANP restrained cell proliferation.26 However, another study indicated that hsa_circ_0000523 was declined in CRC, and its overexpression-retarded cell proliferation.27 The biological processes of tumor progression were complicated in CRC. The controversial results might be caused by the different roles of circRNAs in regulatory networks. In the present study, circ_0007142 was increased in CRC. The silencing of circ_0007142 constrained cell proliferation, colony formation, and metastasis in vitro. The above results of circ_0007142 in CRC were in line with a previous study.28 These data demonstrated that circ_0007142 might play important roles in CRC.
Recent studies demonstrated that miR-122-5p was also implicated in tumor progression. For instance, a study in gastric cancer (GC) unraveled that miR-122-5p was distinctly declined in GC, and its overexpression constrained cell metastasis by targeting DUSP4.11 Another report in hepatocellular carcinoma (HCC) suggested that miR-122-5p was declined in HCC, and the overexpression of it hampered cell viability, growth, and metastasis mediated by ANRIL.12 In this research, circ_0007142 was identified to function as a sponge of miR-122-5p, and miR-122-5p was decreased in CRC. MiR-122-5p mimics retarded cell viability, growth, and mobility in CRC cells, while attenuated by circ_0007142. Although the miR-122-5p was not reported in CRC, its analogue miR-122 in CRC were reported to be decreased in 5-FU-resistant colon cancer cell lines.29 These results suggested that circ_0007142 facilitated CRC progression by sponging miR-122-5p.
Convincing data demonstrated that the aberrant expression of CDC25A was associated with tumor progression. For example, studies in glioma elucidated that CDC25A was notably increased in glioma, and its knockdown refrained cell proliferation while facilitated apoptosis.30,31 Another research in endometrial cancer indicated that miR-449a overexpression blocked cell viability and invaded ability by regulating CDC25A.32 In the current study, CDC25A was a candidate target of miR-122-5p and was significantly increased in CRC. More importantly, CDC25A was negatively regulated by miR-122-5p in CRC cells. CDC25A deletion curbed cell viability, growth, and mobility in CRC cells modulated by miR-122-5p. The effects on cell viability and growth were in agreement with that in other papers.33 Furthermore, circ_0007142 positively regulated CDC25A expression by sponging miR-122-5p in CRC cells. These results demonstrated that circ_0007142 modulated CDC25A expression to regulate cell behaviors in CRC by sponging miR-122-5p in vitro. Apart from that, our data also proved that the knockdown of circ_0007142 could repress CRC tumor growth at least through the miR-122-5p/CDC25A axis in vivo.
In conclusion, circ_0007142, CDC25A were enhanced, and miR-122-5p was reduced in CRC. Circ_0007142 promoted CRC progression by regulating CDC25A expression via miR-122-5p. This circ_0007142/miR-122-5p/CDC25A-induced CRC progression may shed light on the mechanism of CRC and provide novel diagnostic markers for CRC patients.
Disclosure
The authors have no conflict of interest to declare.
References
1. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi:10.1016/S0140-6736(13)61649-9
2. Siegel R, Desantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–241. doi:10.3322/caac.21149
3. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453. doi:10.1038/nbt.2890
4. Chen L, Huang C, Wang X, et al. Circular RNAs in eukaryotic cells. Curr Genomics. 2015;16(5):312–318. doi:10.2174/1389202916666150707161554
5. Jiang L-H, Hou J-C, Zhong S-L, et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342–1353. doi:10.1016/j.biopha.2018.08.030
6. Huang W, Lu Y, Wang F, et al. Downregulation of circular RNA hsa_circ_0000144 inhibits bladder cancer progression via stimulating miR-217 and suppressing RUNX2 expression. Gene. 2018;678:337–342. doi:10.1016/j.gene.2018.08.036
7. Chen Z, Ren R, Wan D, et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene. 2019;38(32):6017–6034. doi:10.1038/s41388-019-0857-8
8. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60(1):167–179. doi:10.1146/annurev.med.59.053006.104707
9. Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. doi:10.1136/gutjnl-2011-301122
10. Xiao R, Li C, Chai B. miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomedicine & Pharmacotherapy. 2015;74:138–144. doi:10.1016/j.biopha.2015.08.006
11. Xu X, Gao F, Wang J, et al. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol Ther. 2018;19(5):427–435. doi:10.1080/15384047.2018.1423925
12. Ma J, Li T, Han X, et al. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144(2):205–214. doi:10.1007/s00432-017-2543-y
13. Liu Y, Liu J, Wang Z, et al. MiR-122-5p suppresses cell proliferation, migration and invasion by targeting SATB1 in nasopharyngeal carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(2):622–629. doi:10.26355/eurrev_201901_16876
14. Maierthaler M, Benner A, Hoffmeister M, et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2017;140:1. doi:10.1002/ijc.30433
15. Qin H, Liu W. MicroRNA‐99a‐5p suppresses breast cancer progression and cell‐cycle pathway through downregulating CDC25A. J Cell Physiol. 2019;234(4):3526–3537. doi:10.1002/jcp.26906
16. Yang B, Du K, Yang C, et al. CircPRMT5 circular RNA promotes proliferation of colorectal cancer through sponging miR-377 to induce E2F3 expression. J Cell Mol Med. 2020. doi:10.1111/jcmm.15019
17. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi:10.1373/clinchem.2008.112797
18. Duan X, Shen N, Chen J, et al. Circular RNA MYLK serves as an oncogene to promote cancer progression via microRNA-195/cyclin D1 axis in laryngeal squamous cell carcinoma. Biosci Rep. 2019;39:9. doi:10.1042/BSR20190227
19. Lei B, Huang Y, Zhou Z, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. J. Cell. Biochem. 2019;120(4):4. doi:10.1002/jcb.27966
20. Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496(2):455–461. doi:10.1016/j.bbrc.2018.01.077
21. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol. 2019;234(8):13807–13819. doi:10.1002/jcp.28061
22. Xu W, Chang J, Du X, et al. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother. 2017;95:1112–1118. doi:10.1016/j.biopha.2017.09.019
23. Yang D, Du G, Xu A, et al. Expression of miR‐149‐3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am J Cancer Res. 2017a;7:
24. Kristensen L, Hansen T, Venø M, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555. doi:10.1038/onc.2017.361
25. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
26. Zhu M, Xu Y, Chen Y, et al. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi:10.1016/j.biopha.2016.12.097
27. Jin Y, Yu L, Zhang B, et al. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Me Biol Res. 2018;51(12):12. doi:10.1590/1414-431X20187811
28. Zhu C-L, Sha X, Wang Y, et al. Circular RNA hsa_circ_0007142 is upregulated and targets miR-103a-2-5p in colorectal cancer. J Oncol. 2019;2019:1–10. doi:10.1155/2019/9836819
29. He J, Xie G, Tong J, et al. Overexpression of microRNA-122 re-sensitizes 5-FU-resistant colon cancer cells to 5-FU through the inhibition of PKM2 in vitro and in vivo. Cell Biochem Biophys. 2014;70(2):1343–1350. doi:10.1007/s12013-014-0062-x
30. Yamashita Y, Kasugai I, Sato M, et al. CDC25A mRNA levels significantly correlate with Ki-67 expression in human glioma samples. J Neurooncol. 2010;100(1):43–49. doi:10.1007/s11060-010-0147-3
31. Yu M, Xue Y, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16(1):110. doi:10.1186/s12943-017-0677-9
32. Ye W, Xue J, Zhang Q, et al. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol Rep. 2014;32(3):1193–1199. doi:10.3892/or.2014.3303
33. Liu W, Wu M, Du H, et al. SIRT6 inhibits colorectal cancer stem cell proliferation by targeting CDC25A. Oncol Lett. 2018;15(4):5368–5374. doi:10.3892/ol.2018.7989
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.