Back to Journals » Cancer Management and Research » Volume 13
Circ_0072995 Promotes Proliferation and Invasion via Regulating miR-1253/EIF4A3 Signaling in HCC
Authors He Q, Tao L, Xu H, Xie X, Cheng S
Received 19 April 2021
Accepted for publication 25 June 2021
Published 3 August 2021 Volume 2021:13 Pages 5981—5987
DOI https://doi.org/10.2147/CMAR.S316559
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Qianggui He, Lijun Tao, Hongbo Xu, Xianhai Xie, Shuibing Cheng
Department of Trauma Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China
Correspondence: Lijun Tao
Department of Trauma Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is a major threat for human health. This work aimed to determine the potential function of circ_0072995 in HCC progression and its molecular mechanism.
Methods: qRT-PCR was conducted to analyze circ_0072995 expression. CCK8 and colony formation assays were utilized to detect cell proliferation. Transwell assay was performed to determine migration and invasion. Interactions among circ_0072995, miR-1253 and EIF4A3 (Eukaryotic Translation Initiation Factor 4A3) were predicted through bioinformatics methods and confirmed via luciferase reporter assay and RNA pulldown assay.
Results: circ_0072995 expression was upregulated in HCC tissues. Circ_0072995 high level was associated with poor prognosis. Circ_0072995 knockdown impaired proliferation, migration, invasion and survival. MiR-1253 was sponged by circ_0072995 and targeted EIF4A3 directly. Circ_0072995 inhibited miR-1253 to upregulate EIF4A3 level.
Conclusion: Circ_0072995 exerted tumorigenic roles to enhance HCC progression through activating EIF4A3 by sponging miR-1253.
Keywords: circ_0072995, miR-1253, EIF4A3, HCC
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers around the world and is a leading cause for cancer-related deaths.1 Surgery combined with chemotherapy or radiotherapy is the main therapeutic strategy for HCC treatment.2,3 However, the prognosis of HCC patients remains unfavorable due to recurrence and distant metastasis.4 Thus, determining the underlying molecular mechanism of HCC progression seems to be critical and urgent.
Circular RNAs (circRNAs), one type of noncoding RNAs, are characterized by a closed-loop structure.5 circRNAs also lack the ability to code protein.5,6 As the development of sequencing technology, more and more numbers of circRNAs have been identified. And the close association between circRNA and cancer is gradually uncovered. Emerging evidence indicates that circRNAs play important roles in regulating the biological processes of cancer cells, such as proliferation, migration and survival.7,8 For example, hsa_circ_0001944 enhances bladder cancer proliferation and invasion through sponging miR-548.9 Hsa_circ_0020123 upregulation promotes lung cancer growth and migration via facilitating HOXC9 expression by inhibiting miR-495.10 circCCDC66 is overexpressed in lung cancer cells and contributes to tumor progression through regulating miR-211/SRCIN1 axis.11 Therefore, dissecting circRNA function will be benefit for the understanding of tumor pathogenesis.
Circ_0072995 has been reported to promote breast cancer progression and ovarian cancer development.12,13 Nevertheless, circ_0072995 role in HCC remains undetermined. In this research, we found that circ_0072995 was highly expressed in HCC tissues and predicted poor prognosis. Circ_0072995 knockdown suppressed HCC growth and invasion while inducing apoptosis. Circ_0072995 was identified to sponge miR-1253 and enhance EIF4A3 expression. Thus, our work discovered that circ_0072995/miR-1253/EIF4A3 axis plays an essential oncogenic role in HCC progression.
Materials and Methods
Clinical Sample
HCC tissues and normal tissues were collected from The First Affiliated Hospital of Wenzhou Medical University. Tissues were not treated by radiotherapy or chemotherapy before collection. Tissues were stored in liquid nitrogen. All patients provided written informed consents. Correlation between circ_0072995 expression and clinicopathologic parameters in HCC samples is analyzed in Table 1. This study was approved by the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University. The experiments were conducted in accordance with the Declaration of Helsinki.
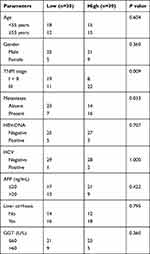 |
Table 1 Correlation Between Circ_0072995 Expression and Clinicopathologic Parameters in HCC Samples |
Cell Lines and Transfection
Cell lines were obtained from Shanghai Institute of Cell Biology (Shanghai, China) and cultured using RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% FBS. siRNAs, miR-1253 mimics, miR-1253 inhibitors and corresponding negative controls were purchased from Ribobio Co. Ltd (Guangzhou, China) and transfected into cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
qRT-PCR
Total RNA was isolated through Trizol reagent (Invitrogen). cDNA was synthesized using First-Strand cDNA Synthesis SuperMix (Transgen Biotech). qPCR was performed using SYBR Green (Solarbio, Beijing, China) and calculated according to the 2−ΔΔCt method. Relative expression was normalized to GAPDH or U6.
CCK8 Assay
Cells were seeded into 96-well plates and cultured for indicated times. Then CCK8 solution (Beyotime, Shanghai, China) was added into each plate and incubated for 4 h. The absorbance at 450 nm was determined using a microplate reader.
Colony Formation Assay
Cells were seeded into 6-well plates and cultured for 14 days. Then colonies were fixed and stained using crystal violet. Colony number was determined finally.
Tumor Xenograft Assay
6-week old female Nude mice were inoculated subcutaneously on the right flank with 2×106 Hep3B cells. Tumor volumes were measured every one week according to the formula: Tumor volume = (length×width2)/2. All mouse experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (People’s Republic of China National Standard GB/T 35892 2018) and were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Wenzhou Medical University.
Transwell Assay
Cell migration and invasion were analyzed by Transwell chambers (Corning Inc., Corning, NY, USA). The cells were seeded into the upper chamber filled with serum-free medium. The lower chamber was filled with serum-containing medium. After cultured for 48 h, the migrated or invaded cells in the lower member was fixed and stained with crystal violet. Then, the number of cells was counted using a microscope.
Dual-Luciferase Reporter Assay
The circ_0072995 or EIF4A3 sequence containing the binding element for miR-1253 was constructed into pmirGLO expression vector (Promega, Madison, WI, USA). For luciferase reporter assay, the luciferase vector and miR-1253 mimics were transfected into cells. After cultured for 48 h, the luciferase activity was measured using the Dual-Luciferase Assay System (Promega).
Statistical Analysis
Results from three independent experiments were analyzed by GraphPad Prism 6 software (San Diego, CA, USA) and expressed as the mean ± standard deviation. Student’s t test or one-way analyses of variance followed by Tukey’s test was used for statistical analyses. P < 0.05 was defined as statistically significant.
Results
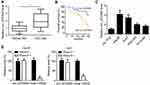
Circ_0072995 Was Upregulated in HCC
Firstly, the expression of circ_0072995 was examined by qRT-PCR in HCC tissues. Results indicated that circ_0072995 level was raised in HCC tissues compared to normal tissues (Figure 1A). Then HCC tissues were divided into circ_0072995 high and circ_0072995 low groups based on the median value. Overall survival rate was plotted. As shown, circ_0072995 high expression was associated with low survival rate (Figure 1B). Afterwards, HCC cell lines were obtained and qRT-PCR was performed. Circ_0072995 expression was also increased in HCC cell lines compared to HL-7702 cells (Figure 1C). To prove circ_0072995 is a circular RNA, we treated RNA with RNase R, followed by qRT-PCR analysis. Circ_0072995 was resistant to RNase R digestion (Figure 1D).
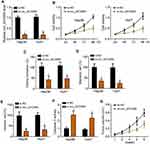
Circ_0072995 Downregulation Inhibited HCC Progression
Because Circ_0072995 was the most highly expressed in Hep3B and Huh7 cells among all detected HCC cell lines (new Figure 1C), we chose hep3B and Huh7 for following experiments. siRNAs were synthesized to knock down circ_0072995 in Hep3B and Huh7 cells (Figure 2A). CCK8 and colony formation assays were carried out and results showed that circ_0072995 knockdown impaired the growth of Hep3B and Huh7 cells (Figure 2B and C). Moreover, transwell assay was performed and we found that circ_0072995 silencing suppressed HCC cell migration and invasion (Figure 2D and E). We also noticed that the caspase3 activity in the cells of sicirc_0072995 group was higher than that in control group (Figure 2F), suggesting that circ_0072995 knockdown promoted apoptosis. Importantly, circ_0072995 silencing also inhibited tumor growth in vivo (Figure 2G).
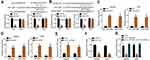
Circ_0072995 Promoted EIF4A3 Expression via Sponging miR-1253
To explore the downstream molecular mechanism, bioinformatics analysis was performed using circinteractome and TargetScan7 tools. Circ_0072995 was found to sponge miR-1253 while EIF4A3 was the most potential target of miR-1253. To prove it, luciferase reporter assays were carried out. MiR-1253 suppressed the luciferase activity of either WT-circ_0072995 or WT-EIF4A3 (Figure 3A and B). miRNAs are often associated with the AGO2 complex. Thus, we used anti-AGO2 to perform RIP assay. Results showed that Ago2 antibody can enrich both circ_0072995 and miR-1253 (Figure 3C). Additionally, Ago2 antibody also enriched miR-1253 and EIF4A3 (Figure 3D). We found that miR-1253 level was increased after circ_0072995 knockdown (Figure 3E). However, miR-1253 suppressed the expression of EIF4A3 (Figure 3F). Interestingly, circ_0072995 knockdown inhibited EIF4A3 expression while miR-1253 inhibition reversed it (Figure 3G). Therefore, circ_0072995 promoted EIF4A3 expression by sponging miR-1253.
Circ_0072995 Promoted HCC Progression Through Regulating miR-1253/EIF4A3 Axis
In HCC tissues, circ_0072995 or EIF4A3 was negatively correlated with miR-1253 (Figure 4A and B) while circ_0072995 was positively correlated with EIF4A3 (Figure 4C). Through TCGA data using ualcan tool, we noticed that EIF4A3 was upregulated in HCC tissues and correlated with poor prognosis (Figure 4C and D), suggesting that EIF4A3 may be an oncogene. To confirm whether circ_0072995 promotes HCC progression through regulating miR-1253/EIF4A3 axis, rescue assays were performed. CCK8 and Transwell assays showed that miR-1253 inhibitors or EIF4A3 overexpression reversed the inhibitory effects of circ_0072995 siRNA on malignant behaviors in HCC cells (Figure 4E–G).
Discussion
HCC is a major public problem for human health.14 However, how HCC development and progression still remains unclear. In this study, we found that circ_0072995 was upregulated in HCC tissues and predicted poor prognosis. Circ_0072995 knockdown resulted in decreased proliferation, migration and invasion. Moreover, circ_0072995 was proven to sponge miR-1253, leading to upregulation of EIF4A3 expression. Thus, our findings supported circ_0072995 is an important oncogene in HCC.
The relationship between circRNA and HCC has been discovered gradually.15 Several circRNAs are demonstrated to regulate several biological processes of HCC cells, such as metastasis and survival.15 For example, circ_0015756 is upregulated in HCC and promotes cell proliferation, migration and invasion.16 CircRNA cIARS affects HCC ferroptosis by binding to ALKBH5.17 Circ_0046599 initiates HCC development and promotes metastasis via miR-1258/RPN2 signaling.18 circ_0072995 was found to promote breast cancer metastasis.12 Another work found that circ_0072995 initiates ovarian cancer development.13 However, whether circ_0072995 affects HCC development is unknown. Our data uncovered that circ_0072995 was upregulated in HCC tissues. And circ_0072995 knockdown repressed proliferation, migration and invasion of HCC tissues. Thus, this study for the first time proved circ_0072995 exerts oncogenic functions in HCC.
CircRNA is a classical type of competing endogenous RNAs (ceRNAs) for microRNAs.10 circRNA-miRNA axis plays crucial roles in the regulation of tumorigenesis.12 For instance, circ-SEC31A is the ceRNA for miR-520a-5p to promote lung cancer progression.19 Hsa_circ_103809 plays an oncogenic function in gastric cancer through interacting with miR-101-3p.20 In addition, circ-CSPP1 contributes to HCC growth and invasion through sponging miR-577.21 circ_0072995 was found to be the ceRNA for miR-147a and miR-30c.12,13 In HCC, we did not observe it. However, we found that circ_0072995 targeted miR-1253. Through luciferase reporter assay and RIP assay, their direct interaction was validated. MiR-1253 expression was also inhibited by circ_0072995. Previously, miR-1253 was found to be a tumor suppressor in lung cancer, osteosarcoma, prostate cancer and pancreatic ductal adenocarcinoma.22–25 Its role in HCC is unclear. Our study showed that miR-1253 inhibitors promoted the proliferation, migration and invasion of HCC cells. Thus, miR-1253 promotes HCC progression.
Next, bioinformatics analysis was conducted to search the downstream target of miR-1253. EIF4A3 was identified. Luciferase report assay and RIP assay confirmed the interaction between miR-1253 and EIF4A3. EIF4A3 is an important oncogene and promotes tumorigenesis in various types of cancer.26 For example, EIF4A3 promotes breast cancer growth and metastasis.27 EIF4A3 promotes circMMP9 expression and induces glioblastoma multiforme tumorigenesis.28 However, there is still no study about EIF4A3 function in HCC. We showed that EIF4A3 was upregulated in HCC and linked with poor prognosis. Moreover, EIF4A3 overexpression enhanced HCC progression. Therefore, EIF4A3 was the downstream signaling of circ_0072995/miR-1253 axis and promoted HCC development.
In sum, this work demonstrates that circ_0072995 was upregulated in HCC and promotes tumor growth and invasion through regulating miR-1253/EIF4A3 axis, which suggests that circ_0072995 may be a new therapeutic target in HCC.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Ochiai T, Ikoma H, Okamoto K, Kokuba Y, Sonoyama T, Otsuji E. Clinicopathologic features and risk factors for extrahepatic recurrences of hepatocellular carcinoma after curative resection. World J Surg. 2012;36(1):136–143. doi:10.1007/s00268-011-1317-y
3. Yang Y, Nagano H, Ota H, et al. Patterns and clinicopathologic features of extrahepatic recurrence of hepatocellular carcinoma after curative resection. Surgery. 2007;141(2):196–202. doi:10.1016/j.surg.2006.06.033
4. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
5. Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
6. Wu J, Qi X, Liu L, et al. Emerging epigenetic regulation of circular rnas in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi:10.1016/j.omtn.2019.04.011
7. Sun Z, Zhang A, Hou M, Jiang T. Circular RNA hsa_circ_0000034 promotes the progression of retinoblastoma via sponging microRNA-361-3p. Bioengineered. 2020;11(1):949–957. doi:10.1080/21655979.2020.1814670
8. Dou D, Ren X, Han M, et al. CircUBE2D2 (hsa_circ_0005728) promotes cell proliferation, metastasis and chemoresistance in triple-negative breast cancer by regulating miR-512-3p/CDCA3 axis. Cancer Cell Int. 2020;20:454. doi:10.1186/s12935-020-01547-7
9. Jin M, Lu S, Wu Y, et al. Hsa_circ_0001944 promotes the growth and metastasis in bladder cancer cells by acting as a competitive endogenous RNA for miR-548. J Exp Clin Cancer Res. 2020;39(1):186. doi:10.1186/s13046-020-01697-6
10. Bi R, Wei W, Lu Y, et al. High hsa_circ_0020123 expression indicates poor progression to non-small cell lung cancer by regulating the miR-495/HOXC9 axis. Aging. 2020;12(17):17343–17352. doi:10.18632/aging.103722
11. Hong W, Yu S, Zhuang Y, Zhang Q, Wang J, Gao X. SRCIN1 regulated by circCCDC66/miR-211 Is upregulated and promotes cell proliferation in non-small-cell lung cancer. Biomed Res Int. 2020;2020:5307641. doi:10.1155/2020/5307641
12. Zhang HD, Jiang LH, Hou JC, et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10(9):1229–1242. doi:10.2217/epi-2018-0002
13. Ding J, Wang Q, Guo N, et al. CircRNA circ_0072995 promotes the progression of epithelial ovarian cancer by modulating miR-147a/CDK6 axis. Aging. 2020;12(17):17209–17223. doi:10.18632/aging.103668
14. Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44(10):1117–1121. doi:10.1038/ng.2391
15. Gao J, Li E, Liu W, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression through the miR29a/KMT5C signaling pathway. Onco Targets Ther. 2020;13:8615–8627. doi:10.2147/OTT.S258715
16. Guo W, Zhao L, Wei G, Liu P, Zhang Y, Fu L. Circ_0015756 aggravates hepatocellular carcinoma development by regulating FGFR1 via Sponging miR-610. Cancer Manag Res. 2020;12:7383–7394. doi:10.2147/CMAR.S262231
17. Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72. doi:10.1038/s41420-020-00306-x
18. Fang Q, Liu H, Zhou A, Zhou H, Zhang Z. Circ_0046599 Promotes the Development of Hepatocellular Carcinoma by Regulating the miR-1258/RPN2 Network. Cancer Manag Res. 2020;12:6849–6860. doi:10.2147/CMAR.S253510
19. Jin M, Shi C, Hua Q, et al. High circ-SEC31A expression predicts unfavorable prognoses in non-small cell lung cancer by regulating the miR-520a-5p/GOT-2 axis. Aging. 2020;12(11):10381–10397. doi:10.18632/aging.103264
20. Huang SS, Guo WX, Ren MS. Circular RNA hsa_circ_103809 promotes cell migration and invasion of gastric cancer cells by binding to microRNA-101-3p. Eur Rev Med Pharmacol Sci. 2020;24(11):6064–6071.
21. Sun Q, Yu R, Wang C, Yao J, Zhang L. Circular RNA circ-CSPP1 regulates CCNE2 to facilitate hepatocellular carcinoma cell growth via sponging miR-577. Cancer Cell Int. 2020;20:202. doi:10.1186/s12935-020-01287-8
22. Liu M, Zhang Y, Zhang J, et al. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis. 2018;9(2):189. doi:10.1038/s41419-017-0218-x
23. Huang L, Chen M, Pan J, Yu W. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 2018;500(2):511–517. doi:10.1016/j.bbrc.2018.04.131
24. Chen Y, Gu M, Liu C, et al. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686:37–42. doi:10.1016/j.gene.2018.10.085
25. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509(1):138–142. doi:10.1016/j.bbrc.2018.12.088
26. Yang H, Yang W, Dai W, Ma Y, Zhang G. LINC00667 promotes the proliferation, migration, and pathological angiogenesis in non-small cell lung cancer through stabilizing VEGFA by EIF4A3. Cell Biol Int. 2020;44(8):1671–1680. doi:10.1002/cbin.11361
27. Zheng X, Huang M, Xing L, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19(1):73. doi:10.1186/s12943-020-01183-9
28. Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166. doi:10.1186/s12943-018-0911-0
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.