Back to Journals » Cancer Management and Research » Volume 13
Circular RNA Circ_0006282 Promotes Cell Proliferation and Metastasis in Gastric Cancer by Regulating MicroRNA-144-5p/Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein β Axis
Authors Hua Y, Wang H, Wang H, Wu X, Yang L, Wang C, Li X, Jin Y, Li M, Wang L, Dong C, Yin F
Received 25 September 2020
Accepted for publication 8 January 2021
Published 27 January 2021 Volume 2021:13 Pages 815—827
DOI https://doi.org/10.2147/CMAR.S283952
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmet Emre Eşkazan
Yunqi Hua,1,* Hailong Wang,2,* Haizhen Wang,3 Xiangming Wu,4 Li Yang,5 Chenlin Wang,1 Xi Li,1 Yunjian Jin,1 Min Li,1 Lina Wang,1 Changcheng Dong,6 Fangrui Yin7
1Department of Digestive Oncology, Baotou Cancer Hospital, Baotou, Inner Mongolia, People’s Republic of China; 2Department of Digestive Minimally Invasive Surgery, The Second Affiliated Hospital of Baotou Medical College, Baotou, Inner Mongolia, People’s Republic of China; 3Center of Digestive Endoscopy, The Second Affiliated Hospital of Baotou Medical College, Baotou, Inner Mongolia, People’s Republic of China; 4Department of General Surgery, The Fourth Hospital of Baotou, Baotou, Inner Mongolia, People’s Republic of China; 5Department of Pathology, The Second Affiliated Hospital of Baotou Medical College, Baotou, Inner Mongolia, People’s Republic of China; 6Department of General Surgery, Inner Mongolia Baogang Hospital, Baotou, Inner Mongolia, People’s Republic of China; 7Department of Central Laboratory, The First Affiliated Hospital of Baotou Medical College, Baotou, Inner Mongolia, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Changcheng Dong
Department of General Surgery, Inner Mongolia Baogang Hospital, No. 20 Shaoxian Road, Kundulun District, Baotou, Inner Mongolia, People’s Republic of China
Tel +86 472-5992765
Email [email protected]
Fangrui Yin
Department of Central Laboratory, The First Affiliated Hospital of Baotou Medical College, No. 41 Linyin Road, Kundulun District, Baotou, People’s Republic of China
Tel +86 472-2178425
Email [email protected]
Background: Circular RNAs (circRNAs) are a class of non-coding RNAs which function as novel regulators in human cancers. In this study, we aimed to investigate the functional roles and related molecular mechanisms of circ_0006282 in gastric cancer (GC) progression.
Methods: Fifty-five GC patients were enrolled in this study. GC cells (AGS and HGC-27) and normal cells (GES-1) were cultured in RPMI1640 added with 10% FBS and 1% penicillin-streptomycin. Quantitative real-time polymerase chain reaction (qRT-PCR) assay was used to determine the expression levels of circ_0006282, transcription elongation factor B subunit 1 (TCEB1) mRNA, miR-144-5p and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein β (YWHAB; also known as 14-3-3β). RNase R assay was used to determine the characteristic of circ_0006282. Cell Counting Kit-8 (CCK-8) assay and colony formation assay were employed for cell proliferation. Transwell assay was conducted for cell migration and invasion. Western blot assay was carried out to measure the protein levels of Cyclin D1, matrix metalloprotein 9 (MMP9) and YWHAB. Dual-luciferase reporter assay, RNA pull-down assay and RIP assay were adopted to analyze the interaction between miR-144-5p and circ_0006282 or YWHAB. Murine xenograft model assay was performed to explore the function of circ_0006282 in vivo.
Results: Circ_0006282 level was increased in GC tissues and cells compared to normal tissues and cells. Silencing of circ_0006282 restrained GC cell proliferation, migration and invasion. For mechanism analysis, circ_0006282 was identified to function as the sponge for miR-144-5p to positively regulate YWHAB expression in GC cells. Moreover, miR-144-5p inhibition or YWHAB overexpression effectively reversed the impacts of circ_0006282 knockdown on GC cell growth and motility. Additionally, circ_0006282 knockdown blocked tumor growth of GC in vivo.
Conclusion: Circ_0006282 facilitated the malignant behaviors of GC cells through circ_0006282/miR-144-5p/YWHAB axis.
Keywords: gastric cancer, circ_0006282, miR-144-5p, YWHAB
Introduction
Gastric cancer (GC) is a usual deadly cancer in the world, which responsible for a large proportion of cancer-associated mortalities.1,2 Tumor metastasis and recurrence are the main obstacles for GC therapy.3,4 Although the methods of surgery, chemotherapy and radiotherapy can alleviate the symptoms and improve the survival of GC patients, the prognosis of GC patients remains unfavorable.5 Therefore, exploring the pathological mechanisms of GC is crucial to search for effective therapeutic targets for this lethal disease.
Circular RNAs (circRNAs) are newly discovered endogenous, circular transcripts with neither 5ʹ-3ʹ polarities nor polyadenylated tails.6,7 CircRNAs can adsorb microRNAs (miRNAs), a sort of non-coding RNAs with ~22 nucleotides, to modulate gene expression.8,9 Up to date, emerging evidence has reported that circRNAs are widely expressed and participate in the development of human malignancies, including GC.10 For instance, circ-SFMBT2 aggravated the malignancy of GC by decreasing miR-182-5p and increasing CREB1.11 Circ_0008035 contributed to GC cell viability and invasion via sponging miR-375 to enhance YBX1 expression.12 Circ_0006282 originated from TCEB1 gene and played an oncogenic role in GC by modulation of miR-155/FBXO22 axis.13 Even so, the molecular mechanism by which circ_0006282 mediating GC progression remains largely unclear. In this study, the functional roles and regulatory mechanisms of circ_0006282 in GC cell growth and metastasis were investigated.
The aberrant expression of miRNAs exerts an outstanding role in regulating the biological roles of tumor progression.14 In GC, multiple miRNAs have been claimed to serve as a tumor inhibitor through targeting the 3ʹ untranslated regions (3ʹUTR) of target genes.15,16 For example, miR-12129 impeded cell growth and cell cycle process in GC by targeting SIRT1.17 MiR-338-3p knockdown facilitated GC cell migration and prevented apoptosis through interaction with PTP1B.18 MiR-144-5p was found to be lowly expressed in GC and linked to the overall survival of GC patients.19 Nevertheless, the exact roles of miR-144-5p and its related mechanisms still need further investigation.
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein β (YWHAB; also termed as 14-3-3β) is a form of 14-3-3 proteins, which can modulate cell proliferation, survival and motility.20 It has been documented that YWHAB plays a crucial role in the carcinogenesis of diverse human cancers, such as papillary thyroid carcinomas,21 glioma22 and hepatocellular carcinoma.23 However, the effects of YWHAB on GC are unknown.
Here, we found that miR-144-5p contained the potential binding sites of circ_0006282 and YWHAB. However, the relationships among circ_0006282, miR-144-5p and YWHAB in GC development remain unclear. Thus, we deciphered the functions of circ_0006282, miR-144-5p and YWHAB in GC and their relationships in regulating GC cell malignant behaviors.
Materials and Methods
Tissues Collection
Fifty-five GC patients were recruited in this study. After the work was allowed by the Ethics Committee of Baotou Cancer Hospital (NeiMengGu, China) and written informed consents were obtained from the patients, the tumor tissue specimens and adjacent non-tumor tissue specimens were harvested from the patients at Baotou Cancer Hospital during April 2015 and January 2019. The inclusion criteria were as follows: (1) The patients were pathologically diagnosed with GC; (2) The patients did not receive any chemotherapy or radiotherapy before surgery. The exclusion criteria were as follows: (1) The patients <18 years old or >80 years old; (2) pregnant or lactating patients; (3) The patients had autoimmune diseases or other malignancies. The harvested specimens were stored at −80°C before use.
Cell Culture
Human normal gastric mucosal epithelial cells (GES-1) and GC cells (AGS and HGC-27) were selected based on the previous literatures24,25 and purchased from Procell (Wuhan, China). These cells were maintained in Roswell Park Memorial Institute 1640 Medium (RPMI1640; Procell) added with 10% fetal bovine serum (FBS; Procell) and 1% penicillin-streptomycin (Procell) at 37°C in a humid incubator with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The extraction of total RNA was done utilizing TRIzol (Beyotime, Shanghai, China). The RNA was quantified with the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For RNase treatment, 20 μg RNA was treated with or without RNase R (3U/μg; Epicenter Biotechnologies, Madison, WI, USA) for 20 min at 37°C. Next, for circ_0006282 and mRNA, PrimeScript™ RT reagent Kit (Takara, Dalian, China) and either Oligo (dt)18 primers or Random primers were used for cDNA synthesis. For miR-144-5p, TaqMan microRNA Assay kit (Applied Biosystems, Foster City, CA, USA) was utilized for cDNA synthesis. Afterwards, qRT-PCR reaction was executed with BeyoFast™ SYBR Green qPCR Mix (Beyotime) and relevant primers (GeneCopoeia, Guangzhou, China) on the StepOnePlus Real-Time PCR System (Applied Biosystems). The primers were: circ_0006282: (F: 5ʹ-AGGCACGATAAAAGCCATGT-3ʹ and R: 5ʹ-GGTCCTTCACAGCCACCATA-3ʹ); TCEB1: (F: 5ʹ-TTATGCATGGGGAAAGAAGC-3ʹ and R: 5ʹ-CTCAAACTCCTGGCCTCAAG-3ʹ); miR-144-5p: (F: 5ʹ-GCGCGAATTCGAGATCTTAACAGACCCTAGCTC-3ʹ and R: 5ʹ-GCGCGGATCCGTGCCCTGGCAGTCAGTAGG-3ʹ); YWHAB: (F: 5ʹ-GTCACCAGGTCTCCCAAGTG-3ʹ and R: 5ʹ-TTCTCCCCACTGCAGTGTTC-3ʹ); glyceraldehyde 3-phosphate dehydrogenase (GAPDH): (F: 5ʹ-ACCCACTCCTCCACCTTTGAC-3ʹ and R: 5ʹ-TGTTGCTGTAGCCAAATTCGTT-3ʹ); U6: (F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ). The expression was computed via the 2−ΔΔCt strategy with GAPDH or U6 as a control.
Subcellular Fraction Assay
The cytoplasmic and nuclear fractions in AGS and HGC-27 cells were separated through the usage of PARIS Kit (Invitrogen, Carlsbad, CA, USA) in line with the manufacturer’s instructions. Then the aforementioned qRT-PCR analysis was adopted for the levels of circ_0006282, U6 and GAPDH. U6 and GAPDH were employed as the controls of nuclear transcript and cytoplasmic transcript, respectively.
Cell Transfection
Circ_0006282 small interfering RNA (si-circ_0006282) and scramble siRNA (si-NC), miR-144-5p mimics (miR-144-5p) and mimics NC (miR-NC), miR-144-5p inhibitors (anti-miR-144-5p) and inhibitors NC (anti-NC), the overexpression vector of YWHAB (YWHAB) and its control (Vector), circ_0006282 short hairpin RNA (sh-circ_0006282) and sh-NC were all bought by GeneCopoeia. AGS and HGC-27 cells were seeded into 12-well plates with 1.0×104 cells/well and then transfected with 50 nM synthetic oligonucleotides or 2 μg vectors using Lipofectamine 2000 (Invitrogen) based on the protocols. Forty-eight-hour later, the transfected cells were harvested for subsequent experiments.
Cell Counting Kit-8 (CCK-8) Assay
The proliferation capacity of AGS and HGC-27 cells was tested by CCK-8 experiment. In brief, the transfected cells (1×104 cells/well) were plated into 96-well plates. After cultivating overnight, 10 μL CCK-8 (Boster, Wuhan, China) was supplemented into each well at 24 h, 48 h and 72 h followed by incubation for a further 4 h. Finally, the optical density (OD) value was examined with a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) at 450 nm. The experiment was run in triplicate.
Colony Formation Assay
Following 48 h of transfection, AGS and HGC-27 cells (500 cells/well) were collected and plated onto 6-well plates in complete medium and cultivated for 2 weeks. The medium was changed every 4 days. Thereafter, the colonies were fixed with 4% paraformaldehyde (Sangon, Shanghai, China) for 20 min and dyed with 0.1% crystal violet (Sangon) for 15 min. Finally, the colonies were photographed and counted.
Transwell Assay
The transwell chambers (BD Bioscience, San Jose, CA, USA) were employed for cell migration and invasion as previously described.26 Briefly, transfected AGS and HGC-27 cells (1×105) were harvested, resuspended in 200 μL serum-free medium and then plated into the top compartment coated with or without Matrigel (BD Bioscience) to assess cell invasion or migration. Six hundred microliter medium added with 10% FBS (Procell) was filled in the lower compartment. Twenty-four-hour later, the cells passed through the membranes were fixed with 4% paraformaldehyde (Sangon) and dyed with 0.1% crystal violet (Sangon). An inverted microscope (Olympus, Tokyo, Japan) was employed to count the invaded/migrated cells at the magnification of 100×.
Western Blot Assay
The protein isolation was finished using Radio Immunoprecipitation Assay (RIPA) buffer (Beyotime) and examined using a BCA Protein Quantification Kit (Vazyme, Nanjing, China). Then the protein samples were electrophoresed on 10% sodium dodecyl sulfonate-polyacrylamide gel (SDS-PAGE; Solarbio, Beijing, China) and blotted on polyvinylidene difluoride membranes (PVDF; Pall Corporation, New York, NYC, USA). Next, the membranes were blocked in 5% defatted milk for 1 h at room temperature. After that, the membranes were incubated overnight with primary antibodies against GAPDH (bs-10900R; Bioss, Beijing, China), Cyclin D1 (bs-20596R; Bioss), matrix metalloprotein 9 (MMP9; bs-4593R; Bioss) or YWHAB (bs-12420; Bioss) at 4°C and then interacted with relevant secondary antibody (bs-0924M; Bioss) for 2 h at room temperature. At last, the protein bands were visualized with an ECL reagent (Vazyme).
Dual-Luciferase Reporter Assay
The potential binding sites between miR-144-5p and circ_0006282 or YWHAB 3ʹUTR were predicted by Starbase V3.0 and TargetScan, respectively. To verify the prediction, the sequences of circ_0006282 (or YWHAB 3ʹUTR) consisting of the mutant (MUT) or wild-type (WT) binding sites of miR-144-5p were cloned into pmirGLO vector (Promega, Fitchburg, WI, USA), constructing circ_0006282-WT, circ_0006282-MUT, YWHAB-3ʹUTR-WT and YWHAB-3ʹUTR-MUT. Then AGS and HGC-27 cells were seeded into 24-well plated at a density of 5.0×104 cells/well. Next, the constructs (100 ng) were co-transfected into in combination with miR-144-5p or miR-NC (50 nM) using Lipofectamine 2000 (Invitrogen). After 48 h of transfection, the Renilla and firefly luciferase activities were examined through the usage of Dual-Luciferase Reporter Assay Kit (Promega). The experiment was done referring to previous research.11
RNA Pull-Down Assay
The Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) was used for RNA pull-down assay.27 In brief, 3ʹ-biotinylated miR-144-5p (Bio-miR-144-5p) or Bio-miR-NC was transfected into AGS and HGC-27 cells. After 48 h, the cells were lysed and incubated with streptavidin-coupled beads (Invitrogen). Next, the biotin-coupled RNA complex was pulled down. Finally, the enrichment of circ_0006282 and YWHAB in bound fractions was detected by qRT-PCR.
RNA Immunoprecipitation (RIP) Assay
The Magna RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) was employed to conduct RIP assay.28 Briefly, the cells were lysed in RIP buffer. Then the collected RIP buffer was incubated with magnetic beads conjugated with anti-Argonaute2 (anti-Ago2) and anti-immunoglobulin G (anti-IgG). The anti-IgG and Input were used as controls. The levels of circ_0006282 and miR-144-5p in the beads were detected by qRT-PCR assay.
Murine Xenograft Model
The 4–6 weeks old nude mice were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Then the mice were divided into 2 groups (n=6/group). AGS cells (1×107) transfected with sh-NC or sh-circ_0006282 were injected into the flank of the nude mice. The length and width of the xenograft tumor were measured weekly and tumor volume was estimated by the equation: (Length×Width2)/2. The mice were euthanized after 5 weeks and the neoplasms were harvested and weighted. The neoplasms were preserved at −80°C for further qRT-PCR and Western blot assays. The animal research was permitted by the Ethics Committee of Animal Research of Baotou Cancer Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. The experiment was done according to previous study.29
Statistical Analysis
The experiments were run in triple times and the collected data were presented as mean ± standard deviation, processed by using GraphPad Prism 7 software. The differences were estimated by Student’s t-test and one-way analysis of variance. P<0.05 was deemed as a significant difference.
Results
Circ_0006282 Was Upregulated in GC Tissues and Cell Lines
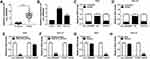
To begin with, we determined the expression level of circ_0006282 in GC tissues (n=55) and adjacent non-tumor tissues (n=55) with qRT-PCR assay. The data showed that compared to non-tumor tissues, circ_0006282 was highly expressed in tumor tissues (Figure 1A). We also found that circ_0006282 level was conspicuously elevated in AGS and HGC-27 cells compared to GES-1 cells (Figure 1B). Subcellular fraction assay showed that circ_0006282 was mainly enriched in the cytoplasm of AGS and HGC-27 cells (Figure 1C and D). Moreover, we found that Oligo (dt)18 primers reversely transcribed cDNA could barely amplify circ_0006282 compared with Random primers reversely transcribed cDNA in both AGS and HGC-27 cells (Figure 1E and F). In addition, the results of RNase R digestion assay indicated that circ_0006282 was resistant to RNase R treatment, whereas TCEB1 mRNA was digested by RNase R treatment (Figure 1G and H). These findings suggested that the dysregulation of circ_0006282 might play a role in GC progression.
Silencing of Circ_0006282 Repressed GC Cell Proliferation and Metastasis
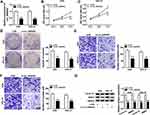
In order to elucidate the precise roles of circ_0006282 in GC development, loss-of-function experiments were conducted by transfecting si-circ_0006282 into AGS and HGC-27 cells. The transfection efficiency was estimated by qRT-PCR assay, exhibiting that si-circ_0006282 evidently decreased the level of circ_0006282 in AGS and HGC-27 cells compared to si-NC control groups (Figure 2A). As demonstrated by CCK-8 assay, circ_0006282 silencing markedly restrained the proliferation of AGS and HGC-27 cells compared to control groups (Figure 2B and C). The colony formation assay showed that circ_0006282 knockdown suppressed AGS and HGC-27 cell colony formation compared to control groups (Figure 2D). Moreover, we conducted transwell assay to evaluate cell migration and invasion abilities, showing that circ_0006282 deficiency drastically suppressed cell migration and invasion in AGS and HGC-27 cells compared to control groups (Figure 2E and F). Additionally, the levels of proliferation-related protein (cyclin D1) and metastasis-related protein (MMP9) were measured by Western blot assay. The results showed that circ_0006282 deficiency led to marked downregulation of Cyclin D1 and MMP9 levels in AGS and HGC-27 cells compared to si-NC control groups (Figure 2G). Collectively, circ_0006282 knockdown hampered the malignant behaviors of GC cells.
Circ_0006282 Functioned as the Sponge for miR-144-5p
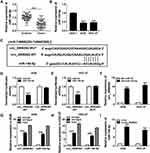
As shown in Figure 3A and B, miR-144-5p was weakly expressed in GC tissues and cells compared to normal tissues and cells. Through analyzing online tool Starbase V3.0, we found miR-144-5p contained the complementary sequences of circ_0006282, indicating that miR-144-5p might be a target of circ_0006282 (Figure 3C). Then dual-luciferase reporter assay exhibited that miR-144-5p transfection caused a remarkable suppression in the luciferase activity of circ_0006282-WT in AGS and HGC-27 cells, while the luciferase activity of circ_0006282-MUT was not affected, demonstrating the interaction between circ_0006282 and miR-144-5p (Figure 3D and E). The results of RNA pull-down assay showed that circ_0006282 level was abundantly pulled down by Bio-miR-144-5p RNA pull-down assay (Figure 3F). RIP assay further confirmed the interaction between circ_0006282 and miR-144-5p for the levels of circ_0006282 and miR-144-5p were enhanced by anti-Ago2 RIP compared to control anti-IgG groups (Figure 3G and H). Moreover, our results indicated that circ_0006282 silencing distinctly increased miR-144-5p level in AGS and HGC-27 cells compared to si-NC groups (Figure 3I). These data indicated that circ_0006282 negatively regulated miR-144-5p expression in GC cells by sponging miR-144-5p.
Circ_0006282 Knockdown Repressed the Malignant Behaviors of GC Cells by Targeting miR-144-5p
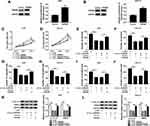
As presented in Figure 4A, anti-miR-144-5p transfection significantly reduced miR-144-5p level in AGS and HGC-27 cells compared to anti-NC groups. To further explore the relationship between circ_0006282 and miR-144-5p in the biological behaviors of GC cells, AGS and HGC-27 cells were divided into 4 groups: si-NC, si-circ_0006282, si-circ_0006282+anti-NC and si-circ_0006282+anti-miR-144-5p. As illustrated by CCK-8 assay, colony formation assay and transwell assay, the inhibitory effects on the proliferation, colony formation, migration and invasion of AGS and HGC-27 cells mediated by circ_0006282 silencing were partially reversed by decreasing miR-144-5p (Figure 4B–I). Moreover, the results of Western blot assay showed that circ_0006282 knockdown markedly reduced the levels of Cyclin D1 and MMP9 in AGS and HGC-27 cells, while miR-144-5p inhibition abrogated the impacts (Figure 4J and K). To sum up, circ_0006282 knockdown decelerated GC cell proliferation and metastasis by targeting miR-144-5p.
YWHAB Was the Target Gene of miR-144-5p
As shown in Figure 5A and B, the mRNA and protein levels of YWHAB were all obviously enhanced in GC tissues compared to adjacent non-tumor tissues. We also found that YWHAB protein level was elevated in AGS and HGC-27 cells in comparison with GES-1 cells (Figure 5C). Then miR-144-5p was successfully transfected into AGS and HGC-27 cells, as demonstrated by the upregulation of miR-144-5p in AGS and HGC-27 cells after miR-144-5p transfection (Figure 5D). By searching online website TargetScan, YWHAB was found to be a direct target gene of miR-144-5p and their potential binding sites were presented in Figure 5E. The dual-luciferase reporter assay was then conducted to confirm the interaction between miR-144-5p and YWHAB. The results showed that miR-144-5p transfection apparently inhibited the luciferase activity of YWHAB-3ʹUTR-WT in AGS and HGC-27 cells, but did not affect the luciferase activity of YWHAB-3ʹUTR-MUT (Figure 5F and G). RNA pull-down assay further demonstrated the interaction between miR-144-5p and YWHAB for the level YWHAB was evidently enriched by Bio-miR-144-5p assay compared to Bio-miR-NC control groups (Figure 5H). Furthermore, we observed that miR-144-5p overexpression strikingly decreased the protein level of YWHAB in both AGS and HGC-27 cells, while miR-144-5p suppression evidently increased the protein level of YWHAB (Figure 5I and J). Additionally, our results showed that circ_0006282 knockdown led to a marked reduction of YWHAB protein level in AGS and HGC-27 cells, while miR-144-5p inhibition abolished the impact (Figure 5K and L). All these data suggested that circ_0006282 positively regulated YWHAB expression by targeting miR-144-5p.
Overexpression of YWHAB Reversed the Suppressive Roles of Circ_0006282 Knockdown in GC Cell Malignant Behaviors
As presented in Figure 6A and B, YWHAB transfection distinctly elevated YWHAB protein level in AGS and HGC-27 cells compared to control groups, indicating the successful transfection of YWHAB. Subsequently, AGS and HGC-27 cells were assigned to 4 groups: si-NC, si-circ_0006282, si-circ_0006282+Vector and si-circ_0006282+YWHAB. The results of CCK-8 assay, colony formation assay and transwell assay indicated that YWHAB overexpression ameliorated the inhibitory effects of circ_0006282 silencing on cell proliferation, colony formation, migration and invasion in AGS and HGC-27 cells (Figure 6C–J). Furthermore, the elevation of YWHAB rescued the downregulation of Cyclin D1 and MMP9 in AGS and HGC-27 cells mediated by circ_0006282 silencing (Figure 6K and L). Taken together, circ_0006282 knockdown hampered GC cell progression by downregulating YWHAB.
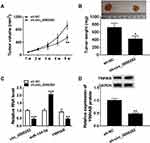
Circ_0006282 Knockdown Blocked Tumorigenesis of GC in vivo
At last, the functional role of circ_0006282 in GC progression in vivo was investigated by murine xenograft model assay. As a result, the mice with sh-circ_0006282 transfected AGS cells showed reduced tumor volume and tumor weight compared to control groups (Figure 7A and B). Moreover, our results showed that circ_0006282, YWHAB mRNA and YWHAB protein levels were all declined and miR-144-5p level was raised in the tissues collected from sh-circ_0006282 groups compared to sh-NC groups (Figure 7C and D). In summary, circ_0006282 silencing repressed tumor growth of GC in vivo.
Discussion
Currently, there is a growing body of evidence exhibited that circRNAs play pivotal roles in the malignancy of GC.30–32 Herein, we focused on the relationship between circ_0006282 and GC cell malignant phenotypes. It was found that circ_0006282 aggravated the progression of GC. Furthermore, the regulatory network of circ_0006282/miR-144-5p/YWHAB axis was discovered in GC.
CircRNAs are key regulators implicated in GC malignant biological behaviors.11,12,33 Shao et al disclosed that circ_0006282 was aberrantly improved in GC tissues.34 He et al reported that circ_0006282 level was closely linked to TNM stage, tumor size and lymph nodes motility, and promoted GC cell growth, migration and invasion by competitively binding to miR-155 and reducing FBXO22.13 Corresponding to these previous reports, circ_0006282 was strikingly elevated in GC tissues and cells. Moreover, interference of circ_0006282 restrained the proliferation, colony formation, migration and invasion capacities of GC cells in vitro and blocked the tumorigenicity of GC in vivo. Cyclin D1 is a critical protein in G1 phase, which can promote the rapid transformation from G1 to S phase through accelerating DNA replication and mitosis.35 MMP9 can degrade extracellular matrix and basement membrane, thereby promoting tumor invasion and metastasis.36,37 Thus, in this study, we investigated the effects of circ_0006282 on the levels of cyclin D1 and MMP9. As a result, cyclin D1 and MMP9 levels were reduced in GC cells following the downregulation of circ_0006282. Taken together, circ_0006282 facilitated GC malignancy.
Subsequently, the regulatory mechanism of circ_0006282 in GC carcinogenesis was further investigated. As a result, circ_0006282 functioned as the sponge for miR-144-5p to positively modulate YWHAB expression in GC cells. Thereafter, our study explored the relationships among circ_0006282, miR-144-5p and YWHAB in regulating GC cell malignant behaviors for the first time. MiR-144-5p level was declined in GC. Moreover, the impacts of circ_0006282 interference on tumor cell growth and metastasis were abolished by miR-144-5p suppression, suggesting that miR-144-5p might act as a tumor inhibitor in GC. In support of our findings, Li et al declared that miR-194 level was decreased in GC, and regulated GC cell proliferation and motility by targeting ZFX.38 Yao et al uncovered that miR-144 upregulation suppressed cell viability and growth, as well as induced cell apoptosis and cell cycle arrest in GC cells by binding to COX-2.39 Additionally, previous reports indicated that YWHAB level was enhanced in GC and accelerated the growth, invasion and migration of GC cells.40,41 Consistently, we demonstrated the enhancement of YWHAB level in GC tissues and cells. Moreover, YWHAB overexpression ameliorated the suppressive roles of circ_0006282 knockdown in GC cell growth and metastasis.
However, there were still some shortcomings in our study. For example, the tumor tissue samples were not enough and the function of circ_0006282 in tumor metastasis in vivo has not been explored. We will validate our results with more tumor samples and explore the role of circ_0006282 in tumor metastasis.
In summary, our present study unveiled that circ_0006282 sponged miR-144-5p to increase YWHAB expression, thereby contributing to GC cell proliferation, migration and invasion, as presented in Figure 8. This research might bring new light to the pathogenesis of GC and offer a target for GC therapy.
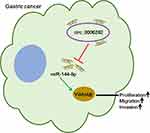 |
Figure 8 The schematic diagram of circ_0006282/miR-144-5p/YWHAB axis in the modulation of GC cell proliferation, migration and invasion. |
Abbreviations
GC, gastric cancer; circRNAs, circular RNAs; QRT-PCR, quantitative real-time polymerase chain reaction; TCEB1, transcription elongation factor B subunit 1; YWHAB, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein β; CCK-8, Cell Counting Kit-8; MMP9, matrix metalloprotein 9; RIP, RNA immunoprecipitation (RIP); MiRNAs, microRNAs; CREB1, cAMP responsive element binding protein 1; YBX1, Y-box binding protein-1; FBXO22, F-box protein 22; 3ʹUTR, 3ʹ untranslated regions; SIRT1, sirtuin 1; PTP1B, protein-tyrosine phosphatase 1B; RPMI1640, Roswell Park Memorial Institute 1640 Medium; FBS, fetal bovine serum; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OD, optical density; RIPA, Radio Immunoprecipitation Assay; SDS-PAGE, sodium dodecyl sulfonate-polyacrylamide gel; PVDF, polyvinylidene difluoride; ZFX, zinc finger protein X-linked; COX-2, cyclooxygenase-2.
Disclosure
The authors declare no conflicts of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
3. Veronese N, Fassan M, Wood LD, et al. Extranodal extension of nodal metastases is a poor prognostic indicator in gastric cancer: a systematic review and meta-analysis. J Gastrointest Surg. 2016;20(10):1692–1698.
4. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386.
5. Blackham AU, Greenleaf E, Yamamoto M, et al. Tumor regression grade in gastric cancer: predictors and impact on outcome. J Surg Oncol. 2016;114(4):434–439.
6. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
7. Wu J, Qi X, Liu L, et al. Emerging Epigenetic Regulation of Circular RNAs in Human Cancer. Mol Ther Nucleic Acids. 2019;16:589–596.
8. Li J, Yang J, Zhou P, et al. RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480.
9. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6.
10. Wang K-W, Dong M. Role of circular RNAs in gastric cancer: recent advances and prospects. World J Gastrointest Oncol. 2019;11(6):459–469. doi:10.4251/wjgo.v11.i6.459
11. Sun H, Xi P, Sun Z, et al. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res. 2018;10:5725–5734. doi:10.2147/CMAR.S172592
12. Huang S, Zhang X, Guan B, et al. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis.. Am J Transl Res. 2019;11(4):2455–2462.
13. He Y, Wang Y, Liu L, et al. <p>Circular RNA circ_0006282 Contributes to the Progression of Gastric Cancer by Sponging miR-155 to Upregulate the Expression of FBXO22. Onco Targets Ther. 2020;13:1001–1010. doi:10.2147/OTT.S228216
14. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004.
15. Zhou W, Ding X, Jin P, Li P. miR-6838-5p Affects Cell Growth, Migration, and Invasion by Targeting GPRIN3 via the Wnt/beta-Catenin Signaling Pathway in Gastric Cancer. Pathobiology. 2020;87(6):327–337.
16. Liu J, Li SM. MiR-484 suppressed proliferation, migration, invasion and induced apoptosis of gastric cancer via targeting CCL-18. Int J Exp Pathol. 2020;101(6):203–214.
17. Zhang W, Liao K, Liu D. MiRNA-12129 Suppresses Cell Proliferation and Block Cell Cycle Progression by Targeting SIRT1 in GASTRIC Cancer. Technol Cancer Res Treat. 2020;19:1533033820928144.
18. Sun F, Yu M, Yu J, et al. miR-338-3p functions as a tumor suppressor in gastric cancer by targeting PTP1B. Cell Death Dis. 2018;9(5):522.
19. Li CY, Liang GY, Yao WZ, et al. Identification and functional characterization of microRNAs reveal a potential role in gastric cancer progression. Clin Transl Oncol. 2017;19(2):162–172.
20. Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19(1):16–23.
21. Musholt TJ, Brehm C, Hanack J, von Wasielewski R, Musholt PB. Identification of differentially expressed genes in papillary thyroid carcinomas with and without rearrangements of the tyrosine kinase receptors RET and/or NTRK1. J Surg Res. 2006;131(1):15–25.
22. Cao L, Lei H, Chang MZ, Liu ZQ, Bie XH. Down-regulation of 14-3-3beta exerts anti-cancer effects through inducing ER stress in human glioma U87 cells: involvement of CHOP-Wnt pathway. Biochem Biophys Res Commun. 2015;462(4):389–395.
23. Liu TA, Jan YJ, Ko BS, et al. Increased expression of 14-3-3beta promotes tumor progression and predicts extrahepatic metastasis and worse survival in hepatocellular carcinoma. Am J Pathol. 2011;179(6):2698–2708.
24. Yang H, Wang L, Tang X, Bai W. miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncol Lett. 2017;14(6):7687–7690.
25. Lu M, Huang Y, Sun W, Li P, Li L, Li L. miR-135b-5p promotes gastric cancer progression by targeting CMTM3. Int J Oncol. 2018;52(2):589–598.
26. Li J, Zhen L, Zhang Y, et al. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529.
27. Subramanian M, Li XL, Hara T, Lal A. A biochemical approach to identify direct microRNA targets. Methods Mol Biol. 2015;1206:29–37.
28. Zhang L, Cheng H, Yue Y, Li S, Zhang D. H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/beta-catenin in ox-LDL -stimulated vascular smooth muscle cells. J Biomed Sci. 2018;25(1):11.
29. Li P, Chen H, Chen S, et al. RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633.
30. Cai X, Nie J, Chen L, Yu F. Circ_0000267 promotes gastric cancer progression via sponging MiR-503-5p and regulating HMGA2 expression. Mol Genet Genomic Med. 2020;8(2):e1093.
31. Du W, Li D, Guo X, et al. promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif Cells Nanomed Biotechnol. 2019;47(1):4120–4130.
32. Zhang Z, Wang C, Zhang Y, Yu S, Zhao G, CircDUSP16 XJ. promotes the tumorigenesis and invasion of gastric cancer by sponging miR-145-5p. Gastric Cancer. 2020;23(3):437–448.
33. Cao C, Han S, Yuan Y, et al. Downregulated Circular RNA hsa_circ_0000291 Suppresses Migration And Proliferation Of Gastric Cancer Via Targeting The miR-183/ITGB1 Axis. Cancer Manag Res. 2019;11:9675–9683.
34. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180.
35. Cadoni G, Boccia S, Petrelli L, et al. A review of genetic epidemiology of head and neck cancer related to polymorphisms in metabolic genes, cell cycle control and alcohol metabolism. Acta Otorhinolaryngol Ital. 2012;32(1):1–11.
36. Iizasa T, Fujisawa T, Suzuki M, et al. Elevated levels of circulating plasma matrix metalloproteinase 9 in non-small cell lung cancer patients. Clin Cancer Res. 1999;5(1):149–153.
37. Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–2392.
38. Li H, Yao G, Zhai J, Hu D, LncRNA FY. FTX Promotes Proliferation and Invasion of Gastric Cancer via miR-144/ZFX Axis. Onco Targets Ther. 2019;12:11701–11713.
39. Yao Q, Gu A, Wang Z, Xue Y. MicroRNA-144 functions as a tumor suppressor in gastric cancer by targeting cyclooxygenase-2. Exp Ther Med. 2018;15(3):3088–3095.
40. Ma Y, Li YF, Wang T, Pang R, Xue YW, Zhao SP. Identification of proteins associated with lymph node metastasis of gastric cancer. J Cancer Res Clin Oncol. 2014;140(10):1739–1749.
41. Tseng CW, Yang JC, Chen CN, et al. Identification of 14-3-3beta in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics. 2011;11(12):2423–2439.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.