Back to Journals » OncoTargets and Therapy » Volume 13
CircHIPK3 Promotes Clear Cell Renal Cell Carcinoma (ccRCC) Cells Proliferation and Metastasis via Altering of miR-508-3p/CXCL13 Signal
Authors Han B, Shaolong E, Luan L , Li N , Liu X
Received 26 February 2020
Accepted for publication 1 June 2020
Published 25 June 2020 Volume 2020:13 Pages 6051—6062
DOI https://doi.org/10.2147/OTT.S251436
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Bin Han,1 E Shaolong,1 Lan Luan,2 Nanyang Li,2 Xuefeng Liu1
1Department of Urology, Shengjing Hospital of China Medical University, Shenyang, People’s Republic of China; 2Department of Pathology, Central Hospital Affiliated to Shenyang Medical College, Shenyang, People’s Republic of China
Correspondence: Xuefeng Liu
Department of Urology, Shengjing Hospital of China Medical University, Shenyang, Liaoning 110004, People’s Republic of China
Email [email protected]
Introduction: Accumulating evidence has demonstrated that circular RNAs (circRNAs) play a key role in the tumorigenesis of various types of cancers, including clear cell renal cell carcinoma (ccRCC).
Materials and Methods: Reverse transcription-quantitative polymerase chain reaction was used to detect the expression of circRNA homeodomain interacting protein kinase 3 (circHIPK3) and microRNAs (miRNAs), including miR-508-3p. The clinical measurement of circHIPK3 was evaluated by Kaplan–Meier survival analysis and receiver operating characteristic analysis. Cell Counting Kit-8 and Transwell chamber assays were performed to determine the changes in the proliferative and metastatic ability of A498 and 786-O cells. C-X-C motif chemokine ligand 13 (CXCL13) protein expression was detected by Western blot analysis. The targeted binding effect between miR-508-3p and circHIPK3 or CXCL13 was confirmed by constructed luciferase and RNA immunoprecipitation (RIP) assays, respectively. Fluorescence in situ hybridization (FISH) assay was used to measure the subcellular localization of circHIPK3 and miR-508-3p.
Results: It was found that circHIPK3 was markedly upregulated in ccRCC tissue and cell lines, and circHIPK3-upregulation was closely correlated with poor clinicopathological features in patients with ccRCC. It was found that both miR-508-3p and circHIPK3 were localized in the cytoplasm of ccRCC cells. The up- and downregulation of circHIPK3 positively regulated ccRCC cell proliferation and metastasis, and this regulatory effect was reversed by miR-508-3p. Through luciferase and RIP assays, it was confirmed that circHIPK3 could interacted with miR-508-3p. Furthermore, it was revealed that CXCL13, which was negatively correlated with miR-508-3p, was upregulated in ccRCC. It was also shown that CXCL13 was a downstream target of miR-508-3p. miR-508-3p suppressed ccRCC cell proliferation and metastasis by targeting CXCL13. Lastly, it was demonstrated that circHIPK3 promoted CXCL13 to facilitate ccRCC cell proliferation and metastasis by decoying miR-508-3p.
Conclusion: In brief, the results of the present study showed that circHIPK3 promoted ccRCC cell proliferation and metastasis by altering miR-5083p/CXCL13 signaling. The present findings might provide a novel target for the molecular treatment of ccRCC.
Keywords: circHIPK3, miR-508-3p, CXCL13, proliferation, metastasis, ccRCC
Introduction
As one of the most common malignancies in adults, renal cell carcinoma (RCC), which is derived from renal tubular epithelial cells, has been estimated to account for ~23.4% deaths in China.1 Among its histological subtypes, clear cell RCC (ccRCC) accounts for 70–75% of all RCC, and is responsible for the majority of kidney cancer-related mortality.2,3 There is still a great challenge in the treatment of ccRCC, particularly in patients with metastatic ccRCC. As the molecular mechanisms of ccRCC are still not completely understood, the incidence rates are on an upward trend. Therefore, it is of great urgency to thoroughly investigate the molecular mechanisms of ccRCC tumorigenesis.
As a class of stable covalently closed RNAs, circular RNA (circRNA) is highly stable and resistant to exonucleases due to their closed-ring structure without an 5`-end cap and 3`-end poly A tail.4 circRNAs are generated by back-splicing of a single pre-mRNA with gene-regulatory potential.5 As a member of the circRNA family, circRNA homeodomain interacting protein kinase 3 (circHIPK3) derives from Exon 2 of its host gene HIPK3, and is implicated in the progression of several types of cancer.6 circHIPK3 serves as an miRNA sponge absorbing several miRNAs in different types of malignancies. Zeng et al reported that circHIPK3 promoted colorectal cancer growth and metastasis by sponging miR-7.7 Through the regulation of microRNA 338-3p (miRNA-338-3p)/ADAM metallopeptidase domain 17 signaling, circHIPK3 overexpression accelerates proliferation and invasion in prostate cancer cells.8 To date, the expression and function of circHIPK3 in ccRCC remains obscure.
C-X-C motif chemokine ligand 13 (CXCL13) is a homeostatic chemokine. Aberrantly active CXCL13 has been closely involved in the development and progression of multiple advanced malignancies.9 Zheng Z reported that CXCL13 was upregulated in ccRCC, and CXCL13 promoted the proliferation and migration of ccRCC cells by binding to CXCR5.10 Through its co-working with some miRNAs, CXCL13 is implicated in certain autoimmune diseases, such as myasthenia gravis or osteonecrosis.11–13 However, whether CXCL13 could interacted with any circRNAs remains largely unknown.
In the present study, the role of circHIPK3 in ccRCC was determined. It was found that circHIPK3 functioned as an oncogene and promoted ccRCC cell proliferation and metastasis through circHIPK3-upregulation by absorbing miR-508-3p.
Materials and Methods
Patients and Tissue Samples
A total of 50 paired ccRCC and adjacent paratumor tissues were collected from patients through surgical resection at the Shengjing Hospital of China Medical University (Shenyang, China). All patients were clinically diagnosed by a definite pathological diagnosis. All patients signed written informed consent, and permission for this study was granted by the Institute Research Medical Ethics Committee of the Shengjing Hospital of the China Medical University. Patients with ccRCC were classified as stage I, II, III and IV, using the TNM staging system (version 8).
Cell Culture, Plasmid Construction and Cell Transfection
Three human ccRCC cell lines (A498, 786-O and 769-P) and a human renal proximal tubular epithelial HK2 cell line (HRPTEpiC) were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) at a constant temperature of 37°C in a humidified atmosphere containing 5% CO2.
In order to silence circHIPK3, shRNA target circHIPK3 or negative control (shNC), was inserted into a pLKO.1 vector (Biosettia). CircHIPK3 overexpression plasmids containing wild-type or mutant miR-508-3p-binding sequences (circHIPK3 and mut-circHIPK3), as well as CXCL13 overexpression plasmids (oeCXCL13), were synthesized by Shanghai GenePharma Co., Ltd. miR-508-3p mimics (miR-508-3p) and negative control (miR-NC), as well as miR-508-3p inhibitors (inh-508-3p) and negative control (inh-NC), were also purchased from Shanghai GenePharma Co., Ltd. All plasmids or oligonucleotides were transfected into A498 and 786-O cells using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer’s instructions.
GEO Database Reanalysis
The differentially expressed miRNA data in ccRCC were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116251). Differentially expressed miRNAs between 18 ccRCC tissues and 18 paired ccRCC normal tissues were reanalyzed using an online software GEO2R. The expression of miR-508-3p in 18 ccRCC tissues and 18 paired ccRCC normal tissues were also reanalyzed in GSE116251.
Actinomycin D Assay and RNase R Treatment
Actinomycin D assay and RNase R were used to detect the stability of circHIPK3. For the Actinomycin D assay, the procedure was performed as previously described.14 A498 cells were exposed to 2 μg/mL actinomycin D (Merck KGaA) at different time points. Total RNA from different time points was extracted, and the expression of circHIPK3 and linear HIPK3 mRNA was determined by reverse transcription quantitative polymerase chain reaction (RT-qPCR). The RNase R treatment procedure was carried out as previously described.7 Total RNA (2 mg) was incubated for 30 min at 37°C with or without 5 U/μg RNase R (Epicentre Technologies Pvt. Ltd), and subsequently purified using an RNeasy MinElute Cleaning Kit (Qiagen, Inc.). Analysis was then performed by RT-qPCR.
RNA Extraction and RT-qPCR
The procedures were performed as previously described.15 Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific, Inc.). miRNAs were extracted using an miRcute miRNA Isolation Kit (Tiangen Biotech Co., Ltd.). Cytoplasmic and nuclear RNAs were separated using a PARIS Kit (Thermo Fisher Scientific, Inc.). An miScript Reverse Transcription Kit (Qiagen, Inc.) was used for reverse transcription with 500 ng total RNA treated by RNase R or not. RT-qPCR was performed with the diluted cDNA products, special primers and SYBR Green mix (Thermo Fisher Scientific, Inc.). GAPDH was used as endogenous control for circRNAs and mRNAs. U6 was used as endogenous control for miRNAs, respectively. Primer sequences are listed in Table 1.
 |
Table 1 Primer Sequences Used for qRT-PCR |
In situ Hybridization
The procedures were carried out as previously described.16 ccRCC tissues slices (5-μm thick) were 10% formalin-fixed, paraffin-embedded, dewaxed and rehydrated. The slices were then digested with 20 μg/mL proteinase K for 30 min; 4% paraformaldehyde-fixed slices were hybridized with 8 ng/μL specific antisense oligonucleotide DNA probe (synthesized by Thermo Fisher Scientific, Inc.) overnight at 55°C. The samples were incubated with horseradish peroxidase (Merck KGaA) at 4°C for 30 min. Hybridization signals were amplified with diaminobenzidine (Merck KGaA), and images were captured using a fluorescent microscope (Leica Microsystems, Inc.).
RNA Fluorescence in situ Hybridization (RNA-FISH)
The procedures were performed as previously described.15 A498 and 786-O cells were first fixed in 4% formaldehyde solution and then incubated with 0.1% Triton X-100. Fluorescence-conjugated circHIPK3 or miR-508-3p probes (Thermo Fisher Scientific, Inc.) were hybridized at 37°C with cells in the dark for 5 h. The cells were then photographed by laser scanning confocal microscopy (Carl Zeiss AG).
Cell Counting Kit-8 (CCK8) Assay
The procedure was carried out as previously described.16 Briefly, A498 and 786-O cells after different interventions were incubated in 96-well plates (2x103), supplemented with 200 µL culture medium and conditioned in 37°C with 5% CO2. On days 1, 2, 3, 4 and 5, 20 μL CCK-8 solution was added into each well, and incubation was performed for 2 h. Absorbance was measured at an optical density of 450 nm using a Microplate reader (Bio-Rad Laboratories, Inc.). Experiments were repeated in triplicate.
Transwell Assay
The procedure was performed as previously reported.17 A498 and 786-O cells (with an incubation density of 5x104) were incubated in the upper chambers (Corning). For the invasion assay, the upper chambers were pre-coated with Matrigel (BD Biosciences). Culture medium without and with 10% FBS was added into the upper and lower chambers, respectively. After 12 h, non-migrated cells were wiped out while migrated or invaded CRC cells were fixed, stained and counted using an inverted microscope (Olympus Corporation).
Western Blot Analysis
The procedure was carried out as previously described.7 Radio immunoprecipitation assay lysis buffer (Merck KGaA) was used to split cells, and total protein was extracted. Samples (20 μg) were separated in 10% SDS-PAGE gels, transferred onto a PVDF membrane (EMD Millipore) and blocked for 1 h at room temperature. The membranes were incubated with primary antibodies (CXCL13 concentration, 0.5 µg/mL; GAPDH dilution rate, 1:500; Abcam) at 4°C overnight. The next day, the membranes were incubated with the secondary antibody (Abcam; dilution rate, 1:2000) at 24°C for 1 h. Signals of targeted proteins were detected using an enhanced chemiluminescence detection system.
Dual Luciferase Reporter Assay
The procedure was performed as previously described.15 Wild-type or mutant circHIPK3 or 3ʹ untranslated region (3ʹ-UTR) of CXCL13 was synthesized and then subcloned into pmirGLO (Promega Corporation). miR-508-3p containing the binding sites of circHIPK3 was also subcloned into pmirGLO luciferase reporter vector. Meanwhile, CXCL13 3ʹ-UTR containing miR-508-3p response elements (MRE-508-3p) was also subcloned into pmirGLO luciferase reporter vector. A498 and 786-O cells were seeded onto 24-well plates at a concentration of 3x104 per well and co-transfected with miR-508-3p mimics (miR-508-3p)/negative control miRNA (miR-NC) and pmirGLO-wt-circHIPK3 (wt)/pmirGLO-mut-circHIPK3 (mut), pmirGLO-wt-CXCL13 (WT)/pmirGLO-mut-CXCL13 (MUT). After 48 h, luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega Corporation), according to the manufacturer’s instructions.
RNA Immunoprecipitation (RIP) Assay
The procedure was performed as previously described.15 786-O or A498 cells were collected and lysed using a Magna RIP Kit (EMD Millipore), and then incubated with protein G Sepharose beads (GE Healthcare) coated with an anti-AGO2 antibody (Abcam) at 4°C overnight; an anti-IgG antibody was used as the negative control. RNA was then isolated for RT-qPCR, as mentioned above.
Statistical Analysis
All results are expressed as the mean ± SEM. Statistical analysis was performed using GraphPad Prism V5.0 (GraphPad Software, Inc.) software and one-way analysis of variance. Overall survival differences between patients with high or low levels of circHIPK3 expression were analyzed using Kaplan–Meier method and Log rank test. *P<0.05, **P<0.01 and ***P<0.001 were considered to indicate a statistically significant difference.
Results
circHIPK3 Is Upregulated and Correlated with Poor Prognosis in ccRCC
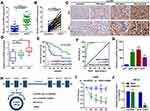
The circHIPK3 expression was detected in 50 paired ccRCC and paratumor tissue specimens. According to the RT-qPCR results displayed in Figure 1A and B, circHIPK3 was clearly upregulated in ccRCC tissues (n=50; P<0.0001). Meanwhile, an RNA ISH assay showed that circHIPK3 was gradually elevated in patients with an advanced TNM staging (Figure 1C). In addition, circHIPK3 was found to be upregulated in patients with lymph-node metastasis (Figure 1D). Furthermore, the high expression of circHIPK3 was closely associated with a shorter overall survival in patients with ccRCC (n=25 per group; P=0.0191; Figure 1E). In addition, receiver operating characteristic curve analysis demonstrated that circHIPK3 was a valuable biomarker in ccRCC (area under the curve, 0.95322; Figure 1F). The elevated expression of circHIPK3 and the clinicopathological features of patients with ccRCC were also analyzed. As shown in Table 2, the high expression of circHIPK3 was closely correlated with a high TNM grade (P=0.001), lymph node metastasis (P=0.037), distant metastasis (P=0.022), a bigger tumor size (P=0.021) and a higher Fuhrman grade (P=0.004). Expression of circHIPK3 at the cellular level was also measured by RT-qPCR. As shown in Figure 1G, cicrHIPK3 was significantly upregulated in three human ccRCC cell lines (A498, 786-O and 769-P) as compared with that in a human renal proximal tubular epithelial HK2 cell line. Since circHIPK3 was more highly expressed in A498 and 786-O cells, these two cell lines were selected in the following cellular detections. In addition, it was demonstrated that circHIPK3 was derived from exon 2 of linear HIPK3, as previously reported (Figure 1H). Finally, it was verified that circHIPK3 was more stable than linear HIPK3 mRNA, as determined by the Actinomycin D (Figure 1I) and RNase R (Figure 1J) assays.
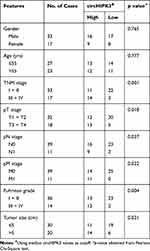 |
Table 2 Association of circHIPK3 Expression with Clinicopathological Features of Patients with ccRCC |
circHIPK3 Promotes Proliferation, Migration and Invasion in A498 and 786-O Cells
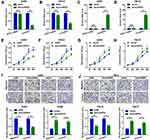
First, circHIPK3-shRNA and circHIPK3-overexpression plasmids (oecircHIPK3) were transfected into A498 and 786-O cells to knock down or overexpress circHIPK3. The transfection efficiency was detected by RT-qPCR, as shown in Figure 2A–D. Next, CCK8 assay was performed to measure the proliferation ability changes in A498 and 786-O cells. As shown Figure 2E and F, circHIPK3-knockdown (shcircHIPK3) suppressed A498 and 786-O cell proliferation and vice versa (Figure 2G and H). Finally, circHIPK3-overexpression promoted migration and invasion in A498 and 786-O cells, as determined by the Transwell assay (Figure 2I and J).
circHIPK3 Sponges miR-508-3p in A498 and 786-O Cells
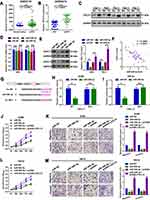
circRNAs are mainly located in the cytoplasm and serve as miRNA sponges in cells. It was primarily found that circHIPK3 was mainly located in the cytoplasm of A498 and 786-O cells (Figure 3A–C). Two online prediction software Circular RNA interactome and circBank (http://www.circbank.cn/) were used to infiltrate potential miRNAs that might interact with circHIPK3.18 A total of 5 miRNAs that overlapped among aforementioned databases were identified, including miR-508-3p (Figure 3D). The expression levels of the 5 miRNAs in ccRCC tissues were determined by a RT-qPCR. miR-508-3p was selected for further study, as it was stably downregulated in the collected tissue samples and cells lines (Figure 3E and G). An in-depth analysis of GEO datasets also indicated that miR-508-3p was downregulated in ccRCC (GSE116251; probe ID of hsa-miR-508-3p; Figure 3F). It was then verified by RNA FISH assay that circHIPK3 and miR-508-3p were co-localized in the cytoplasm in A498 and 786-O cells (Figure 3H). Furthermore, it was shown that circHIPK3 was inversely correlated with miR-508-3p (Figure 3I). Functionally, it was revealed that the up- and downregulation of circHIPK3 negatively regulated the miR-508-3p expression (Figure 3J). Finally, it was verified that miR-508-3p targeted circHIPK3 via directly binding (Figure 3K–M).
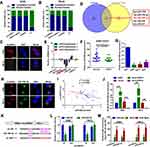 |
Figure 3 circHIPK3 sponges miR-508-3p in A498 and 786-O cells. (A and B) circHIPK3 was mainly expressed in the cytoplasm rather than the nucleus in 786-O and A498 cells, as detected by RT-qPCR. (C) RNA-FISH assay confirmed that circHIPK3 was mainly located in the cytoplasm. (D) miRNAs that might interact with circHIPK3 were predicted using two online prediction software [circBank (http://www.circbank.cn/) and CircularRNA Interactome (https://circinteractome.nia.nih.gov/)], and 5 miRNAs, including miR-508-3p, were primarily filtered out. (E) The expression of miRNAs in 3 collected tissue samples was qualified by RT-qPCR. (F) The expression of miR-508-3p in GSE116251 was analyzed using an online software (GEO2R). *P<0.05. (G) circHIPK3 at the cellular level was explored by RT-qPCR. **P<0.01. (H) Subcellular localization of circHIPK3 and miR-508-3p in A498 and 786-O cells was verified by RNA-FISH assay. (I) circHIPK3 was inversely correlated with miR-508-3p, as determined by a Spearman correlation analysis. r2=0.1243 and P=0.0120. (J) circHIPK3 negatively regulated miR-508-3p expression in A498 and 786-O cells, as determined by RT-qPCR. ***P<0.001. (K) Diagram of constructed luciferase reporter plasmids. (L) Compared with miR-NC, the co-transfection of wild reporter plasmids (WT) and miR-508-3p led to a marked cancellation of fluorescence. *P<0.05. n.s, P>0.05. (M) Enrichment of circHIPK3 and miR-508-3p in Ago2-containing beads of 786-O and A498 cells, as determined by RIP assay and subsequent RT-qPCR. **P<0.01. All data are presented as the mean ± SD from three independent experiments. miRNA, microRNA; circHIPK3, circRNA homeodomain interacting protein kinase 3; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; RNA-FISH, RNA fluorescence in situ hybridization; RIP, RNA immunoprecipitation. |
miR-508-3p Suppresses Proliferation, Migration and Invasion by Targeting CXCL13 in A498 and 786-O Cells
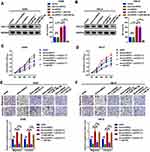
CXCL13 is implicated in the proliferation and metastasis of multiple types of cancer, including ccRCC. First, the expression of CXCL13 in ccRCC was determined by analyzing GEO datasets GSE53757 and GSE66272. As shown in Figure 4A and B, CXCL13 was significantly upregulated in ccRCC, as compared with that in normal kidney and adjacent nontumor tissues. CXCL13 was also found to be upregulated in ccRCC tissues (Figure 4C). It was then verified that miR-508-3p inversely regulated CXCL13 at the protein level (Figure 4D and E). Meanwhile, it was found that miR-508-3p was negatively correlated with the mRNA expression of CXCL13 (Figure 4F). Furthermore, through a luciferase assay, it was demonstrated that miR-508-3p targeted CXCL13 directly (Figure 4G–I). Functionally, it was found that miR-508-3p suppressed proliferation, migration and invasion in A498 and 786-O cells, and that that suppressive effect was reversed by the upregulation of CXCL13 (miR-508-3p + pcDNA-CXCL13) (Figure 4J–M).
circHIPK3 Promotes CXCL13 Expression by Decoying miR-508-3p in A498 and 786-O Cells
In the current section, we attempted to determine whether circHIPK3 could regulate CXCL13 by decoying miR-508-3p. Constructed wild-type and mutant (with mutated MRE-508-3p) circHIPK3-overexpression plasmids (circHIPK3 and mut-circHIPK3), as well as miR-508-3p mimics (miR-508-3p) were transfected into A498 and 786-O cells. miR-NC was used as a control. As shown in Figure 5A and B, wild-type (circHIPK3) circHIPK3-overexpression plasmids promoted CXCL13 protein expression, but mutant (mut-circHUIPK3) ones did not. In addition, the facilitating effect of circHIPK3 was significantly attenuated by the upregulation of miR-508-3p (co-transfection of circHIPK3 and miR-508-3p). Finally, functional CCK8 and Transwell assays were used to detect the proliferative and metastatic ability changes in ccRCC cells. shcircHIPK3 suppressed proliferation in ccRCC, and that suppressive effect was reversed by the upregulation of CXCL13 (shcircHIPK3 + oeCXCL13). More convincingly, when miR-508-3p (shcircHIPK3 + oeCXCL13 + miR-508-3p) was further upregulated, the proliferation ability was again weakened (Figure 5C and D). The Transwell assay results suggested a similar tendency (Figure 5E and F). These phenomena indicated that circHIPK3 promoted ccRCC cell proliferation, migration and invasion through circHIPK3-upregulation by decoying miR-508-3p.
Discussion
circRNA is a category of RNA transcript with covalently closed continuous loops of single-stranded structures and limited protein coding abilities.4,19 circRNAs are broadly expressed in the cytoplasm of eukaryotic cells.6 Recent studies have reported the dysregulation of circRNAs in ccRCC and their association with malignant behavior.20,21 As a member of the circRNA family, circHIPK3 is a recently identified circRNA, shown to be implicated in the progression of various types of cancer and relevant to human pathologies.6 In the present study, the biological and clinical role of circHIPK3 in ccRCC was explored. First, it was found that the expression of circHIPK3 was markedly upregulated in the 50 paired ccRCC tissue specimens that were collected. A further clinical study indicated that circHIPK3 was closely correlated with bad clinicopathological features in patients with ccRCC. Functional CCK8 and Transwell chamber assays suggested that circHIPK3 played a key role in the regulation of proliferation, migration and invasion in ccRCC cells. Next, the underlying mechanism of circHIPK3 in ccRCC cells was determined. A previous study has suggested that most circRNAs exert their various biological functions through their co-working with certain miRNAs, by acting as competing endogenous RNAs.22 Through an online bioinformatics prediction, it was theoretically predicted that miR-508-3p might be a potential target of circHIPK3.
miRNAs are small non-coding RNAs measuring 22–25 nucleotides in length. miRNAs are widely expressed and play crucial roles in various biological processes, including cell growth, apoptosis, cell-cycle control, cell differentiation and metastasis.23,24 miR-508-3p, which contains one exon, is located at human chromosome Xq27.3. miR-508-3p acts as a tumor suppressor by targeting diverse downstream genes in multiple malignancies, including breast, cervical, colorectal and ovarian cancer.25–28 Zhai et al reported that miR-508-3p was closely associated with cell invasion and migration and involved in the apoptosis of ccRCC.29 In the present study, it was primarily found that miR-508-3p was downregulated in ccRCC by analyzing the collected tissue samples and GEO dataset GSE116251. It was then verified that circHIPK3 and miR-508-3p were co-located in the cytoplasm in ccRCC cells. Through a luciferase and RIP assay, it was proven that circHIPK3 could interacted with miR-508-3p via directly targeting.
Meanwhile, the association between miR-508-3p and CXCL13, a downstream target of miR-508-3p, was detected. Aberrantly active CXCL13 and its downstream CXCR5 signaling has been implicated in the development and progression of several types of advanced human cancer.9 Previous studies have indicated that CXCL13 has good diagnostic and prognostic value and may become a candidate biomarker and therapeutic target for ccRCC.30,31 It was also demonstrated herein that CXCL13 was upregulated in ccRCC. miR-508-3p was found to target CXCL13 at the post-transcriptional level. Functionally, it was showed that miR-508-3p suppressed ccRCC cell proliferation and metastasis via the inhibition of CXCL13. Since a circRNA might sponge one or more miRNAs and further regulate multiple mRNAs, we further explored whether circHIPK3 could regulate CXCL13 by absorbing miR-508-3p. The theoretical MRE-508-3p was mutated in circHIPK3, and the CXCL13 protein expression was measured. It was found that circHIPK3 promoted CXCL13 protein expression. Both the mutation of MRE-508-3p in circHIPK3 and the upregulation of miR-508-3p were found to cripple the facilitating effect of circHIPK3 on CXCL13, which strongly supported our hypothesis that circHIPK3 regulated CXCL13 via miR-508-3p sponging.
In conclusion, the findings of the present study suggested that circHIPK3 promoted proliferation, migration and invasion through circHIPK3-upregulation by miR-508-3p sponging. The circHIPK3/miR-508-3p/CXCL13 axis might be a novel therapeutic target for the molecular treatment of ccRCC.
Acknowledgments
This work was supported by grants from the Technological Innovation fund of the Shenyang Technology Division (grant no. 18-014-4-06).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3(1):17009. doi:10.1038/nrdp.2017.9
3. Vera-Badillo FE, Conde E, Duran I. Chromophobe renal cell carcinoma: a review of an uncommon entity. Int J Urol. 2012;19(10):894–900. doi:10.1111/j.1442-2042.2012.03079.x
4. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
5. Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi:10.1016/j.molcel.2015.03.027
6. Xie Y, Yuan X, Zhou W, et al. The circular RNA HIPK3 (circHIPK3) and its regulation in cancer progression: review. Life Sci. 2020;117252.
7. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
8. Cai C, Zhi Y, Wang K, et al. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi:10.2147/OTT.S196931
9. Hussain M, Adah D, Tariq M, Lu Y, Zhang J, Liu J. CXCL13/CXCR5 signaling axis in cancer. Life Sci. 2019;227:175–186. doi:10.1016/j.lfs.2019.04.053
10. Zheng Z, Cai Y, Chen H, et al. CXCL13/CXCR5 axis predicts poor prognosis and promotes progression through PI3K/AKT/mTOR pathway in clear cell renal cell carcinoma. Front Oncol. 2018;8:682. doi:10.3389/fonc.2018.00682
11. Li J, Qiu D, Chen Z, Du W, Liu J, Mo X. miR-548k regulates CXCL13 expression in myasthenia gravis patients with thymic hyperplasia and in Jurkat cells. J Neuroimmunol. 2018;320:125–132. doi:10.1016/j.jneuroim.2018.03.021
12. Wu DM, Wen X, Han XR, et al. Micro-RNA-143 inhibits proliferation and promotes apoptosis of thymocytes by targeting CXCL13 in a myasthenia gravis mouse model. Am J Physiol Cell Physiol. 2019;316(1):C70–C80. doi:10.1152/ajpcell.00090.2018
13. Xu W, Li J, Tian H, et al. MicroRNA1865p mediates osteoblastic differentiation and cell viability by targeting CXCL13 in nontraumatic osteonecrosis. Mol Med Rep. 2019;20(5):4594–4602. doi:10.3892/mmr.2019.10710
14. Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10(1):3200.
15. Chen Z, Xiao K, Chen S, Huang Z, Ye Y, Chen T. Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 2020;111(2):713–726.
16. Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi:10.1186/s12943-018-0837-6
17. Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108(5):859–867.
18. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
19. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
20. Feng J, Chen K, Dong X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18(1):35. doi:10.1186/s12943-019-0996-0
21. Ma C, Qin J, Zhang J, Wang X, Wu D, Li X. Construction and analysis of circular RNA molecular regulatory networks in clear cell renal cell carcinoma. Mol Med Rep. 2020;21(1):141–150. doi:10.3892/mmr.2019.10811
22. Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149. doi:10.1038/s41467-017-01216-w
23. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
24. Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10.
25. Guo SJ, Zeng HX, Huang P, Wang S, Xie CH, Li SJ. MiR-508-3p inhibits cell invasion and epithelial-mesenchymal transition by targeting ZEB1 in triple-negative breast cancer. Eur Rev Med Pharmacol Sci. 2018;22(19):6379–6385. doi:10.26355/eurrev_201810_16050
26. Hu P, Zhou G, Zhang X, Song G, Zhan L, Cao Y. Long non-coding RNA Linc00483 accelerated tumorigenesis of cervical cancer by regulating miR-508-3p/RGS17 axis. Life Sci. 2019;234:116789. doi:10.1016/j.lfs.2019.116789
27. Wang Z, Jin J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging. 2019;11(14):4876–4889. doi:10.18632/aging.102081
28. Zhao L, Wang W, Xu L, et al. Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer. Oncogene. 2019;38(13):2305–2319.
29. Zhai Q, Zhou L, Zhao C, et al. Identification of miR-508-3p and miR-509-3p that are associated with cell invasion and migration and involved in the apoptosis of renal cell carcinoma. Biochem Biophys Res Commun. 2012;419(4):621–626. doi:10.1016/j.bbrc.2012.02.060
30. Wang Y, Zheng B, Xu M, et al. Prediction and analysis of hub genes in renal cell carcinoma based on CFS gene selection method combined with Adaboost algorithm. Med Chem. 2019;15. doi:10.2174/1573406415666191004100744
31. Xu T, Ruan H, Song Z, et al. Identification of CXCL13 as a potential biomarker in clear cell renal cell carcinoma via comprehensive bioinformatics analysis. Biomed Pharmacother. 2019;118:109264. doi:10.1016/j.biopha.2019.109264
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.