Back to Journals » Cancer Management and Research » Volume 12
CircCFL1/MiR-107 Axis Targeting HMGB1 Promotes the Malignant Progression of Diffuse Large B-Cell Lymphoma Tumors
Received 28 May 2020
Accepted for publication 30 July 2020
Published 30 September 2020 Volume 2020:12 Pages 9351—9362
DOI https://doi.org/10.2147/CMAR.S263222
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Xiaowei Chen,1,* Xiaobin Xie,2,* Wei Zhou1
1Department of Hematology, Guangzhou First People’s Hospital, South China University of Technology, Guangzhou 510080, Guangdong, People’s Republic of China; 2Department of Pathology, School of Basic Medical Science, Guangzhou Medical University, Guangzhou 511436, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Zhou Department of Hematology
Guangzhou First People’s Hospital, South China University of Technology, No. 1 Panfu Road, Guangzhou 510080, Guangdong, People’s Republic of China
Tel +86-20-81048386
Email [email protected]
Objective: The pathogenesis of diffuse large B-cell lymphoma (DLBCL) has not yet been fully elucidated. An increasing number of studies have shown that circular RNAs (circRNAs) play an important role in tumorigenesis and development. The aim of this study was to investigate the effect of CircCFL1 on the malignant progression of DLBCL.
Methods: RT-qPCR was used to detect the expression levels of CircCFL1 and miR-107. A dual-luciferase reporter gene experiment was conducted to verify that CircCFL1 targeted miR-107 and the miR-107 target gene HMGB1. BrdU, transwell, and MTT tests were performed to detect cell invasion and proliferation. Western blot analysis was used to detect the phosphorylation of proteins. Xenograft models were established to confirm the effect of CircCFL1 on DLBCL tumor growth in vivo.
Results: The expression of CircCFL1 in cells transfected with the CircCFL1 overexpression vector was higher than that in the control group. After overexpressing CircCFL1, the expression of miR-107 in cells decreased significantly, and the protein level of HMGB1 increased. The dual-luciferase reporter gene experiment showed that CircCFL1 directly bound to miR-107 and reduced the inhibition of the target gene HMGB1. After CircCFL1 was overexpressed, cell migration and proliferation were enhanced. The tumor volume and weight in the lentivirus CircCFL1 group were higher than those in the lentivirus NC group.
Conclusion: Results showed that the circRNA CircCFL1 could regulate the expression of HMGB1 through miR-107 to promote the proliferation and migration of DLBCL.
Keywords: circular RNA, CircCFL1, lymphoma, HMGB1, proliferation, migration
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin B-cell lymphoma in adults.1,2 DLBCL has considerable heterogeneity in morphologies, immunophenotypes, genetic characteristics, and clinical manifestations. In accordance with gene expression profiles, DLBCL can be divided into germinal center B-cell type (GCB type) with good prognosis and activated B-cell type (ABC type).3,4 The difference in prognosis between GCB and ABC is due to their different pathways. The difference in pathways is subsequently attributed to the abnormal expression of proteins or the broken rearrangement, amplification, and translocation of key genes. However, the pathogenesis of DLBCL has not yet been fully elucidated.5
Competitive endogenous RNA (ceRNA) is a transcript that achieves mutual regulation by miRNA response elements that are competing to bind to common miRNAs.6,7 Circular RNAs (circRNAs) are a type of noncoding RNAs that are produced by special selective shearing. Given their closed circular structures, circRNAs lack free ends and are unaffected by RNA exonucleases and are thus more stable than linear RNAs. Few studies have been conducted on circRNAs, and their underlying mechanism has not been elucidated. Nevertheless, experiments have revealed that some circRNAs are rich in miRNA-binding sites and play a competitive role in binding to microRNAs (miRNAs), which are a class of highly efficient ceRNAs in cells. MiRNAs are a class of endogenous single-stranded noncoding regulatory small RNA molecules; they have wide distribution and highly conserved sequences and are approximately 18–25 nucleotides in length.8,9 MiRNAs regulate gene expression and participate in a series of important biological processes, such as cell development, proliferation, differentiation, and apoptosis. MiRNAs have specific expression levels and patterns in DLBCL. Changes in miRNA expression can cause the abnormal expression of target genes, thereby affecting the occurrence and development of DLBCL.10
HMGB1 is an ubiquitous DNA-binding protein that is involved in maintaining gene stability and regulating DNA transcription and repair.11,12 Under the influence of injury, infection, chemotherapy, and other factors, HMGB1 can be used as a DAMP to bind to RPR, TLR, CXCL12, and other PRR receptors by activating CDC42, MyD88/TRAF6, NF-κB, MAPK, and the antiapoptotic protein c-IAP; this phenomenon leads to the mediation of inflammatory responses, angiogenesis, tumor cell growth, and metastasis and consequently promotes tumor development.13 The current study found that HMGB1 played an important role in lymphoma development. Kang et al.14,15 conducted immunohistochemical tests and found that the expression level of HMGB1 in the lymph nodes of patients with non-Hodgkin’s lymphoma is significantly higher than that in normal lymph nodes.
In this study, the expression of CircCFL1 was up-regulated by transfecting a circular RNA overexpression vector. The role of CircCFL1 in the proliferation and migration of DLBCL was investigated to provide a new target for clinical treatment.
Methods
Cell Lines and Cell Cultures
The human DLBCL cell lines OCI-Ly7 and OCI-Ly3 were purchased from the ATCC. The cell lines were routinely cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 2.0 mM L-glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin. All cells were cultured in a humidified incubator at 37 °C with 5% CO2. The cells were transfected by using the transfection reagent Lip2000 in accordance with the manufacturer’s instructions. After transfection for 48 h, the cells were harvested for subsequent experiments.16
Cell Transfection
Si-CircCFL1, vector-CircCFL1 (pc-DNA3.1-CircCFL1), miR-107 mimics, si-HMGB1, and corresponding negative control groups were all purchased from Ruibo (Shanghai, China). Lentivirus CircCFL1 overexpression vector and lentivirus-NC were constructed by OBiO Technology Corp., Ltd. (Shanghai, China). OCI-Ly7 and OCI-Ly3 cell lines were transfected with these above agents by using Lipofectamine 3000 reagent (Invitrogen). The OCI-Ly7 and OCI-Ly3 cells were divided into four groups: si-NC group, si-CircCFL1 group, NC vector group, and vector-CircCFL1 group. Furthermore, the OCI-Ly7 cell line was divided into the NC vector, CircCFL1, CircCFL1+miR-NC, CircCFL1+miR-107, CircCFL1+si-NC, and CircCFL1+si-HMGB1 groups. Transfection efficiency was detected via RT-qPCR.
Quantitative PCR
TRIzol was used to extract total RNA from the cells. The integrity of the extracted RNA was determined via agarose gel electrophoresis. RNA concentration and purity were determined by using a microplate reader. cDNA synthesis was performed in accordance with the instructions of the reverse transcription kit, and qPCR was completed by using a Bio-Rad real-time fluorescence quantitative PCR instrument. The primer sequences of HMGB1 are as follows: HMGB1F: 5ʹ-TGCAGATGACAAGCAGCCTT-3ʹ and R: 5ʹ-GCTGCATCAGGCTTTCCTTT-3ʹ. The primer sequences of β-actin are F: 5ʹ-CCTGTACGCCAACACAGTGC-3ʹ and R: 5ʹ-ATACTCCTGCTTGCTGATCC-3ʹ. The primer sequences of miR-107 are F: 5ʹ-GGAGCAGCATTGTACAGG-3ʹ and R: 5ʹ-CAGTGCGTGTCGTGGA-3ʹ. The U6 primer sequences are F: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and R: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ. For the quantification of circRNA and miRNA, β-actin and U6 were used as internal controls, respectively. The reaction conditions were as follows: predenaturation at 95 °C for 10 min; 95 °C for 10 s, and 60 °C for 60 s. A total of 40 cycles were performed. Three replicate wells were set for each gene, and gene expression was calculated by using the 2–ΔΔCt method. The experiment was repeated three times.
Western Blot Analysis
9The cells were cultured for 48 h after transfection. Total protein was extracted from the cells. Approximately 25 μg of the sample was processed through 12% polyacrylamide gel electrophoresis and then transferred onto a PVDF membrane. The membrane was placed in blocking solution and shaken slowly at 37 °C for 2 h. After blocking, the membranes were incubated with the primary antibodies AKT (Abcam, 1: 1000), p-AKT (Abcam, 1: 500), ERK (Abcam, 1: 1000), p-ERK (Abcam, 1: 500), STAT3 (Abcam, 1: 1000), p-STAT3 (Abcam, 1: 500), and actin (Abcam, 1: 2000) at 4 °C overnight. The membrane was then incubated with the secondary antibody IgG–HRP (Abcam, 1: 1000) at 37 °C for 2 h. ECL chemiluminescence was developed, and the gel image was processed for analysis. The relative expression of the target protein was obtained by calculating the ratio of the absorbance of the target protein to the internal reference. The experiment was repeated three times.
Dual-Luciferase Reporter Gene Experiment
Specially designed upstream and downstream circularization frameworks were inserted into the circRNA overexpression vector pCDHCMV-MCS-EF1-copGFP. The upstream circularization framework contained endogenous flanking genomic sequences and EcoRI restriction sites. The downstream circularization framework contained partially inverted upstream sequences and BamHI restriction sites. The CircCFL1 sequence was amplified, and the amplified fragment was connected between the upstream and downstream circularization frameworks of the vector. An empty vector without the CircCFL1 sequence was constructed. The psiCHECK2 dual-luciferase reporter gene vector was purchased from Shanghai Hanheng Biotechnology Co., Ltd. The CircCFL1 sequence was cloned into the psiCHECK2 vector, which was named as psiCHECK2-CircCFL1-WT. The CircCFL1 sequence with the miR-107 binding site mutation was synthesized and cloned into the psiCHECK2 vector. This construct was designated as psiCHECK2-CircCFL1-Mut. The HMGB1 3ʹUTR containing the miR-107 binding site was cloned into the psiCHECK2 vector. This construct was named as psiCHECK2-HMGB1-3ʹUTR-WT. The HMGB1 3ʹUTR with the mutated miR-107 binding site was cloned into the psiCHECK2 vector. This construct was designated as psiCHECK2-HMGB1-3ʹUTR-Mut. This vector used the Renilla luciferase gene as the reporter gene and the firefly luciferase gene as the internal reference gene. After vector construction was completed, the vector was identified by sequencing. The cells were transfected in groups by using Lip2000 in accordance with the instructions. Luciferase activity was detected in accordance with the manufacturer’s instructions. The luciferase gene luminescence value/firefly luciferase gene luminescence value (Rluc/Fluc) was used to represent relative luciferase activity.
Invasion Experiment
The cells were cultured in 300 μL of serum-free medium. A total of 30 μL (or 50 μg/well) of Matrigel was added to the cells. The mixture was mixed well on an ice bath at 4 °C. Approximately 100 µL (3 rooms) of the mixture was placed in the upper chamber and incubated at 37 °C for 4–5 h (>5 h). The cells were digested, washed three times with serum-free medium, counted, and prepared into a cell suspension. Then, they were washed with serum-free medium. Approximately 100 μL of the cell suspension was added to each well in the lower chamber. The chamber was filled with 600 μL of serum-free conditioned medium and incubated at 37 °C for 24 h. The transwell was removed. The cells were washed twice with PBS, fixed with 5% glutaraldehyde at 4 °C, washed twice with PBS, and stained with crystal violet (0.1%) at room temperature for 0.5 h. The stained cells were washed twice with PBS, and the upper surface cells were wiped with a cotton ball. The cells were observed under microscopy.
Scratch Test to Detect Cell Migration Capability
The cells were cultured for 48 h after transfection. When cell confluence reached 95%, a 200 μL pipette was used to create scratches. After changing the medium to serum-free medium for 24 h, the cells were observed under an inverted microscope. The relative migration capacity of the cells was determined. Three visual fields were taken for analysis in each group.
MTT Test to Detect Cell Proliferation Capability
At 48 h after transfection, the cells were seeded in 24-well plates to a cell confluence of 60%. A total of 200 µL of diluted MTT solution was added to each well, and the cells were incubated for 2 h in accordance with the instructions of the MTT kit. Absorbance was recorded by using a microplate reader.17
Establishment of the Nude Mouse Model
Ten female BALB/c nude mice (4 weeks old) were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and randomly divided into the lentivirus CircCFL1 and lentivirus NC groups (5 in each group). OCI-Ly7 cell lines (1 × 107 cells) transfected with lentivirus CircCFL1 or lentivirus NC were injected into the backs of the mice. The tumor volume was tested every week until the mice were sacrificed. Finally, xenograft tumors were removed and weighed. All animal experiments were approved by the animal ethics committee of Guangzhou First People’s Hospital. The guidelines for the welfare of experimental animals are GB/T 35892–2018 standard issued by the General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China.
Statistical Analysis
SPSS19.0 was used for statistical analysis. T-test was used to compare the means of two samples, and ANOVA was conducted to compare multiple groups. P < 0.05 means that the difference was statistically significant.18
Results
CircCFL1 Promoted the Malignant Behavior of DLBCL
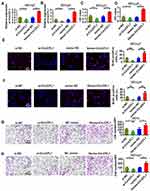
We constructed an overexpression plasmid for CircCFL1 and applied siRNA to knock down the expression of CircCFL1 to study the effect of CircCFL1 on DLBCL. The experimental groups were divided into four groups, namely, the si-NC, si-CircCFL1, vector-NC, and vector-CircCFL1 groups. CircCFL1-overexpressing vector and empty vector were transfected into the cells. Relative CircCFL1 expression was detected. The results showed that the relative expression of CircCFL1 in cells transfected with the expression vector was significantly higher than that in the control cells. CircCFL1 expression was effectively reduced in the knockdown group (Figure 1A and B). The proliferation capabilities of OCI-Ly7 and OCI-Ly3 were promoted by the overexpression of CircCFL1 and inhibited by the knockdown of CircCFL1 (Figure 1C and D). Cell proliferation and transwell tests were also conducted. The proliferation and invasion of OCI-Ly7 and OCI-Ly3 were promoted by the overexpression of CircCFL1 and the knockdown of CircCFL1 (Figure 1E–H).
Overexpression of CircTCF25 Activated the AKT/ERK Pathway
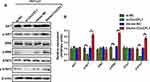
The AKT, ERK, and STAT3 signaling pathways control cell proliferation, and these molecules are actively phosphorylated. Therefore, we tested whether CircCFL1 promotes cell proliferation via the above signaling pathways. Protein phosphorylation changed after we overexpressed and knocked down CircCFL1. The overexpression of CircCFL1 up-regulated p-AKT, p-ERK, and p-STAT3, whereas the knockdown of CircCFL1 suppressed p-AKT, p-ERK, and p-STAT3 expression (Figure 2A and B).
CircCFL1 Sponged MiR-107 to Inhibit MiR-107 Expression
We conducted a predictive analysis of miRNAs bound by CircCFL1 to further verify the target gene of CircCFL1. The prediction results showed that CircCFL1 could bind to miR-107 (Figure 3A). The dual-luciferase assay results revealed that CircCFL1 directly bound to miR-107 (Figure 3B). The miR-107 expression test also proved that the overexpression of CircCFL1 inhibited the expression of miR-107. The inhibition of CircCFL1 promoted the expression of miR-107 (Figure 3C). The RIP experiment (AGO antibody probe) also confirmed that CircCFL1 bound to miR-107 (Figure 3D). The RNA pull-down experiment (miR-107 probe) showed that miR-107 was bound by CircCFL1. Compared with the control treatment, the biotin-miR-107 probe could bind to more CircCFL1 (Figure 3E).
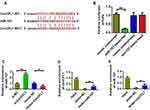 |
Figure 3 CircCFL1 sponged miR-107. (A) Information on binding sites between CircCFL1 and miR-107. Prediction was performed by using the Starbase (http://starbase.sysu.edu.cn/) website. (B) Double luciferase reporter assay confirmed that CircCFL1 bound to miR-107. (C) MiR-107 expression after CircCFL1 overexpression and knockdown was detected through qPCR. (D) RNA pull-down assay confirmed the binding of CircCFL1 to miR-107. (CircCFL1 as a probe). (E) RNA pull-down assay confirmed the binding of CircCFL1 to miR-107 (miR-107 probe). **p < 0.01, ***p < 0.001. |
MiR-107 Bound to HMGB1 to Inhibit the Expression of HMGB1
We first predicted the genes that could be bound and determined potential binding target genes through TargetScan and database prediction to study the target genes of miR-107 (Figure 4A). The dual-luciferase experimental results showed that miR-107 directly bound to HMGB1 (Figure 4B). The results of RNA pull-down experiments showed that biotin-miR-107 directly bound with HMGB1 (Figure 4C). The qPCR expression analysis results indicated that miR-107 could inhibit the expression of HMGB1. However, miR-107 inhibition promoted the expression of HMGB1 (Figure 4D). Further research showed that the overexpression of CircCFL1 promoted the expression of HMGB1, and the inhibition of CircCFL1 reduced the expression of HMGB1 (Figure 4E).
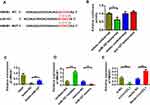 |
Figure 4 MiR-107 bound to HMGB1. (A) HMGB1 site information on miR-107 targets. Prediction was performed by using the TargetScan (http://www.targetscan.org/vert_72/) website. (B) Detection of the luciferase activities of CircCFL1-wt and CircCFL1-mut by dual-luciferase reporter gene experiment. (C) RNA pull-down assay was performed to detect whether miR-107 bound to HMGB1. (D) MiR-107 inhibited the expression of HMGB1. (E) HMGB1 expression after CircCFL1 overexpression and knockdown was detected through qPCR. *p < 0.05, **p < 0.01, ***p < 0.001. |
Overexpression of MiR-107 and Knockdown of HMGB1 Reversed the Function of CircCFL1
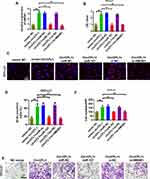
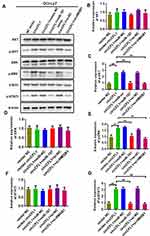
We conducted cell proliferation, migration, and invasion experiments to further study the biological role of CircCFL1. The overexpression of CircCFL1 up-regulated the expression of HMGB1, whereas miR-107 inhibited the expression of HMGB1 (Figure 5A). Cell proliferation experiments showed that the overexpression of CircCFL1 promoted cell growth, whereas the overexpression of miR-1107 and the knockdown of HMGB1 reversed the promoting effect of CircCFL1 on cell proliferation (Figure 5B). The results of cell proliferation and invasion experiments showed that CircCFL1 overexpression promoted cell proliferation and invasion capability, whereas excessive miR-107 and HMGB1 knockdown reversed CircCFL1-enhanced cell proliferation and invasion capability (Figure 5C–F). The protein expression test results also showed that CircCFL1 overexpression up-regulated the expression of p-AKT, p-ERK, and p-STAT3. By contrast, the overexpression of miR-107 or the knockdown of HMGB1 reversed the enhanced protein expression of CircCFL1 (Figure 6A–G).
Overexpression of CircCFL1 Promoted the Tumor Growth of DLBCL
We established xenograft models to further verify the role of CircCFL1 in the malignant progression of DLBCL. In vivo experiments showed that CircCFL1 overexpression enhanced the tumor growth of DLBCL (Figure 7A and B). The CircCFL1 level in the lentivirus CircCFL1 group was higher than that in the lentivirus NC group, indicating that transfection was effective. In addition, CircCFL1 overexpression could inhibit the expression of miR-107 in vivo (Figure 7C). All results indicated that the overexpression of CircCFL1 enhanced the tumor growth of DLBCL.
Discussion
DLBCL is the most common aggressive lymphoma and accounts for 30–40% of all non-Hodgkin lymphomas.19–22 Genetic abnormalities and biological changes are important factors that affect the occurrence and evolution of DLBCL. In recent years, the role of circRNAs in the course of DLBCL disease has received increasing attention. Finding new molecular markers and discovering new mechanisms are of great significance in the early diagnosis, treatment, and prognosis of DLBCL.
Although the ceRNA hypothesis has not been proposed for a long time, scholars have confirmed the role of ceRNA regulatory mechanisms in tumorigenesis in prostate, colorectal cancer, melanoma, malignant glioma, liver cancer, and breast cancers.23 CircRNAs are usually circular RNAs that are composed of exons and a new and mysterious endogenous RNA. In recent years, some studies have shown that circRNAs contain MREs with a large number of miRNAs, which can play the role of ceRNAs and become a new ceRNA member.24 With the continuous exploration and research of circRNAs, their role in tumorigenesis has been gradually recognized. CircRNA ciRS-7 can competitively bind to miR-7 to relieve or reduce the inhibition of carcinogenic factors (including the EGF receptors IRS-1, IRS-2, Pakl, Rafl, Ackl, and PIK3CD) by miR-7, increase the expression of oncogenic factors, and promote cancer progression. In rectal cancer, ciRS-7 binds to miR-7, promotes the expression of the oncogenic transcript YY1, inhibits the expression of P53, promotes cancer cell proliferation and metastasis, and inhibits cancer cell apoptosis.25,26
In this study, we first reported the function of CircCFL1 in DLBCL. The role of CircCFL1 in other tumors has not been studied. We conducted bioinformatics analysis prior to this study. Predictions were made for circRNAs. The ceRNA mechanism of CircCFL1/miR-107/HMGB1 that might exist in large B-cell lymphoma tumors was first constructed on the basis of references and by applying TargetScan (http://www.targetscan.org/vert_72/) and Starbase (http://starbase.sysu.edu.cn/index.php). Furthermore, we confirmed the role of CircCSNK1G1 in promoting cell proliferation and invasion at the cell level through pre-experiments. We further studied the function and regulatory mechanism of CircCSNK1G1 in large B-cell lymphoma tumors. We successfully constructed the CircCFL1 overexpression vector and transfected it into DLBCL cells. Up-regulating CircCFL1 could significantly inhibit the expression of miR-107, increase the protein level of HMGB1, and promote the proliferation and migration of cells. By using the dual-luciferase reporter gene system, we verified that CircCFL1 targeted the miR-107 and miR-107 target gene HMGB1. HMGB1, together with Cyclin D, phosphorylates and deactivates RB1, thereby guiding the cell cycle.27 MiR-107 negatively regulates HMGB1 and other carcinogenic factors.28,29 Our experimental results were consistent with the results of previous studies. According to bioinformatics prediction results and our experimental results, CircCFL1 can play an important regulatory function through miR-103a-3p and miR-107 in DLBCL.
HMGB1 plays an important role in lymphoma development. Staratschek et al30 found that 11 out of 18 patients with non-Hodgkin’s lymphoma have significantly higher levels of HMGB1 mRNA expression than normal controls. They speculated that HMGB1 released by necrotic cells promotes lymphoma cell growth and angiogenesis through paracrine pathways. Dejean et al31 found that HMGB1 can also induce IL-8 release by activating the MMP-9, PAR-2, and NF-κB pathways by binding to CXCR1 and CXCR2 on the surfaces of ALK-positive lymphoma cells to promote the growth and metastasis of lymphoma cells; after treatment with the HMGB1 inhibitor glycyrrhizin, the invasion and metastatic capabilities of lymphoma cells are significantly reduced.32,33 In the present study, we found that CircCFL1 can promote the expression of HMGB1 by targeting miR-107, thereby activating the HMGB1 signaling pathway and up-regulating the phosphorylation levels of p-AKT, p-ERK, and p-STAT3.
Conclusion
We initially confirmed the regulatory function of the CircCFL1/miR-107/HMGB1 pathway in DLBCL. Further studies on the function and mechanism of CircCFL1 in the development of DLBCL could provide potential new molecular targets for treatment.
Funding
Guangdong Medical Science and Technology Research Fund (A2020361)
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sauter CS, Matasar MJ, Schoder H, et al. A phase I study of ibrutinib in combination with R-ICE in patients with relapsed or primary refractory DLBCL. Blood. 2018;131(16):1805–1808.
2. Ming H, Trevino J, Yang L, Cao D, Lai JJAR. Primary gastric EBV-positive diffuse large B cell lymphoma (DLBCL) of the elderly with plasmablastic differentiation. In Vivo (Brooklyn). 2018;32(2):413–417. doi:10.21873/invivo.11255
3. Ludmir EB, Milgrom SA, Pinnix CC, Gunther JR, Nastoupil LJJLL. Primary breast diffuse large B-cell lymphoma: treatment strategies and patterns of failure. Leuk Lymphoma. 2018;59(12):2896–2903.
4. Li L, Zhang X, Zhang T, et al. Prognostic significance of BCL-2 and BCL-6 expression in MYC-positive DLBCL. Clin Lymphoma Myeloma Leuk. 2018;18(10):e381–e389. doi:10.1016/j.clml.2018.06.010
5. Lang J, Ma K, Guo J, Zhang J, Wang Q, Sun H. Clinical significance of elevated antinuclear antibodies in patients with diffuse large B-cell lymphoma: a single center study. J Cancer Res Ther. 2018;14(1):213. doi:10.4103/0973-1482.183559
6. Liu W, Ma W, Yuan Y, Zhang Y, Sun SJB. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Communications BR. 2018;500:4.
7. Jiang X, Wu X, Chen F, et al. The profiles and networks of miRNA, lncRNA, mRNA, and circRNA in benzo(a)pyrene-transformed bronchial epithelial cells. J Toxicol Sci. 2018;43(4):281–289. doi:10.2131/jts.43.281
8. Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(3):743–754. doi:10.3892/ijo.2018.4265
9. Lin F, Zhao GA, Chen ZG, et al. [Network correlation of circRNA-miRNA and the possible regulatory mechanism in acute myocardial infarction]. Zhonghua Yi Xue Za Zhi. 2018;98(11):851. Chinese.
10. Bradshaw G, Sutherland HG, Camilleri ET, Lea RA, Haupt LM, Griffiths L. Genetic and epigenetic variants in the MTHFR gene are not associated with non-Hodgkin lymphoma. Meta Gene. 2015;6:91–95.
11. Wu D, Liu J, Chen J, He H, Ma H, Lv X. miR-449a Suppresses Tumor Growth, Migration, and Invasion in non-small cell lung cancer by targeting a HMGB1-mediated NF-κB signaling pathway. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2019;27(2):227–235.
12. Tang D, Kang R, Cheh CW, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29(8):5299–5310.
13. Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2009;28(1):367–388.
14. Kang R, Tang D, Cao L, Yu Y, Zhang G, Xiao X. High mobility group box 1 is increased in children with acute lymphocytic leukemia and stimulates the release of tumor necrosis factor-alpha in leukemic cell. Zhonghua Er Ke Za Zhi= Chin J Pediatr. 2007;45(5):329–333.
15. Meyer A, Eberle N, Bullerdiek J, Nolte I, Simon DJV. High‐mobility group B1 proteins in canine lymphoma: prognostic value of initial and sequential serum levels in treatment outcome following combination chemotherapy. Pediatr Blood Cancer. 2010;8(2):127–137.
16. Zhong W, Sun B, Lu C, et al. Problems and solutions in click chemistry applied to drug probes. Sci Rep. 2016;6(1):35579. doi:10.1038/srep35579
17. Xi X, Liu N, Wang Q, et al. ACT001, a novel PAI-1 inhibitor, exerts synergistic effects in combination with cisplatin by inhibiting PI3K/AKT pathway in glioma. Cell Death Dis. 2019;10(10):757. doi:10.1038/s41419-019-1986-2
18. Zhong W, Sun B, Gao W, et al. Salvianolic acid A targeting the transgelin-actin complex to enhance vasoconstriction. EBioMedicine. 2018;37:246–258. doi:10.1016/j.ebiom.2018.10.041
19. Yang H, Qiu B, Chen S, et al. Soluble CXCL16 promotes TNF-α-induced apoptosis in DLBCL via the AMAD10-NF-κB regulatory feedback loop. Cell Biol Int. 2019;43(8):863.
20. Fuerst ML. CAR T-cell therapy induces sustained responses in DLBCL. Oncology Times. 2018;40(3):29.
21. Dunleavy K, Erdmann T, Lenz GJCTR. Targeting the B-cell receptor pathway in diffuse large B-cell lymphoma. Cancer Treat Rev. 2018;65:41.
22. Zhong W, Yang W, Qin Y, et al. 6-Gingerol stabilized the p-VEGFR2/VE-cadherin/beta-catenin/actin complex promotes microvessel normalization and suppresses tumor progression. J Exp Clin Cancer Res. 2019;38(1):285. doi:10.1186/s13046-019-1291-z
23. Xie Y, Chen T, Luo J, et al. Mechanism of circRNA and its effect on development of animal muscles. 2018;8:2270–2275.
24. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
25. Hansen TB, Kjems J, Damgaard C. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612.
26. Zhou B, Zong S, Zhong W, et al. Interaction between laminin-5gamma2 and integrin beta1 promotes the tumor budding of colorectal cancer via the activation of Yes-associated proteins. Oncogene. 2019.
27. Choi YJ, Anders LJO. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33(15):1890–1903.
28. Kim DS, Lee SY, Lee JH, Bae YC, Jung JSJE. MicroRNA-103a-3p controls proliferation and osteogenic differentiation of human adipose tissue-derived stromal cells. Medicine. 2015;47(7):e172.
29. Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. J Clin Oncol. 2012;29(2):856–863.
30. Meyer A, Staratschek-Jox A, Springwald A, et al. Non-Hodgkin lymphoma expressing high levels of the danger-signalling protein HMGB1. Leuk Lymphoma. 2008;49(6):1184–1189.
31. Dejean E, Foisseau M, Lagarrigue F, et al. ALK+ ALCLs induce cutaneous, HMGB-1–dependent IL-8/CXCL8 production by keratinocytes through NF-κB activation. Blood. 2012;119(20):4698–4707.
32. Lotze MT, Demarco RAJCOID. Dealing with death: HMGB1 as a novel target for cancer therapy. Curr Opin Investig Drugs. 2004;4(12):1405–1409.
33. Chung HW, Lee SG, Kim H, et al. Serum high mobility group box-1 (HMGB1) is closely associated with the clinical and pathologic features of gastric cancer. J Transl Med. 2009;7(1):38.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.