Back to Journals » Cancer Management and Research » Volume 13
CircRNA ZNF609 Knockdown Represses the Development of Non-Small Cell Lung Cancer via miR-623/FOXM1 Axis
Authors Wang F, Li X, Jia X, Geng L
Received 15 September 2020
Accepted for publication 7 November 2020
Published 4 February 2021 Volume 2021:13 Pages 1029—1039
DOI https://doi.org/10.2147/CMAR.S282162
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Fanghan Wang,1 Xiangfeng Li,2 Xigao Jia,3 Luxin Geng1
1Department of Oncology, 4th People’s Hospital of Zibo, Zibo, Shandong, 255000, People’s Republic of China; 2Department of Radiology, 4th People’s Hospital of Zibo, Zibo, Shandong, 255000, People’s Republic of China; 3Department of Medicine, 4th People’s Hospital of Zibo, Zibo, Shandong, 255000, People’s Republic of China
Correspondence: Fanghan Wang
Department of Oncology, 4th People’s Hospital of Zibo, No. 210, Shanquan Road, Zhangdian District, Zibo, Shandong, 255000, People’s Republic of China
Tel +86-533-2982547
Email [email protected]
Background: The dysregulated circular RNAs (circRNAs) are relevant to the development of non-small cell lung cancer (NSCLC). Nevertheless, the function and mechanism of circRNA zinc finger protein 609 (circZNF609) in NSCLC development remain uncertain.
Methods: Sixty-two NSCLC patients were recruited. circZNF609, microRNA-623 (miR-623) and forkhead box M1 (FOXM1) abundances were measured via quantitative reverse transcription polymerase chain reaction or Western blot. Cell viability, apoptosis, migration and invasion were analyzed via cell counting kit-8 (CCK8), flow cytometry, caspase3 activity, transwell assay and Western blot. The interaction between miR-623 and circZNF609 or FOXM1 was analyzed via dual-luciferase reporter analysis, RNA immunoprecipitation and pull-down. The function of circZNF609 on cell growth in vivo was tested via xenograft model.
Results: circZNF609 abundance was enhanced in NSCLC tissues and cells. High expression of circZNF609 indicated the lower overall survival. circZNF609 interference restrained cell viability, migration and invasion and increased apoptosis. miR-623 was targeted via circZNF609. FOXM1 was targeted via miR-623 and regulated via circZNF609. miR-623 knockdown or FOXM1 overexpression mitigated the role of circZNF609 silence in NSCLC development. circZNF609 knockdown decreased NSCLC xenograft tumor growth.
Conclusion: circZNF609 knockdown repressed NSCLC development via regulating miR-623 and FOXM1.
Keywords: non-small cell lung cancer, circZNF609, FOXM1, miR-623
Introduction
Non-small cell lung cancer (NSCLC) accounts for a large proportion of lung cancer which is the leading cause of cancer death.1 Although the research on NSCLC has made great progress, the outcomes of patients remain poor.2 Thus, exploring new target for the therapy of NSCLC is of importance.
Circular RNAs (circRNAs) are a group noncoding RNAs formed via back-splicing, which are relevant to multiple cancer cell processes.3 CircRNAs have vital roles in pathogenesis, diagnosis and treatment of NSCLC.4 The dysregulated circRNAs are relevant to NSCLC cell proliferation, migration and invasion.5 The circRNA zinc finger protein 609 (circZNF609) is derived from ZNF609 gene, and functions as an oncogene via promoting cell growth and metastasis in multiple cancers, including renal carcinoma, breast cancer and nasopharyngeal carcinoma.6–8 Nevertheless, how and whether circZNF609 participates in NSCLC development remain poorly understood.
The crosstalk of circRNA/microRNA (miRNA)/mRNA in competing endogenous RNA (ceRNA) network is an important mechanism in NSCLC.9 The aberrantly expressed miRNAs are associated with the development and treatment of NSCLC.10 MiR-623 has been reported that could reduce tumor growth and metastasis in human cancers, such as gastric cancer, pancreatic cancer and hepatocellular carcinoma.11–13 Moreover, miR-623 could repress NSCLC progression via targeting Ku80 and inactivating the extracellular signal-related kinase (ERK)/c-Jun N-terminal kinase (JNK) pathway.14 Nevertheless, whether miR-623 is required for circZNF609 in NSCLC development is unclear.
Forkhead box M1 (FOXM1) is required for cell proliferation and usually overexpressed in various cancers.15 FOXM1 plays key roles in the regulation of many pathological processes in pulmonary disorders, including lung cancer.16,17 The overexpressed FOXM1 predicts the poor survival of NSCLC patients.18 Furthermore, FOXM1 facilitates cell proliferation, migration and invasion in NSCLC.19 The bioinformatics analysis predicts miR-623 might act as a crosstalk for circZNF609 and FOXM1. Hence, we hypothesize that circZNF609 might regulate miR-623/FOXM1 axis to participate in NSCLC development.
In this research, we detected circZNF609 expression in NSCLC and assessed the function of circZNF609 on NSCLC development. Additionally, we explored the interaction network of circZNF609/miR-623/FOXM1 axis in NSCLC cells.
Materials and Methods
Samples Acquisition
Sixty-two NSCLC patients were recruited from 4th People’s Hospital of Zibo. The inclusion criteria were as follows: all subjects received the surgical resection at our hospital and were pathological diagnosed with NSCLC; patients did not receive other therapy before tissue resection; patients had complete clinicopathological data, which are shown in Table 1. The NSCLC tumor tissues and para-tumor normal control (NC) tissues (5-cm away from tumors) were collected and stored at −80°C. A 60-month follow-up was performed, and the overall survival was analyzed according to the median value of circZNF609. Every patient signed written informed consent. The research was permitted via the ethics committee of 4th People’s Hospital of Zibo and executed on the principles of the Helsinki Declaration.
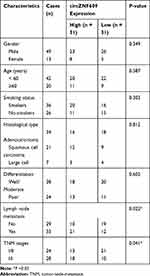 |
Table 1 Correlation Between circZNF609 Expression and the Clinicopathological Features of NSCLC Patients |
Cell Culture
Human NSCLC cell lines Calu-3, Calu-6, A549 and H1299 cells and the human bronchial cell line HBE1 cells were purchased from Procell (Wuhan, China) and grown in RPMI-1640 medium (Thermo Fisher, Waltham, MA, USA) plus 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin (Thermo Fisher) at 37°C and 5% CO2.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The RNA was isolated and extracted by Trizol reagent (Thermo Fisher) following the instructions as previously reported.20 Next, the RNA was applied to cDNA generation via the specific reverse transcription kit (Thermo Fisher). Subsequently, the generated cDNA was utilized for qRT-PCR. The primers (Genscript, Nanjing, China) included: circZNF609 (hsa_circ_0000615) (sense, 5’-TGAGTGTCGCCTGCTAAAGA-3’; antisense, 5’-CCCCCAGCTTTCCTATTTTC-3’), FOXM1 (sense, 5’-CCTTCTGGACCATTCACCCC-3’; antisense, 5’-TTCGGTCGTTTCTGCTGTGA-3’), miR-623 (sense, 5’-ATCCCTTGCAGGGGCTGTTGGGT-3’; antisense, 5’-AACGCTTCACGAATTTGCGT-3’), U6 (sense, 5’-AATTGGAACGATACAGAGAAGATTAGC-3’; antisense, 5’-TATGGAACGCTTCACGAATTTG-3’), and GAPDH (sense, 5’-GAATGGGCAGCCGTTAGGAA-3’; antisense, 5’-AAAAGCATCACCCGGAGGAG-3’). U6 or GAPDH was used as a reference. The relative RNA abundance was calculated via 2−ΔΔCt method.21
Vector and Oligonucleotide Construct and Cell Transfection
The FOXM1 overexpression vector was constructed by inserting the full-length sequence of FOXM1 into pcDNA3.1 vector (Thermo Fisher), with the empty vector as corresponding control. The shRNA for circZNF609 (sh-circZNF609#1, 5’-AGUCUGAAAAGCAAUGAUGUU-3’; sh-circZNF609#2, 5’-GUCUGAAAAGCAAUGAUGUUG-3’; sh-circZNF609#3, 5’-CAAGUCUGAAAAGCAAUGAUG-3’), miR-623 mimic (5’-AUCCCUUGCAUUUUCUGUUGGGU-3’), miR-623 inhibitor (anti-miR-623, 5’-ACCCAACAGCCCCUGCAAGGGAU-3’), corresponding controls (sh-NC, 5’-AAGACAUUGUGUGUCCGCCTT-3’; miR-NC, 5’-UUCUCCGAACGUGUCACGUTT-3’; anti-NC, 5’-UGCAGUAUGCAGAGUUAGGUAUA-3’) were formed from GenePharma (Shanghai, China). Lipofectamine 2000 (Thermo Fisher) was enforced to transfect.
Cell Viability
Cell viability was checked via cell counting kit-8 (CCK8). 1 × 104 A549 and H1299 cells were added into 96-well plates and maintained for 72 h. Then, 10 μL of CCK8 solution (Beyotime, Shanghai, China) was infused and cultured for 3 h. The wavelength was detected at 450 nm through a microplate reader (Bio-Rad, Hercules, CA, USA). The cell viability was displayed as a percentage relative to the control group.
Cell Apoptosis
Cell apoptosis was assessed via apoptotic rate and caspase3 activity. Cell apoptotic rate was discerned via flow cytometry with Annexin V-FITC apoptosis kit (Thermo Fisher). 2 × 105 A549 and H1299 cells were added in 6-well plates and maintained for 72 h. Subsequently, cells were interacted with Annexin V-FITC binding buffer and dyed with Annexin V-FITC and PI. The apoptotic rate was estimated through a flow cytometer (Agilent, Hangzhou, China).
For caspase3 activity assay, 1 × 104 A549 and H1299 cells were moved into 96-well plates and nurtured for 72 h. Then, collected cells were used for detection of caspase3 activity via the caspase3 activity assay kit (Sigma, St. Louis, MO, USA) following the instructions.
Transwell Analysis
Cell migration and invasion were tested via transwell analysis using transwell chamber (BD, Franklin Lakes, NJ, USA). For migration analysis, 1 × 105 A549 and H1299 cells in serum-free medium were placed in the upper chambers, and the medium plus 10% serum was added into lower chambers. Cells were incubated for 24 h, and then dyed with 0.1% crystal violet (Beyotime), followed via observation under a microscope (magnification ×100; Nikon, Tokyo, Japan) with 5 random fields. For invasion analysis, the upper chambers were precoated with Matrigel (BD). 5 × 105 A549 and H1299 cells in serum-free medium were added in the upper chambers, and the other procedures were like the migration analysis.
Western Blot
The protein was dissociated by RIPA buffer (Beyotime), and the quality was analyzed via a BCA kit (Solarbio). The proteins were loaded on SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). After blocking with 5% bovine serum albumin (Solarbio), the membranes were interacted with primary and secondary antibodies (Abcam, Cambridge, MA, USA), including: anti-matrix metalloproteinase 9 (MMP9) (ab38898, 1:300 dilution), anti-MMP2 (ab97779, 1:2000 dilution), anti-FOXM1 (ab226928, 1:3000 dilution), GAPDH (ab9485, 1:2000 dilution) and IgG conjugated via HRP (ab205718, 1:10000 dilution). GAPDH acted as a loading reference. After exposing to ECL reagent (Beyotime), the blots were visualized and then analyzed by Image J software (NIH, Bethesda, MD, USA).
Dual-Luciferase Reporter Analysis, RNA Immunoprecipitation and Pull-Down
The target correlation of miR-623 and circZNF609 or FOXM1 was predicted via CircInteractome or TargetScan. The wild-type (circZNF609-WT or FOXM1-WT) or mutant luciferase reporter vectors (circZNF609-MUT or FOXM1-MUT) were generated via inserting the corresponding sequence of circZNF609 or FOXM1 containing wild-type or mutant complementary sites of miR-623 in the pmirGlO vectors (Promega, Madison, WI, USA) via endonuclease sites Nhe I and Xba I. These constructed vectors and miR-623 mimic or miR-NC were transfected into A549 and H1299 cells for 24 h. The luciferase activity was detected via dual-luciferase analysis system (Promega) and a GloMax 20/20 Luminometer (Promega).
RIP analysis was performed using a Magna RIP kit (Sigma). 1 × 107 A549 and H1299 cells were lysed and interacted with anti-Ago2-conjugated magnetic beads for 8 h. IgG was used as a negative control, and input acted as a positive control. The immunoprecipitated RNA was extracted, and abundance levels of miR-623 and circZNF609 were tested via qRT-PCR.
RNA pull-down analysis was administrated with a Magnetic RNA-Protein Pull-Down kit (Thermo Fisher). A549 and H1299 cells were transfected with the biotinylated miR-623 (Bio-miR-623) or negative control (Bio-NC) generated via GenePharma for 24 h. Next, cells were lysed and interacted with magnetic beads conjugated via streptavidin for 8 h. The RNA on the beads was extracted, and the abundance of the enriched RNA was tested via qRT-PCR.
Xenograft Experiment
BALB/c nude mice (5-week-old, male) were provided via Charles River (Beijing, China), and arbitrarily divided into two groups (n=6/group): sh-circZNF609#1 group (subcutaneous injection with 5 × 106 H1299 cells with stable transfection of sh-circZNF609#1) or sh-NC group (subcutaneous injection with 5 × 106 H1299 cells with stable transfection of sh-NC). The tumor size was monitored every 7 days, and volume was counted via 1/2 × length × width2. After 35 days, mice were euthanized via 5% isoflurane. Tumor tissues were weighed and then used for detection of circZNF609, miR-623 and FOXM1 expression. The experiment was performed in line with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978), and assented via the Animal Ethical Committee of 4th People’s Hospital of Zibo in line with the National Institutes of Health.
Statistical Analysis
The experiments were performed 3 times, unless otherwise indicated. The overall survival of patients was generated and analyzed via Kaplan-Meier plot and Log-rank test. The results were expressed as mean ± standard deviation (SD). The association between circZNF609 expression and clinical data was investigated by χ2 test. The prognostic indicators were analyzed by a multivariate Cox regression analysis. The difference was compared via Student’s t-test or ANOVA with Tukey’s test by GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). P<0.05 was regarded as significant.
Results
circZNF609 Expression is Increased in NSCLC
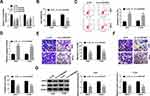
circZNF609 level was examined in NSCLC tissues (n=62). Results exhibited that circZNF609 abundance was evidently up-regulated in NSCLC tissues compared with NC samples (Figure 1A). Furthermore, circZNF609 level was detected in NSCLC cell lines and control HBE1 cells. As displayed in Figure 1B, circZNF609 expression was higher in Calu-3, Calu-6, A549 and H1299 cells than HBE1 cells. Besides, the overall survival of patients was analyzed according to the median value of circZNF609 expression. circZNF609 high expression group (n = 31) exhibited the lower survival than low expression group (P<0.01) (Figure 1C). Moreover, high expression of circZNF609 was associated with lymph node metastasis and tumor-node-metastasis (TNM) (P<0.05), but not with gender, age, smoking, histological type and differentiation (Table 1). Additionally, lymph node metastasis [P= 0.037; hazard ratio (HR)= 1.816; 95% confidence interval (CI)= 1.356–2.432] and high expression of circZNF609 (P= 0.023; HR= 2.120; 95% CI= 1.121–4.123) were independent predictors of poor overall survival of NSCLC (Table 2). A549 and H1299 cells with relative higher expression of circZNF609 were used for further experiments.
 |
Table 2 Multivariate Analysis for Factors Related to Overall Survival Using the Cox Proportional Hazard Model |
circZNF609 Knockdown Suppresses Cell Viability, Migration and Invasion and Promotes Apoptosis
To analyze the function of circZNF609 in NSCLC, A549 and H1299 cells were transfected with sh-circZNF609#1-#3 or sh-NC. The transfection efficacy of sh-circZNF609#1-#3 was confirmed in Figure 2A, and sh-circZNF609#1 with the relative highest efficacy was used for further experiments. circZNF609 knockdown markedly decreased the viability of A549 and H1299 cells (Figure 2B). Furthermore, circZNF609 silence obviously promoted cell apoptosis via increasing apoptotic rate and caspase3 activity (Figure 2C and D). In addition, circZNF609 interference markedly restrained the abilities of migration and invasion (Figure 2E and F). Besides, circZNF609 down-regulation significantly declined the protein levels of MMP9 and MMP2 in the two cell lines (Figure 2G). These data suggested that circZNF609 silence repressed NSCLC development.
miR-623 is Targeted via circZNF609
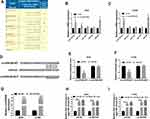
To explore the potential mechanism, the targets of circZNF609 were predicted by CircInteractome. The top 5 miRNAs (miR-1224-3p, miR-1231, miR-1233, miR-149, miR-623) with highest predicted scores were selected (Figure 3A). Moreover, the influence of circZNF609 knockdown on their expression was investigated in A549 and H1299 cells. miR-623 expression was changed most by circZNF609 silence (Figure 3B and C). Hence, miR-623 was selected for further experiments. The predicted binding sequence of circZNF609 and miR-623 was shown in Figure 3D. To identify this prediction, the luciferase reporter vectors circZNF609-WT and circZNF609-MUT were constructed. The data of dual-luciferase reporter analysis showed that miR-623 overexpression evidently declined the luciferase activity of circZNF609-WT, while it showed little impact on the luciferase activity of circZNF609-MUT (Figure 3E and F). Furthermore, the results of RNA pull-down displayed that there were amount of circZNF609 enriched via miR-623 (Figure 3G). Additionally, both circZNF609 and miR-623 were enriched in the same complex via Ago2 (Figure 3H and I). These data indicated that miR-623 was targeted via circZNF609.
FOXM1 is Regulated via miR-623 and circZNF609
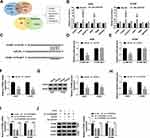
To further analyze the potential mechanism, the targets of miR-623 were predicted. After the analyses of the top 100 mRNAs that were upregulated in NSCLC via GEPIA (http://gepia.cancer-pku.cn/), we found there were 34 mRNAs that were predicted both in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Moreover, only 6 mRNAs might be targeted by miR-623 according to the prediction of TargetScan (Figure 4A). In addition, the results of RNA pull-down analysis showed that FOXM1 had the highest enrichment level (Figure 4B). Hence, FOXM1 was selected as a potential of miR-623 for further experiments. The predicted complementary sequence of miR-623 and FOXM1 is exhibited in Figure 4C. To validate this prediction, the luciferase reporter vectors FOXM1-WT and FOXM1-MUT were constructed. The results of dual-luciferase reporter analysis displayed that miR-623 addition markedly declined the luciferase activity of FOXM1-WT, but it did not change the luciferase activity of FOXM1-MUT (Figure 4D and E). In addition, FOXM1 expression was evidently decreased via miR-623 overexpression (Figure 4F and G). Besides, the influence of circZNF609 and miR-623 on FOXM1 abundance was assessed. The transfection effectivity of anti-miR-623 was confirmed in Figure 4H. Moreover, FOXM1 abundance in A549 and H1299 cells was significantly reduced via circZNF609 silence, which was restored by miR-623 knockdown (Figure 4I and J). These results indicated that circZNF609 could target FOXM1 via miR-623.
miR-623 Knockdown or FOXM1 Overexpression Weakens the Influence of circZNF609 Interference on Cell Viability, Apoptosis, Migration and Invasion
To explore whether miR-623 and FOXM1 were required for circZNF609 in NSCLC development, A549 and H1299 cells were transfected with sh-NC, sh-circZNF609#1, sh-circZNF609#1 + anti-NC, anti-miR-623, vector or FOXM1 overexpression vector. The transfection efficacy of FOXM1 overexpression vector was validated in Figure 5A. Moreover, miR-623 knockdown or FOXM1 overexpression attenuated silence of circZNF609-mediated viability reduction in A549 and H1299 cells (Figure 5B). Additionally, miR-623 knockdown or FOXM1 overexpression weakened interference of circZNF609-induced cell apoptosis via reducing the apoptotic rate and caspase3 activity (Figure 5C and D). Furthermore, miR-623 knockdown or FOXM1 overexpression mitigated knockdown of circZNF609-mediated inhibition of migration and invasion (Figure 5E and F). Besides, miR-623 knockdown or FOXM1 overexpression abolished down-regulation of circZNF609-mediated reduction of MMP9 and MMP2 expression in the two cell lines (Figure 5G-I). These results indicated that circZNF609 knockdown repressed NSCLC development via regulating miR-623 and FOXM1.
circZNF609 Knockdown Reduces NSCLC Xenograft Tumor Growth
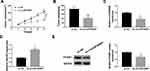
To test the function of circZNF609 in NSCLC development in vivo, H1299 cells with transfection of sh-circZNF609#1 or sh-NC were used to establish the xenograft model, and then the mice were divided into sh-circZNF609#1 or sh-NC group. At 35 days after cell injection, tumor volume and weight were evidently decreased in sh-circZNF609#1 group in comparison to sh-NC group (Figure 6A and B). In addition, the abundances of circZNF609, miR-623 and FOXM1 in tumor tissues were detected. As displayed in Figure 6C-E, circZNF609 and FOXM1 levels were markedly reduced, and miR-623 expression was increased in sh-circZNF609#1 group in comparison to sh-NC group. These data suggested that circZNF609 silence decreased NSCLC cell growth in vivo.
Discussion
NSCLC is the major subtype of lung cancer, and the prognosis of patients remains poor.22 The circRNAs act as important biomarkers in the progression of NSCLC.23 Our study aims to test the function of mechanism of circZNF609 in NSCLC development and found that circZNF609 knockdown plays a tumor-suppressive role in NSCLC, which might be associated with miR-623/FOXM1 axis.
In this research, we first detected the high level of circZNF609 in NSCLC, and found that it was associated with the lower overall survival. These findings elucidated that circZNF609 was dedicated to the malignancy of NSCLC. To explore the function of circZNF609 on NSCLC development, we performed the loss-of-function experiments using the shRNA for circZNF609. We found that circZNF609 knockdown suppressed NSCLC cell development via inhibiting cell viability, migration and invasion and promoting apoptosis, which was also in agreement with the carcinogenic role of circZNF609 in renal carcinoma, breast cancer and nasopharyngeal carcinoma.6–8 This implied that circZNF609 might be used as an important target for treatment of NSCLC.
The ceRNA regulatory network is a major mechanism addressed by circRNA via competing with miRNA in NSCLC.24 Hence, our study analyzed and explored the targeted miRNA of circZNF609. The increasing reports have validated that miR-138-5p, miR-145-5p and miR-150-5p could function as the crosstalk of circZNF609 and the targeted genes.6–8 In this research, we identified an additional target miR-623. Previous study disclosed that miR-623 could suppress cell proliferation via targeting cyclin D1 in gastric cancer.11 Moreover, miR-623 could inhibit cell migration and invasion via targeting MMP1 in pancreatic cancer.12 In addition, miR-623 could repress cell proliferation and invasion via decreasing X-ray repair cross complementing 5 (XRCC5) in hepatocellular carcinoma.13 These reports all indicated the anti-tumor role of miR-623 in many cancers. Importantly, Wei et al reported that miR-623 could reduce cell proliferation and metastasis via decreasing Ku80 in lung adenocarcinoma, a major type of NSCLC.14 Similarly, we identified the role of miR-623 in NSCLC via reversing the role of circZNF609, which also indicated that miR-623 was responsible for the regulatory effect of circZNF609 on NSCLC development.
Next, we further explored the potential circRNA/miRNA/mRNA axis in this study and validated that FOXM1 was targeted via miR-623. Besides, we found that FOXM1 abundance was regulated via circZNF609 and reversed via miR-623, indicating that circZNF609 could target FOXM1 via competitively binding with miR-623 in NSCLC cells. Previous studies reported that FOXM1 was highly expressed in NSCLC and associated with the poor survival.18,25,26 A former report using integrated network analysis proposed FOXM1 contributed to NSCLC cell proliferation.27 Zhao et al suggested that FOXM1 could promote NSCLC cell growth and metastasis via regulating its downstream cyclin D1 and MMP2.28 Yuan et al showed that FOXM1 regulated NSCLC cell proliferation and metastasis via target of LINC00339/miR-145 axis.29 Moreover, Wang et al showed that FOXM1 targeted via miR-216b was participated in the proliferation, migration and invasion of NSCLC.19 These reports all suggested the oncogenic role of FOXM1 in NSCLC. Similarly, we also found this function, revealed via abolishing the suppressive effect of circZNF609 knockdown on NSCLC development. Thus, we concluded the importance of circZNF609/miR-623/FOXM1 network in NSCLC development. The animal model is significant to mimic the complexity of microenvironment in human cancers, including NSCLC.30,31 To explore the potential function of circZNF609 in NSCLC development in vivo, H1299 cells with relative highest expression of circZNF609 were used to establish the xenograft model. By establishing this model, we found that circZNF609 knockdown could attenuate NSCLC cell growth in vivo. This further unraveled the anti-cancer function of circZNF609 silence in NSCLC.
Overall, circZNF609 interference restrained NSCLC development, possibly via regulating miR-623 and FOXM1 in a ceRNA crosstalk. This study indicates a new target for treatment of NSCLC.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
2. Duma N, Santana-Davila R, Molina JR. Non–small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94(8):1623–1640. doi:10.1016/j.mayocp.2019.01.013
3. Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005.
4. Zhang C, Ma L, Niu Y, et al. Circular RNA in Lung Cancer Research: biogenesis, Functions, and Roles. Int J Biol Sci. 2020;16(5):803–814.
5. Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889.
6. Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654.
7. Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res. 2018;10:
8. Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23(7):2817–2826.
9. Jin X, Guan Y, Sheng H, et al. Crosstalk in competing endogenous RNA network reveals the complex molecular mechanism underlying lung cancer. Oncotarget. 2017;8(53):91270–91280.
10. Zhou Q, Huang S-X, Zhang F, et al. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017;50(6):54. doi:10.1111/cpr.12394
11. Jiang L, Yang W, Bian W, et al. MicroRNA-623 targets cyclin d1 to inhibit cell proliferation and enhance the chemosensitivity of cells to 5-fluorouracil in gastric cancer. Oncol Res. 2018;27(1):19–27. doi:10.3727/096504018X15193469240508
12. Chen Y, Peng S, Cen H, et al. MicroRNA hsa-miR-623 directly suppresses MMP1 and attenuates IL-8-induced metastasis in pancreatic cancer. Int J Oncol. 2019;55(1):142–156. doi:10.3892/ijo.2019.4803
13. Ren F, Su H, Jiang H, et al. Overexpression of miR-623 suppresses progression of hepatocellular carcinoma via regulating the PI3K/Akt signaling pathway by targeting XRCC5. J Cell Biochem. 2020;121(1):213–223. doi:10.1002/jcb.29117
14. Wei S, Zhang Z-Y, Fu S-L, et al. RETRACTED ARTICLE: hsa-miR-623 suppresses tumor progression in human lung adenocarcinoma. Cell Death Dis. 2016;7(9):e2388. doi:10.1038/cddis.2016.260
15. Gartel AL. FOXM1 in Cancer: interactions and Vulnerabilities. Cancer Res. 2017;77(12):3135–3139. doi:10.1158/0008-5472.CAN-16-3566
16. Li Y, Wu F, Tan Q, et al. The multifaceted roles of FOXM1 in pulmonary disease. Cell Commun Signal. 2019;17(1):35. doi:10.1186/s12964-019-0347-1
17. Zhang J, Zhang J, Cui X, et al. FoxM1: a novel tumor biomarker of lung cancer. Int J Clin Exp Med. 2015;8(3):3136–3140.
18. Liu B, Su F, Lin R, et al. Overexpression of forkhead box M1 is associated poor survival in patients with nonsmall cell lung cancer. J Cancer Res Ther. 2018;14(7):S1121–S1123. doi:10.4103/0973-1482.203605
19. Wang L, Wang Y, Du X, et al. <p>MiR-216b suppresses cell proliferation, migration, invasion, and epithelial–mesenchymal transition by regulating FOXM1 expression in human non-small cell lung cancer. Onco Targets Ther. 2019;12:
20. Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581–585. doi:10.1038/nprot.2006.83
21. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
22. Balata H, Fong KM, Hendriks LE, et al. Prevention and Early Detection for NSCLC: advances in Thoracic Oncology 2018. J Thorac Oncol. 2019;14(9):1513–1527. doi:10.1016/j.jtho.2019.06.011
23. Drula R, Braicu C, Harangus A, et al. Critical function of circular RNAs in lung cancer. Wiley Interdiscip Rev RNA. 2020;11(5):e1592. doi:10.1002/wrna.1592
24. Hu W, Bi Z-Y, Chen Z-L, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:
25. Zhang Y, Qiao WB, Shan L. Expression and functional characterization of FOXM1 in non-small cell lung cancer. Onco Targets Ther. 2018;11:
26. Li R, Wang X, Zhao X, et al. Centromere protein F and Forkhead box M1 correlation with prognosis of non-small cell lung cancer. Oncol Lett. 2020;19(2):1368–1374.
27. Integrated Network AF. Analysis Reveals FOXM1 and MYBL2 as key regulators of cell proliferation in non-small cell lung cancer. Front Oncol. 2019;9:
28. Zhao L, Liu L, Dong Z, et al. miR-149 suppresses human non-small cell lung cancer growth and metastasis by inhibiting the FOXM1/cyclin D1/MMP2 axis. Oncol Rep. 2017;38(6):3522–3530.
29. Yuan Y, Haiying G, Zhuo L, et al. Long non-coding RNA LINC00339 facilitates the tumorigenesis of non-small cell lung cancer by sponging miR-145 through targeting FOXM1. Biomed Pharmacother. 2018;105:
30. Langdon SP. Animal modeling of cancer pathology and studying tumor response to therapy. Curr Drug Targets. 2012;13(12):1535–1547.
31. Singh AP, Adrianzen Herrera D, Zhang Y, et al. Mouse models in squamous cell lung cancer: impact for drug discovery. Expert Opin Drug Discov. 2018;13(4):347–358.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.