Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 16
Circ-OSBPL2 Contributes to Smoke-Related Chronic Obstructive Pulmonary Disease by Targeting miR-193a-5p/BRD4 Axis
Authors Zheng C, Zhang Y, Zhao Y, Duan Y, Mu Q, Wang X
Received 21 December 2020
Accepted for publication 11 March 2021
Published 7 April 2021 Volume 2021:16 Pages 919—931
DOI https://doi.org/10.2147/COPD.S298465
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Caifen Zheng,1 Yongping Zhang,2 Yingchun Zhao,3 Yuanfang Duan,1 Qianghua Mu,1 Xinying Wang1
1Department of Respiratory and Critical Care Medicine, The First People’s Hospital of Lianyungang, Lianyungang, People’s Republic of China; 2Blood Purifying Center, The First People’s Hospital of Lianyungang, Lianyungang, People’s Republic of China; 3Department of Cardiovascular Medicine, The First People’s Hospital of Lianyungang, Lianyungang, People’s Republic of China
Correspondence: Yingchun Zhao
Department of Cardiovascular Medicine, The First People’s Hospital of Lianyungang, No. 6 East Zhenhua Road, Haizhou, Lianyungang, Jiangsu, 222061, People’s Republic of China
Tel +86-0518-85767023
Email [email protected]
Background: Circular RNAs (circRNAs) have been identified to play roles in the respiratory diseases. Here, this study aimed to elucidate the function of circRNA oxysterol binding protein like 2 (circOSBPL2) in the development of smoke-related chronic obstructive pulmonary diseases (COPD).
Methods: The expression of circ-OSBPL2, microRNA (miR)-193a-5p, and bromodomain-containing protein 4 (BRD4) was detected using qRT-PCR and Western blot assays. Cigarette smoke extract (CSE)-induced human bronchial epithelial cells (HBECs) was applied to mimic smoke-related COPD in vitro. Flow cytometric analysis of cell apoptosis and ELISA analysis of interleukins (IL)-6, IL-8, tumor necrosis factor-α (TNF-α) levels were performed. The malondialdehyde (MDA) and superoxide dismutase (SOD) production levels were analyzed according to the kit instructions. The binding interaction between miR-193a-5p and circ-OSBPL2 or BRD4 was confirmed by dual-luciferase reporter assay and RNA immunoprecipitation assays.
Results: Circ-OSBPL2 was highly expressed in lung tissues of smokers without or with COPD, particularly in smokers with COPD. Also, the expression of circ-OSBPL2 was dose and time-dependently elevated in CSE-induced HBECs. Circ-OSBPL2 down-regulation in HBECs attenuated CSE-evoked cell proliferation arrest, and cell apoptosis, inflammation and oxidative stress promotion. Mechanistically, circ-OSBPL2 served as a sponge for miR-193a-5p, and miR-193a-5p inhibition reversed the effects of circ-OSBPL2 knockdown on CSE-mediated HBECs. Besides that, miR-193a-5p directly targeted BRD4, and miR-193a-5p re-expression in HBECs abolished CSE-induced HBEC injury, which was reverted by BRD4 up-regulation. Additionally, we also found circ-OSBPL2 could indirectly regulate BRD4 via miR-193a-5p.
Conclusion: Circ-OSBPL2 contributed to the apoptosis, inflammation, and oxidative stress of HBECs in smoke-related COPD by miR-193a-5p/BRD4 axis, suggesting a novel insight on the pathogenesis of COPD and a potential therapeutic strategy for future clinic intervention in COPD.
Keywords: circ-OSBPL2, miR-193a-5p, BRD4, COPD
Introduction
Chronic obstructive pulmonary disease (COPD) is a progressive degenerative airway disease, mainly characterized by progressive airflow limitation and sustained respiratory symptoms.1 With the fast increasing tendency in morbidity, COPD is estimated to rank as the third leading cause of death worldwide by 2020.2 Cigarette smoking (CS) is recognized as the major aetiological factor, accounting for around 80–90% of the occurrence of COPD cases.3,4 The airway epithelium is the primary defense against inhaled harmful particles, the injury of endothelial cells can lead to acute oxidative injury, epithelial apoptosis, epithelial barrier formation reduction, disruption of cilia function, and chemokine production, ultimately causing the narrow of small airways, producing obstruction and reducing lung compliance.5,6 Therefore, further investigations on the mechanisms underlying the CS-induced endothelial cell injury in the progression of COPD are necessary.
Circular RNAs (circRNAs) are a class of noncoding RNAs characterized by continuous cycle of covalent closures, which protects them from the degradation by RNA exonuclease, so circRNAs are abundantly and stably expressed in eukaryotic cells.7,8 With the development of next-generation sequencing technologies, circRNAs have been confirmed to play specific biological roles in crucial biological processes, like cellular proliferation, apoptosis, metastasis and inflammation.9–11 Importantly, a number of circRNAs have been identified as functional molecules in regulating the progression of different respiratory diseases.12 In COPD, several studies have also indicated that circRNAs might implicate in the response to CS in COPD. For example, Ma et al revealed that circ0061052 promoted CS-triggered EMT of human bronchial epithelial cells (HBECs), involving in airway remodeling in COPD.13 Li et al demonstrated that circRNA circBbs9 enhanced PM 2.5-induced lung inflammation in COPD model mice, which induced COPD exacerbation.14 CircRNA oxysterol binding protein like 2 (circ-OSBPL2, hsa_circ_0061052) is a novel identified circRNA, arises from the back-splicing of OSBPL2 gene, an important regulator in cellular lipid metabolism and transport that regulates intracellular cholesterol homeostasis.15 Circ-OSBPL2 is located in chr20: 60861638–60864386 with the genomic length of 253 bp. A recent study showed that circ-OSBPL2 was elevated in COPD.16 However, the detailed function and molecular mechanisms underlying circ-OSBPL2 in CS-induced COPD remain largely unclear.
Herein, through using a cell model of COPD in HBECs treated with or without cigarette smoke extract (CSE), we aimed to detect the role and mechanisms of circ-OSBPL2 in cell apoptosis, inflammation and oxidative stress, investigated the potential involvement of circ-OSBPL2 in the development of CS-related COPD.
Materials and Methods
Clinical Samples
Lung specimens (non-tumor tissues from at least 5 cm from the lesion) were collected from 81 lung cancer patients with solitary pulmonary tumors by pneumonectomy at The First People’s Hospital of Lianyungang. The enrolled individuals were classified into three groups: non-smokers without COPD (n = 21), smokers without COPD (n = 27), smokers with COPD (n = 33). The patients were diagnosed as COPD in line with the criteria of the Global Initiative for Chronic Obstruct Lung Disease (GOLD) and all subjects had no other respiratory diseases. This study was conducted under the permission of the Ethics Committee of The First People’s Hospital of Lianyungang according to the Declaration of Helsinki, and every enrolled individual and their families were informed of the study and signed informed consent. The information on the demographics of the subjects has been provided in Table 1.
 |
Table 1 The Information on the Demographics of All Subjects |
CSE Preparation
For the preparation of CSE, two commercial cigarettes were combusted with a reformative syringe-driven apparatus. The smoke was bubbled through 30 mL media. Afterwards, a 0.2 μm pore-size filter was employed to filter the resulting collection to eliminate large bacteria and particles, and resulting suspension was collected and defined as “100% CSE” solution. Thereafter, 100% CSE was diluted with phosphate-buffered saline (PBS) to obtain working concentrations of CSE (0.5%, 1%, 2%, and 4%) before used. Cells without any treatment were regarded as the control group (Control).
Cell Culture and Treatment
16HBE, purchased from BioVector NTCC Inc. (Beijing, China), was cultivated in RMPI-1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco) and 1% streptomycin/penicillin (Gibco) at 37°C with 5% CO2. 16HBE cells were then exposed to different concentrations of CSE at 0%, 0.5%, 1%, 2% and 4% for 24 h.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol® Reagent (Life Technologies, Carlsbad, CA, USA) was employed to extract total RNA from frozen patient samples and in vitro cultured cells. Subsequently, reverse transcription was executed to produce complementary DNAs (cDNAs) with the Reverse Transcription System Kit (Takara, Kusatsu, Japan), and then synthesized cDNAs were qualified using the SYBR green PCR kit (Takara). The relative fold changes were evaluated using the 2−ΔΔCt method with U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal reference. Primers for qRT-PCR were listed:
circ-OSBPL2: F 5ʹ-CCGAGGTGCCCATTCCTG-3ʹ, R 5ʹ-TTGGTGCAGTTGCCTTTCG-3ʹ;
OSBPL2: F 5ʹ-CAGAGGCAAATCAGAAAGTCACG-3ʹ, R 5ʹ-TCCCCAGTTTTCCCAATCCTA-3ʹ;
miR-193a-5p: F 5ʹ-TATATGGGTCTTTGCGGGCG-3ʹ, R 5ʹ-GTGCAGGGTCCGAGGT-3ʹ;
GADPH: F 5ʹ-CCCACATGGCCTCCAAGGAGTA-3ʹ, R 5ʹ-GTGTACATGGCAACTGTGAGGAGG-3ʹ;
Bromodomain-containing protein 4 (BRD4): F 5ʹ-ACCTCCAACCCTAACAAGCC-3ʹ, R 5ʹ-TTTCCATAGTGTCTTGAGCACC-3ʹ;
U6: F 5ʹ-AAAGCAAATCATCGGACGACC-3ʹ, R 5ʹ-GTACAACACATTGTTTCCTCGGA-3ʹ.
RNase R Assay
Total RNA (5 μg) was incubated with 3 U/μg RNase R (Sigma-Aldrich, St. Louis, MO, USA) for 15 min at 37°C, then the resulting RNAs were purified using the RNeasy MinElute cleaning Kit (Life Technologies). Finally, qRT-PCR assay was applied to detect the abundance of circ-OSBPL2 and linear OSBPL2 mRNA.
Subcellular Fractionation
The isolation and purification of nuclear and cytoplasm fractions were conducted employing Cytoplasmic and Nuclear RNA Purification Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the expression levels of circ-OSBPL2, GAPDH and U6 were tested by qRT-PCR.
Cell Transfection
The circ-OSBPL2 or BRD4 overexpressing vector (circ-OSBPL2 or BRD4) and corresponding mock vector (pCD5-ciR or pcDNA), siRNAs targeting the back-splice junction site of circ-OSBPL2 (si-circ-OSBPL2) and siRNA negative control (si-NC) were synthesized by Geneseed (Guangzhou, China). The mimic and inhibitor of miR-193a-5p (miR-193a-5p or anti-miR-193a-5p) and their negative controls (miR-NC or anti-miR-NC) were procured from GenePharma (Shanghai, China). Then, the transfection was conducted using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA).
Cell Viability Analysis
Following the assigned transfection and/or treatment, 16HBE cells (5 × 104) were cultured in 96-well plates overnight, and then incubated with 10 μL cell counting kit-8 (CCK-8) solution (5 mg/mL, Sigma-Aldrich) for 4 h. Finally, the optical density (OD) at 450 nm was evaluated using a microplate reader.
Flow Cytometry
After transfection and/or treatment, 16HBE cells were resuspended with 1 × buffer, with the concentration adjusted to 5 × 105 cells/mL. Afterwards, cells were stained with Annexin V-fluorescein isothiocyanate (FITC) (5 μL) and propidium iodide (PI) (5 μL) (BD Biosciences, San Jose, CA, USA) for 15 min in dark, and then cell apoptosis was assessed by the flow cytometry (BD Biosciences) within 1 h.
Enzyme-Linked Immunosorbent Assay (ELISA)
Referring to the instructions of protocol, the concentrations of interleukin-1β (IL-1β), IL-8 and tumor necrosis factor-α (TNF-α) from the culture supernatants of 16HBE cells were evaluated using commercial the ELISA kits (R&D Systems, Minneapolis, Minnesota, USA) after appropriate transfection and/or treatment.
Malondialdehyde (MDA) and Superoxide Dismutase (SOD) Measurement
The production of MDA and SOD was detected using the Lipid Peroxidation MDA Assay Kit or SOD Activity Assay Kit (Nanjing Institute of Biomedical Engineering cohesion, Nanjing, China) following the kit protocols.
Dual-Luciferase Reporter Assay
The wild-type (WT) sequences of circ-OSBPL2 or BRD4 3ʹUTR possessing miR-193a-5p binding sites and their corresponding mutant (MUT) versions without miR-193a-5p binding sites were amplified and cloned into the psiCHECK-2 vectors (Promega, Madison, WI, USA). Then, these vectors (WT-circ-OSBPL2, MUT-circ-OSBPL2, BRD4 3ʹUTR-WT or BRD4 3ʹUTR-MUT) together with miR-193a-5p mimic or miR-NC were co-transfected into 16HBE cells. After 48 h of transfection, the dual luciferase assay kit (Promega) was applied to detect luciferase activities.
RNA Immunoprecipitation (RIP) Assay
16HBE cells were lysed using RIP buffer, then the lysate was incubated with magnetic beads conjugated with human anti-Ago2 (Millipore) or normal mouse anti-IgG (Millipore). After interaction with proteinase K, the immunoprecipitated RNA was eluted and purified RNA was tested using qRT-PCR assay.
Western Blot
Proteins were isolated from tissues and cells using RIPA lysis buffer (Beyotime, Shanghai, China), followed by the qualification of protein concentration with the bicinchoninic acid assay. Thereafter, approximately 30 μg of extracted protein was loaded to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis for separation, and then transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% skimmed milk for 1 h, and then probed with primary antibodies BRD4 (1:3000, ab75898) and GAPDH (1:10000, ab181602) overnight at 4°C, followed by HRP-labeled goat anti-mouse (1:2000, ab205719) for 2 h at 37°C. Protein bands were visualized by enhanced chemiluminescence reagent kit (Millipore). All antibodies were obtained from Abcam (Cambridge, MA, USA).
Statistical Analysis
Each experiment was repeated at least three times. GraphPad Prism 7 software was used to process data, and mean ± standard deviation (SD) was adopted for data representing. Correlation analysis was conducted using Pearson correlation analysis. The comparison was performed using one way analysis of variance (ANOVA) or Student’s t-test as appropriate. P values <0.05 indicated statistically significant.
Results
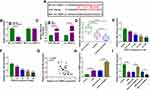
Circ-OSBPL2 is Up-Regulated in COPD Tissues and CSE-Induced HBECs
The expression profiles of circ-OSBPL2 in 81 cases of lung tissue specimens were investigated using qRT-PCR. Results showed that circ-OSBPL2 expression was higher in lung tissues of smokers without or with COPD, particularly in smokers with COPD group, than that in samples from non-smoker without COPD group (Figure 1A). Identically, circ-OSBPL2 was highly expressed in 16HBE cells in a dose-dependent manner after varying concentration of CSE exposure (0, 0.5, 1, 2, 4%) for 24 h (Figure 1B). Moreover, the expression of circ-OSBPL2 was also time-dependently elevated in 16HBE cells by 2% CSE exposure at 0, 6, 12, 24 and 48 h (Figure 1C). Then, the RNase R assay was performed, it was found that circ-OSBPL2 was resistant to RNase R digestion while the linear transcript was remarkably digested by RNase R treatment (Figure 1D). Besides that, the data of subcellular localization assay suggested that circ-OSBPL2 was highly enriched in the cytoplasm fraction in 16HBE cells (Figure 1E). All these data suggested that circ-OSBPL2 was an abundant, circular and stable transcript in HBECs, and aberrant expression of circ-OSBPL2 might be associated with the pathogenesis of COPD.
CSE Suppresses the Viability and Induces the Apoptosis, Inflammation and Oxidative Stress in HBECs
To explore the detailed functions of circ-OSBPL2 in occurrence and progression of COPD, firstly, CSE-mediated HBECs were applied to mimic smoke-related COPD in vitro. 16HBE cells were treated with varying concentration of CSE exposure (0, 0.5, 1, 2, 4%) for 24 h, then we found CSE exposure repressed the viability (Figure 2A) but induced the apoptosis in 16HBE cells with increasing doses of CSE (Figure 2B and C). Moreover, results of ELISA exhibited that when treated with increasing doses of CSE, the levels of IL-8, IL-1β, and TNF-α were increased gradually (Figure 2D). In addition, it was also showed that CSE exposure promoted the production of MDA (Figure 2E) while inhibited the generation of SOD with increasing doses of CSE (Figure 2F). Taken together, CSE induced cell apoptosis, inflammation and oxidative stress in HBECs, suggesting the successful establishment of smoke-related COPD cell model in vitro.
Circ-OSBPL2 Knockdown Reverses CSE-Induced Apoptosis, Inflammation and Oxidative Stress in HBECs
Next, si-circ-OSBPL2 was designed to further explore the potential biological functions of circ-OSBPL2 in CSE-mediated HBECs. 16HBE cells were transfected with si-circ-OSBPL2 or si-NC, followed by treatment with 2% CSE, then the transfection efficiency was demonstrated and qRT-PCR analysis suggested that circ-OSBPL2 knockdown reduced CSE-induced elevation of circ-OSBPL2 expression in 16HBE cells (Figure 3A). After that, it was proved that knockdown of circ-OSBPL2 markedly reversed CSE-evoked cell viability arrest cytometry (Figure 3B) and apoptosis promotion (Figure 3C) in 16HBE cells as CCK-8 assay and flow cytometry indicated. Furthermore, we also observed that circ-OSBPL2 down-regulation abolished CSE-induced elevation of IL-8, IL-1β, TNF-α (Figure 3D) and MDA (Figure 3E) levels and decrease of SOD level (Figure 3F) in 16HBE cells. In short, these results revealed that circ-OSBPL2 knockdown could suppress CSE-induced HBEC injury.
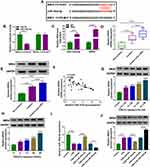
MiR-193a-5p is a Target of circ-OSBPL2
Considering that circ-OSBPL2 was predominantly located in the cytoplasm and displayed high stability, we hypothesized that it might be a competing endogenous RNA (ceRNA) to exert the biological functions. According to the predicted bioinformatics on starBase v.2.0, miR-193a-5p was found to have complementary sequences on circ-OSBPL2 (Figure 4A). Next, the dual-luciferase reporter assay was carried out, results showed that miR-193a-5p overexpression in 16HBE cells significantly weaken the luciferase activity of the reporter vectors containing circ-OSBPL2-WT, while there was no impact on the reporter vectors containing circ-OSBPL2-MUT (Figure 4B). Besides that, a RIP assay showed that circ-OSBPL2 and miR-193a-5p were enriched in Ago2 immunoprecipitates compared with control IgG immunoprecipitates (Figure 4C). These findings demonstrated that circ-OSBPL2 served as a sponge of miR-193a-5p. Converse to circ-OSBPL2, miR-193a-5p was decreased in lung tissues of smokers without or with COPD, especially in smokers with COPD group, in comparison with non-smoker without COPD group (Figure 4D); also, the expression of miR-193a-5p was dose and time-dependently down-regulated in 16HBE cells exposed with CSE (Figure 4E and F); moreover, a negative correlation between circ-OSBPL2 and miR-193a-5p expression in patients with COPD was observed (Figure 4G). Additionally, when we confirmed the transfection efficiency of circ-OSBPL2 overexpressing vectors (Figure 4H), it was found that circ-OSBPL2 overexpression inhibited miR-193a-5p expression, while circ-OSBPL2 deletion promoted miR-193a-5p expression in CSE-mediated 16HBE cells (Figure 4I). Altogether, circ-OSBPL2 directly targeted miR-193a-5p and negatively regulated its expression.
Circ-OSBPL2 Knockdown Reverses CSE-Induced Apoptosis, Inflammation and Oxidative Stress in HBECs via Targeting miR-193a-5p
We then detected whether miR-193a-5p involved in the action of circ-OSBPL2 on CSE-induced HBECs. 16HBE cells were transfected with si-NC, si-circ-OSBPL2, si-circ-OSBPL2 + anti-miR-NC or si-circ-OSBPL2 + anti-miR-193a-5p, and then exposure with 2% CSE, as expected, the introduction of miR-193a-5p inhibitor reduced circ-OSBPL2 silencing induced elevation of circ-OSBPL2 expression (Figure 5A). Thereafter, cell viability, apoptosis and inflammation were determined. Inhibition of miR-193a-5p was proved to evidently impair circ-OSBPL2 knockdown-induced promotion of cell viability (Figure 5B) and suppression of cell apoptosis (Figure 5C) and inflammation (Figure 5D) in CSE-mediated 16HBE cells. Meanwhile, circ-OSBPL2-knockdown triggered decrease of MDA level and increase of SOD level in CSE-mediated 16HBE cells were also attenuated by miR-193a-5p inhibitor (Figure 5E and F). The above discovery implied that miR-193a-5p participated in the injury process regulated by circ-OSBPL2 in HBECs.
BRD4 is a Target of miR-193a-5p
The specific mechanism of miR-193a-5p in mediating HBEC injury was then researched. Through the prediction of starBase v2.0 online database, there were potential base-complementary binding sites between miR-193a-5p and the BRD4 3ʹUTR (Figure 6A). Results from the dual-luciferase reporter assay suggested that miR-193a-5p overexpression remarkably reduced the luciferase activity of the BRD4 ’UTR-WT reporter compared to the control, but did not impact that of the mutant reporter in 16HBE cells (Figure 6B). Further RIP assay showed that BRD4 and miR-193a-5p were all efficiently pulled down by AGO2 antibodies compared with IgG (Figure 6C). Therefore, we verified that miR-193a-5p directly targeted BRD4. Converse to miR-193a-5p, BRD4 expression was increased in lung tissues of smokers without or with COPD, especially in smokers with COPD group (Figure 6D, E), and negatively correlated with miR-193a-5p expression in the tissues of COPD patients (Figure 6F); in the meanwhile, BRD4 expression in 16HBE cells was also discovered to be increased in a dose- or time-dependent manner (Figure 6G and H). In addition, when we up-regulated or down-regulated the level of miR-193a-5p in CSE-induced 16HBE cells (Figure 6I), we found the expression of BRD4 was decreased by miR-193a-5p overexpression, but was increased by miR-193a-5p down-regulation (Figure 6J). Their results confirmed that miR-193a-5p targetedly suppressed BRD4 expression.
MiR-193a-5p Attenuates CSE-Induced Apoptosis, Inflammation and Oxidative Stress in HBECs via Targeting BRD4
Whether miR-193a-5p/BRD4 regulatory loop impacted CSE-induced HBEC injury was further explored. BRD4 overexpressing vector was constructed and transfected into miR-193a-5p-increased 16HBE cells, followed by treatment with 2% CSE. Transfection of miR-193a-5p markedly down-regulated BRD4 level in CSE-induced 16HBE cells, which was rescued by co-transfection of BRD4 (Figure 7A). Then, a series of rescue experiments were designed, we found that miR-193a-5p overexpression abolished CSE-evoked suppression of cell viability (Figure 7B), as well as promotion of cell apoptosis (Figure 7C), inflammation (Figure 7D) and oxidative stress (Figure 7E and F) in 16HBE cells, which were partially overturned by BRD4 up-regulation (Figure 7B–F). Collectively, miR-193a-5p suppressed CSE-induced HBEC injury by regulating BRD4.
Circ-OSBPL2 Can Regulate BRD4 Expression Through Serving as a Sponge for miR-193a-5p
Whether circ-OSBPL2 could regulate the expression of BRD4 in HBECs was further validated. As shown in Figure 8A and B, we found that knockdown of circ-OSBPL2 markedly decreased the expressions of BRD4 in CSE-mediated 16HBE cells, as expected, the decrease of BRD4 induced by circ-OSBPL2 silencing could be reverted by miR-193a-5p inhibition. Therefore, these data suggested that circ-OSBPL2 competitively bound to miR-193a-5p, resulting in up-regulation of BRD4, subsequently contributing to CSE-stimulated HBEC injury (Figure 8C).
Discussion
COPD is a common airway disease, featured by defective tissue repair and chronic inflammation, causing irreversible airflow limitation and lung emphysema.3 COPD affects about 328 million individuals, along with 3.5–4 million deaths through the world annually,16 which has become a huge challenge to global public health. CS is the leading risk factor of COPD, the inflammatory responses due to exposure of CS, followed by tissue repair processes, ultimately results in airway remodeling.17 Airway epithelial cells, the first line of defense against harmful stimuli, are a source of diverse cytokines and chemokines, like IL-8, TNF-α, and IL-1β, which recruit inflammatory cells in response to CS, impacting airway structure and airway remodeling.18,19 Apoptosis of pulmonary structural cells has also been identified in COPD and may contribute to pulmonary structure destruction and pulmonary function impairment.20 Importantly, it has been revealed that apoptosis of bronchial epithelial cells has significant roles in the pathogenesis of COPD.20,21 Recently, there is increasing evidence that oxidative stress is a major predisposing factor in COPD occurrence,22 systemic markers of oxidative stress such as MDA are elevated in COPD patients, and the SOD, which can remove lipid peroxides, plays a vital role in reducing lipid peroxidation by regulating the balance of body’s oxidation and antioxidant,23 has been found to be reduced in COPD patients.24
In this study, we found circ-OSBPL2 expression was higher in lung tissues of smokers with COPD. Then, a cell model of COPD in HBECs treated with or without cigarette smoke extract (CSE) was applied to explore the role of circ-OSBPL2 in CS-related COPD. Circ-OSBPL2 expression was dose and time-dependently elevated in CSE-induced HBECs, more importantly, down-regulation of circ-OSBPL2 in HBECs reversed CSE-evoked cell proliferation arrest, cell apoptosis enhancement, elevated IL-1, IL-8, TNF-α and MDA levels, as well as decreased SOD levels. Taken together, circ-OSBPL2 knockdown suppressed cell apoptosis, inflammation and oxidative stress in CSE-mediated HBECs, suggesting that inhibitory role of circ-OSBPL2 knockdown in CS-related COPD.
Numerous studies have documented that circRNAs can bind with the corresponding miRNAs via acting as sponges of miRNAs to modulate the mRNA expression.25,26 Thus, we hypothesized that circ-OSBPL2 might be a ceRNA to exert the biological functions. The underlying mechanism suggested that functioning as a ceRNA, circ-OSBPL2 directly targeted miR-193a-5p and positively regulated the expression of its target BRD4. MiR-193a-5p is a well-recognized tumor suppressor in different types of cancer.27–29 A previous study suggested that miR-193a was low in blood samples of COPD, predicted decreased COPD susceptibility and acute exacerbation risk, and negatively associated with disease severity and inflammation in COPD patients.30 Nevertheless, the action of miR-193a-5p in COPD remains unclear. In the current work, miR-193a-5p was found to be lowly expressed in lung tissues of smokers with COPD; also, its expression was decreased in CSE-induced HBECs in a dose or time-dependent pattern. Then, it was proved that miR-193a-5p re-expression protected HBECs against CSE-induced apoptosis, inflammation, and oxidative stress. Importantly, we demonstrated that the inhibition of miR-193a-5p reversed the effects of circ-OSBPL2 knockdown on CSE-mediated HBECs. Therefore, the circ-OSBPL2/miR-193a-5p axis is engaged in the pathogenesis of CS-induced COPD.
BRD4 belongs to the family of the bromodomain and extra-terminal domain (BET) proteins, which deregulation has been involved in the development of cancers and non-cancer diseases such as inflammatory diseases.31,32 BRD4 is an emerging actor in NF-κB signaling, which drives the transition to a committed mesenchymal phenotype, leading to the expression of fibronectin (FN1) and collagen I (COL1), a fibrotic response characteristic of airway remodeling.33 BRD4-NFκB pathway plays important roles in airway remodeling and inflammation in COPD, and BRD4 inhibitors are potent anti-remodeling and anti-inflammatory agents.6,34 In this study, we identified that circ-OSBPL2 acted as a sponge for miR-193a-5p to modulate BRD4 expression. Rescue assay analysis exhibited that up-regulation of BRD4 abolished the inhibitory action of miR-193a-5p on CSE-induced HBEC injury.
In short, this study showed that circ-OSBPL2 contributed to CSE-induced HBEC apoptosis, inflammation, and oxidative stress by targeting miR-193a-5p/BRD4 axis, implying that the circ-OSBPL2/miR-193a-5p/BRD4 axis might involve in the pathogenesis of CS-related COPD, which provided a potential therapeutic strategy for COPD treatment.
Abbreviations
COPD, chronic obstructive pulmonary diseases; OSBPL2, oxysterol binding protein like 2; BRD4, bromodomain-containing protein 4; CSE, cigarette smoke extract; HBECs, human bronchial epithelial cells; TNF-α, tumor necrosis factor-α; MDA, malondialdehyde; SOD, superoxide dismutase; PBS, phosphate-buffered saline; cDNA, complementary DNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CCK-8, cell counting kit-8; FITC, fluorescein isothiocyanate; PI, propidium iodide; RIP assay, RNA immunoprecipitation assay; qRT-PCR, quantitative real-time polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of The First People’s Hospital of Lianyungang according to the Declaration of Helsinki. Written informed consent was obtained from all enrolled individual and their families.
Consent for Publication
Patients agree to participate in this work.
Disclosure
The authors declare that they have no competing interests.
References
1. Singh D, Agusti A, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: the GOLD science committee report 2019. Eur Respir J. 2019;53(5):1900164. doi:10.1183/13993003.00164-2019
2. López-Campos JL, Tan W, Soriano JB. Global burden of COPD. Respirology. 2016;21(1):14–23. doi:10.1111/resp.12660
3. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389(10082):1931–1940. doi:10.1016/S0140-6736(17)31222-9
4. Rab A, Rowe SM, Raju SV, Bebok Z, Matalon S, Collawn JF. Cigarette smoke and CFTR: implications in the pathogenesis of COPD. Am J Physiol Lung Cell Mol Physiol. 2013;305(8):L530–L541. doi:10.1152/ajplung.00039.2013
5. Chen Y, Luo H, Kang N, et al. Beraprost sodium attenuates cigarette smoke extract-induced apoptosis in vascular endothelial cells. Mol Biol Rep. 2012;39(12):10447–10457. doi:10.1007/s11033-012-1924-1
6. Song J, Wang Q, Zong L. LncRNA MIR155HG contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-128-5p/BRD4 axis. Biosci Rep. 2020;40(3). doi:10.1042/BSR20192567
7. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
8. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. doi:10.1126/science.1238522
9. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
10. Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90. doi:10.1186/s13045-019-0776-8
11. Hua X, Sun Y, Chen J, et al. Circular RNAs in drug resistant tumors. Biomed Pharmacother. 2019;118:109233. doi:10.1016/j.biopha.2019.109233
12. Wang J, Zhu M, Pan J, Chen C, Xia S, Song Y. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20(1):3. doi:10.1186/s12931-018-0962-1
13. Ma H, Lu L, Xia H, et al. Circ0061052 regulation of FoxC1/Snail pathway via miR-515-5p is involved in the epithelial-mesenchymal transition of epithelial cells during cigarette smoke-induced airway remodeling. Sci Total Environ. 2020;746:141181. doi:10.1016/j.scitotenv.2020.141181
14. Li M, Hua Q, Shao Y, et al. Circular RNA circBbs9 promotes PM(2.5)-induced lung inflammation in mice via NLRP3 inflammasome activation. Environ Int. 2020;143:105976. doi:10.1016/j.envint.2020.105976
15. Yao J, Zeng H, Zhang M, et al. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia. J Genet Genomics. 2019;46(8):379–387. doi:10.1016/j.jgg.2019.06.006
16. Zeng N, Wang T, Chen M, et al. Cigarette smoke extract alters genome-wide profiles of circular RNAs and mRNAs in primary human small airway epithelial cells. J Cell Mol Med. 2019;23(8):5532–5541. doi:10.1111/jcmm.14436
17. Hogg JC, McDonough JE, Suzuki M. Small airway obstruction in COPD: new insights based on micro-CT imaging and MRI imaging. Chest. 2013;143(5):1436–1443. doi:10.1378/chest.12-1766
18. Gao W, Li L, Wang Y, et al. Bronchial epithelial cells: the key effector cells in the pathogenesis of chronic obstructive pulmonary disease? Respirology. 2015;20(5):722–729. doi:10.1111/resp.12542
19. Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2008;8(3):183–192. doi:10.1038/nri2254
20. Min T, Bodas M, Mazur S, Vij N. Critical role of proteostasis-imbalance in pathogenesis of COPD and severe emphysema. J Mol Med. 2011;89(6):577–593. doi:10.1007/s00109-011-0732-8
21. Tsai MJ, Chang WA, Jian SF, Chang KF, Sheu CC, Kuo PL. Possible mechanisms mediating apoptosis of bronchial epithelial cells in chronic obstructive pulmonary disease - A next-generation sequencing approach. Pathol Res Pract. 2018;214(9):1489–1496. doi:10.1016/j.prp.2018.08.002
22. Kirkham PA, Barnes PJ. Oxidative stress in COPD. Chest. 2013;144(1):266–273. doi:10.1378/chest.12-2664
23. Su X, He Y, Yang W, Wang Y, Zhang W, Wang Y. Effect of Dan Hong injection on PON1, SOD activity and MDA levels in elderly patients with coronary heart disease. Int J Clin Exp Med. 2014;7(12):5886–5889.
24. Austin V, Crack PJ, Bozinovski S, Miller AA, Vlahos R. COPD and stroke: are systemic inflammation and oxidative stress the missing links? Clin Sci. 2016;130(13):1039–1050. doi:10.1042/CS20160043
25. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi:10.7150/thno.21299
26. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
27. Li P, Xiao Z, Luo J, Zhang Y, Lin L. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23(4):2475–2488. doi:10.1111/jcmm.14121
28. Pu Y, Zhao F, Cai W, Meng X, Li Y, Cai S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis. 2016;33(4):359–372. doi:10.1007/s10585-016-9783-0
29. Yang Z, Chen JS, Wen JK, et al. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J Exp Clin Can Res. 2017;36(1):178. doi:10.1186/s13046-017-0649-3
30. Ming X, Duan W, Yi W. Long non-coding RNA NEAT1 predicts elevated chronic obstructive pulmonary disease (COPD) susceptibility and acute exacerbation risk, and correlates with higher disease severity, inflammation, and lower miR-193a in COPD patients. Int J Clin Exp Pathol. 2019;12(8):2837–2848.
31. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi:10.1016/j.cell.2011.08.017
32. Tian B, Hosoki K, Liu Z, et al. Mucosal bromodomain-containing protein 4 mediates aeroallergen-induced inflammation and remodeling. J Allergy Clin Immunol. 2019;143(4):1380–1394.e1389. doi:10.1016/j.jaci.2018.09.029
33. Tian B, Zhao Y, Sun H, Zhang Y, Yang J, Brasier AR. BRD4 mediates NF-κB-dependent epithelial-mesenchymal transition and pulmonary fibrosis via transcriptional elongation. Am J Physiol Lung Cell Mol Physiol. 2016;311(6):L1183–l1201. doi:10.1152/ajplung.00224.2016
34. Brasier AR. Therapeutic targets for inflammation-mediated airway remodeling in chronic lung disease. Expert Rev Respir Med. 2018;12(11):931–939. doi:10.1080/17476348.2018.1526677
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.