Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 16
Circ-HACE1 Aggravates Cigarette Smoke Extract-Induced Injury in Human Bronchial Epithelial Cells via Regulating Toll-Like Receptor 4 by Sponging miR-485-3p
Authors Zhou F, Cao C, Chai H, Hong J, Zhu M
Received 1 February 2021
Accepted for publication 19 April 2021
Published 1 June 2021 Volume 2021:16 Pages 1535—1547
DOI https://doi.org/10.2147/COPD.S304859
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Richard Russell
Fujun Zhou,1,* Cheng Cao,2,* Huiping Chai,2 Jingfang Hong,3 Min Zhu1
1Department of Health and Nursing, Anhui Vocational College of City Management, Hefei City, Anhui Province, People’s Republic of China; 2Department of Thoracic Surgery, 4th Affiliated Hospital of Anhui Medical University, Hefei City, Anhui Province, People’s Republic of China; 3School of Nursing, Anhui Medical University, Hefei City, Anhui Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Cheng Cao
Department of Thoracic Surgery, 4th Affiliated Hospital of Anhui Medical University, No. 100 Huaihai Avenue, Hefei, 230022, Anhui Province, People’s Republic of China
Tel/Fax +86 0551-66331040 Email [email protected]
Background: Smoking is the most common cause of chronic obstructive pulmonary disease (COPD), and the early diagnosis for COPD remains poor. Exploring the molecular mechanism and finding feasible biomarkers will be beneficial for clinical management of COPD. Circular RNAs (circRNAs) are noncoding RNAs that act as miRNA sponges to regulate the expression levels of genes, leading to the changes of cellular phenotypes and disease progression. CircRNA HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 (circ-HACE1) was abnormally expressed after the induction of cigarette smoke extract (CSE) in cell model. This study was performed to explore its function and mechanism in COPD.
Methods: Circ-HACE1, microRNA-485-3p (miR-485-3p) and toll-like receptor 4 (TLR4) detection was performed by quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability and apoptosis/cell cycle were respectively examined using cell counting kit-8 (CCK-8) and flow cytometry. Inflammatory cytokines were determined by enzyme-linked immunosorbent assay (ELISA). Oxidative stress was evaluated through the measurement of malondialdehyde (MDA) and superoxide dismutase (SOD). The target binding analysis was conducted via dual-luciferase reporter assay. Western blot was employed for protein expression detection of TLR4.
Results: Circ-HACE1 was overexpressed in smokers or smokers with COPD and CSE upregulated circ-HACE1 expression in 16HBE cells. Knockdown of circ-HACE1 attenuated CSE-stimulated cell viability and cell cycle repression, as well as the enhancement of cell apoptosis, inflammatory response and oxidative stress. MiR-485-3p was a target of circ-HACE1. Circ-HACE1 regulated CSE-induced cell injury via targeting miR-485-3p. TLR4 was a downstream target of miR-485-3p, and miR-485-3p inhibited the CSE-induced cell damages by directly downregulating the level of TLR4. Circ-HACE1/miR-485-3p regulated TLR4 expression in CSE-treated 16HBE cells, and TLR4 overexpression also reversed all effects of si-circ-HACE1 on CSE-treated 16HBE cells.
Conclusion: These findings elucidated that circ-HACE1 contributed to the CSE-induced cell damages in COPD cell models via regulating TLR4 by acting as the sponge of miR-485-3p.
Keywords: circ-HACE1, chronic obstructive pulmonary disease, cigarette smoke extract, miR-485-3p, TLR4
Introduction
Chronic obstructive pulmonary disease (COPD) is an inflammatory disease in the respiratory system with airflow limitation and pulmonary dysfunction.1 Cigarette smoke can cause airway inflammation in COPD, which is progressive and irreversible after smoking cessation.2,3 COPD remains under-diagnosed and under-treated until now, particularly for those mild and moderate patients.4 Exploring promising biomarkers of COPD progression may improve the treatment for early COPD.4
COPD has been found to be linked to dysregulated expression of noncoding RNAs (ncRNAs), including circular RNAs (circRNAs), long noncoding RNAs (lncRNAs) and microRNAs (miRNAs).5 CircRNAs are covalently closed loops different from those linear RNAs and function as gene regulators in mammals via the sponge effects on microRNAs (miRNAs).6 Disease progression in human life is largely affected by circRNAs.7,8 CircRNA HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 (circ-HACE1, hsa_circ_0077520) was shown to be upregulated in cigarette smoke extract (CSE)-induced COPD cell model,9 but its function and underlying mechanism in COPD are still unclear.
Regulatory miRNAs are also tractable makers for the diagnosis and treatment of COPD.10,11 By binding to the 3ʹ-untranslated regions (3ʹ-UTRs) of messenger RNAs (mRNAs), miRNAs can modulate the expression of target genes.12 The miRNA expression profiles in the previous study have indicated that microRNA-485-3p (miR-485-3p) was downregulated in patients with COPD.13 However, it is unknown what role miR-485-3p plays in COPD and whether it can be used as a miRNA target of circ-HACE1. In addition, CSE upregulated the expression of toll-like receptor 4 (TLR4) in THP-1 cells and miR-149-3p targeted TLR4 to downregulate its level.14 Jia et al also reported that miR-497 repressed inflammatory response via targeting TLR4 in CSE-treated human bronchial epithelial (HBE) cells.15 The potential of TLR4 as a target of miR-485-3p needs to be explored.
Currently, the effects of circ-HACE1 and miR-485-3p on CSE-stimulated cell damage were investigated. Moreover, the regulation of circ-HACE1 on TLR4 via targeting miR-485-3p was verified.
Materials and Methods
Blood Samples and Serum Preparation
This study was approved by the Ethics Committee of 4th Affiliated Hospital of Anhui Medical University and performed following the Declaration of Helsinki. The whole blood samples were collected from non-smokers, smokers and smokers with COPD (n=24 per group) at the 4th Affiliated Hospital of Anhui Medical University. All blood samples were centrifuged at 1200 × g at room temperature for 10 min, and the supernatant sera were saved in liquid nitrogen before RNA extraction. All subjects have signed the written informed consent forms for sample use in this research. The demographic and clinicopathological characteristics of these subjects are shown in Table 1.
 |
Table 1 Demographic and Clinicopathological Characteristics of the Subjects in This Study |
Preparation of CSE
CSE was prepared as previously reported.16 Briefly, four commercial cigarettes were combusted with a constant speed vacuum pump and the burning time of each cigarette was controlled to 5 min. Five hundred-milliliter smoke was bubbled into a flask with 25-mL Roswell Park Memorial Institute-1640 (RPMI-1640) medium that was pre-warmed at 37°C. The suspension was titrated to pH 7.4 and filtered through a 0.22-μm filter (Millipore, Bedford, MA, USA) to remove large particles. The solution was sterilized and the optical density (OD) was determined. CSE was qualified when ΔOD (A320-A540) was between 0.9 and 1.2. The final solution was considered as 100% CSE. One hundred percent CSE was diluted with serum-free cell medium to 1%, 2% and 4% working concentrations.
Cell Culture and Treatment
Human bronchial epithelial cell line 16HBE (Sigma-Aldrich, St. Louis, MO, USA) was cultured in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 100 U/mL penicillin and 100 µg/mL streptomycin (Gibco) in a 5% CO2 incubator at 37°C. 16HBE cells were treated with the different concentrations of CSE within 30 min after dilution and cell collection was performed after 24 h.
Cell Transfection
The sequences of circ-HACE1 and TLR4 were inserted into pCE-RB-Mam vector (RIBOBIO, Guangzhou, China) and pcDNA vector (Invitrogen, Carlsbad, CA, USA), respectively. The recombinant vectors pCE-RB-Mam-circ-HACE1 and pcDNA-TLR4 were named as circ-HACE1 and TLR4. The used oligonucleotides (RIBOBIO) are listed in Table 2. 16HBE cells were treated with 2% CSE for 24 h and transfected with oligonucleotides or vectors using Lipofectamine™ 3000 (Invitrogen), referring to the transfection steps provided by manufacturer.
 |
Table 2 The Used Oligonucleotides for Cell Transfection |
The Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from serum samples and cells by TRIzol Reagent (Invitrogen), then 2 µg RNA was applied for synthesizing the complementary DNA (cDNA) by ReverTra Ace® qPCR RT Kit (Toyobo, Kita-Ku, Osaka, Japan). The qRT-PCR was performed through the reaction system by cDNA, primers and SYBR® Green Realtime PCR Master Mix (Toyobo) on the ABI PRISM® 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The primer sequences for each molecule were as follows: forward (F), 5ʹ-GACCTTGTGCAGCATGTCAG-3ʹ and reverse (R), 5ʹ-AGCAAAACCAAGCATTCCAC-3ʹ for circ-HACE1; F, 5ʹ-GGTGTACTGTAGGTGGTCGG-3ʹ and R, 5ʹ-AGAGCCCTACATCTCGCCTG-3ʹ for HACE1; F, 5ʹ-GCCGAGGTCATACACGGCTCTCCTCT-3ʹ and R, 5ʹ-TGTCGTGGAGTCGGCAATTC-3ʹ for miR-485-3p; F, 5ʹ-GGCCATTGCTGCCAACAT-3ʹ and R, 5ʹ-CAACAATCACCTTTCGGCTTTT-3ʹ for TLR4; 5ʹ-ATTCCATGGCACCGTCAAGGCTGA-3ʹ and R, 5ʹ-TTCTCCATGGTGGTGAAGACGCCA-3ʹ for glyceraldehyde-phosphate dehydrogenase (GAPDH); 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ for U6. The internal controls GAPDH and U6 were used for expression standardization. The 2−∆∆Ct method was adopted in this study for data analysis.
Identification of circRNA
12 U Ribonuclease R (RNase R; Epicentre Technologies, Madison, WI, USA) was added to 3 μg total RNA for 60 min at 37°C and the stability of circ-HACE1 was assessed by qRT-PCR detection compared with linear HACE1 expression. In addition, the location of circ-HACE1 in 16HBE cells was analyzed by qRT-PCR after nuclear and cytoplastic RNA extraction using PARIS™ Kit (Invitrogen), with U6 and GAPDH as the respective control for nucleus and cytoplasm.
Cell Counting Kit-8 (CCK-8) Assay
After CSE treatment for 24 h or transfection for 48 h in 16HBE cells, 10 μL CCK-8 solution (Sigma-Aldrich) was pipetted to cells for 4 h-incubation. The viable cell number can be quantitated through the absorbance (450 nm) detection under the microplate reader (Bio-Rad, Hercules, CA, USA).
Flow Cytometry
For cell apoptosis, 1×105 harvested 16HBE cells were stained by Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) via Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA) as per the manufacturer’s guidelines. After cell determination via the flow cytometer (BD Biosciences), we can distinguish the apoptotic cells (Annexin V+/PI-, Annexin V+/PI+) from the viable cells. The apoptotic rate was calculated using the formula: apoptotic cells/total cells × 100%. For cell cycle, 5×105 cells were incubated with ethanol (Sigma-Aldrich) and stained with PI according to the specification of Cell Cycle Assay Kit (Dojindo, Kumamoto, Japan), followed by the detection under the flow cytometer.
Enzyme-Linked Immunosorbent Assay (ELISA)
Inflammatory response was assessed through detecting the common inflammatory cytokines levels via ELISA. The examination of interleukin-1beta (IL-1β) in cell culture supernatant and tumor necrosis factor-alpha (TNF-α) in cell lysate was performed using Human IL-1 β ELISA Kit and Human TNF-alpha ELISA Kit (Sigma-Aldrich), respectively.
Detection of Malondialdehyde (MDA) and Superoxide Dismutase (SOD)
To evaluate oxidative stress in 16HBE cells, MDA and SOD levels were examined by Lipid Peroxidation (MDA) Assay Kit and SOD Assay Kit (Sigma-Aldrich) in accordance with the appendant instruction books.
Dual-Luciferase Reporter Assay
The bioinformatics analysis in this study was completed using online Circinteractome and Targetscan. The sequences of circ-HACE1 and TLR4 3ʹUTR that harbored the miR-485-3p binding sites were respectively cloned into the luciferase expression vector pmirGLO (Promega, Madison, WI, USA) to obtain the wild-type (WT) plasmids WT-circ-HACE1 and TLR4 3ʹUTR-WT. The mutant-type (MUT) plasmids MUT-circ-HACE1 and TLR4 3ʹUTR-MUT were also formed after circ-HACE1 and TLR4 3ʹUTR sequences were mutated in the binding sites for miR-485-3p. The abbreviations of these constructs are shown in Table 3. Each construct was transfected into 16HBE cells along with miR-485-3p or miR-NC, followed by the measurement of relative luciferase activity through the Dual-Luciferase Reporter Assay System (Promega).
 |
Table 3 The Abbreviations of Constructs in Dual-Luciferase Reporter Assay |
Western Blot
Protein isolation in 16HBE cells was carried out by radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich), followed by the separation of equal proteins (40 μg) via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). After protein transferring onto the polyvinylidene difluoride (PVDF) membranes (Millipore) and non-specific binding prevention in 5% skim milk (Invitrogen), PVDF membranes were incubated with anti-TLR4 (Abcam, Cambridge, UK, ab13556, 1:500) or reference gene anti-GAPDH (Abcam, ab181603, 1:10,000) at 4°C overnight and Goat Anti-Rabbit IgG H&L secondary antibody (Abcam, ab205718, 1:3000) at 25°C for 1 h. The bands of proteins were shown by enhanced chemiluminescence (ECL) Western HRP Substrate (Millipore). Image Lab software (Bio-Rad) was used to analyze the grey level of each band.
Statistical Analysis
Pearson’s correlation coefficient was exploited for analyzing the linear relation. Data expressed as the mean ± standard deviation (SD) were analyzed via SPSS 24.0. For difference analysis, Student t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were respectively performed to compare the difference of two groups and multiple groups. P<0.05 represented a significant level in statistical difference.
Results
Upregulation of Circ-HACE1 Was Found in Smokers or Smokers with COPD and CSE-Treated 16HBE Cells
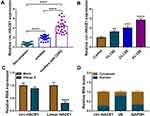
Firstly, we analyzed the expression level of circ-HACE1 in serum samples from three groups. As shown in Figure 1A, circ-HACE1 was upregulated in smokers compared to Non-smokers and its level was higher in smokers with COPD than Non-smokers or smokers. The non-toxic level of CSE was tested by treatment of 0.2% CSE, 0.4% CSE or 0.6% CSE in 16HBE cells. The results indicated that no significance of cell viability was detected in three groups and we defined the non-toxic level of CSE was 0.6% (Supplementary Figure 1A). Circ-HACE1 level was increased in 0.6% CSE group, but the difference was not quite significant (Supplementary Figure 1B). Furthermore, the expression of circ-HACE1 was determined after higher concentrations of CSE treatment. As shown in Figure 1B, circ-HACE1 was elevated by different concentrations of CSE (1%, 2%, 4%) in a dose-dependent way. 2% CSE was used in the subsequent assays. RNase R treatment significantly reduced HACE1 level but had no notable effect on circ-HACE1 expression (Figure 1C), suggesting the high stability of circ-HACE1 relative to its linear form. Through the expression comparison with nuclear U6 and cytoplastic GAPDH, we identified that circ-HACE1 was localized in the cytoplasm of 16HBE cells (Figure 1D). Smoke induced the upregulation of circ-HACE1 in COPD patients and 16HBE cells.
CSE-Induced Cell Viability Reduction, Apoptosis Acceleration, Inflammatory and Oxidative Injuries Were Attenuated After Knockdown of Circ-HACE1 in 16HBE Cells
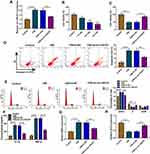
CSE-treated 16HBE cells were transfected with si-circ-HACE1, and the qRT-PCR results indicated that transfection of si-circ-HACE1 had great interfering efficiency to abolish the CSE-mediated circ-HACE1 upregulation contraposed to transfection of si-NC (Figure 2A). CCK-8 assay revealed that cell viability was inhibited by CSE in a dose-dependent way (Figure 2B) while the inhibition was countervailed by si-circ-HACE1 (Figure 2C). The analysis of apoptosis and cell cycle by flow cytometry demonstrated that the promotion of cell apoptosis (Figure 2D) and inhibition of cell cycle progression (Figure 2E) by CSE were offset after circ-HACE1 expression downregulation. The detection of inflammatory cytokines (IL-1β and TNF-α) by ELISA also showed that knockdown of circ-HACE1 abolished the stimulative influences of CSE on inflammatory response (Figure 2F). Oxidative stress is a peroxidation which can increase MDA and decrease SOD, with fundamental association in diverse diseases.17,18 MDA upregulation (Figure 2G) and SOD downregulation (Figure 2H) were triggered by CSE in 16HBE cells contrast to control cells, while this oxidative stress was reverted when CSE-treated cells were transfected with si-circ-HACE1. CSE-induced cell injury was returned by repressing circ-HACE1 expression.
Circ-HACE1 Regulated miR-485-3p as a Sponge
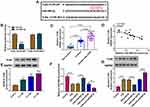
In the Circinteractome software, circ-HACE1 was predicted to have the binding sites of miR-485-3p (Figure 3A). Then, we performed the dual-luciferase reporter assay to prove the combination between circ-HACE1 and miR-485-3p. As Figure 3B displayed, luciferase signal inhibition was observed in WT-circ-HACE1 group rather than MUT-circ-HACE1 group after miR-485-3p transfection in 16HBE cells (relative to miR-NC transfection). The expression of miR-485-3p was downregulated in serum samples from smokers or smokers with COPD by contrast with serum samples from non-smokers (Figure 3C). The same downregulation of miR-485-3p was detected in 16HBE cells after 1%, 2% or 4% CSE treatment relative to the control group (Figure 3D). After the linear analysis by Pearson’s correlation coefficient, a negative relationship (r=−0.7632, p<0.001) was found between circ-HACE1 and miR-485-3p expression (Figure 3E). The level of circ-HACE1 of CSE+circ-HACE1 group was higher than that of CSE-alone group in 16HBE cells (Figure 3F), showing the successful overexpression of circ-HACE1 vector. In CSE-treated 16HBE cells, miR-485-3p expression was upregulated by knockdown of circ-HACE1 while decreased by overexpression of circ-HACE1 (Figure 3G). Circ-HACE1 can negatively regulate miR-485-3p as a molecular sponge.
CSE-Mediated Injury in 16HBE Cells Was Regulated by the Circ-HACE1/miR-485-3p Axis
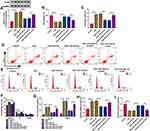
Transfection of anti-miR-485-3p abolished the si-circ-HACE1-induced miR-485-3p upregulation in CSE-treated 16HBE cells, indicating that the inhibitory efficiency of anti-miR-485-3p was great (Figure 4A). We further investigated the relation of miR-485-3p with circ-HACE1 in regulating cell injury. The results manifested that the introduction of anti-miR-485-3p in CSE-treated 16HBE cells mitigated the cell viability enhancement (Figure 4B), apoptosis inhibition (Figure 4C-D), cell cycle promotion (Figure 4E), inflammatory response (Figure 4F-G) and oxidative stress (Figure 4H-I) retardment evoked by si-circ-HACE1. Thus, the circ-HACE1/miR-485-3p axis could regulate 16HBE cell injury by CSE.
TLR4 Acted as a Target of miR-485-3p
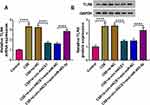
Online Targetscan predicted that TLR4 3ʹUTR sequence was able to bind to miR-485-3p (Figure 5A). The overexpression of miR-485-3p was also exhibited to reduce the relative luciferase activity in 16HBE cells transfected with TLR4 3ʹUTR-WT while luciferase activity in cells transfected with TLR4 3ʹUTR-MUT was not obviously changed (Figure 5B). The qRT-PCR presented the high expression of TLR4 mRNA in serum samples from smokers or smokers with COPD (relative to Non-smokers) (Figure 5C) and there was also a negative correlation (r=−0.7223, p<0.001) between miR-485-3p and TLR4 mRNA expression (Figure 5D). The same overexpression was found in TLR4 protein level after CSE treatment in 16HBE cells (Figure 5E). After 16HBE cells were exposed to CSE, transfection of miR-NC, miR-485-3p, anti-miR-NC or anti-miR-485-3p was performed. The transfection efficiencies of miR-485-3p for overexpression and anti-miR-485-3p for inhibition on miR-485-3p expression were excellent through qRT-PCR analysis (Figure 5F). TLR4 protein expression inhibition by miR-485-3p and promotion by anti-miR-485-3p in CSE-treated 16HBE cells indicated the negative effect of miR-485-3p on the level of TLR4 (Figure 5G). TLR4 was a target gene in the downstream of miR-485-3p.
MiR-485-3p Delayed the CSE-Induced Cell Damage via Downregulating TLR4
The functional mechanism between miR-485-3p and TLR4 was researched in CSE-treated 16HBE cells transfected with miR-NC, miR-485-3p, miR-485-3p+pcDNA or miR-485-3p+TLR4. Western blot has revealed that the TLR4 protein expression inhibition caused by miR-485-3p was reversed after transfection of TLR4 (Figure 6A). Cellular experiments indicated that transfection of TLR4 partly abrogated the miR-485-3p-mediated cell viability promotion (Figure 6B) cell apoptosis inhibition (Figure 6C-D) and cell cycle acceleration (Figure 6E) in CSE-treated 16HBE cells. As for inflammation and oxidative stress, the overexpression of TLR4 also lightened the miR-485-3p-induced concentration decline of IL-1β and TNF-α (Figure 6F-G), as well as MDA downregulation and SOD elevation (Figure 6H-I). MiR-485-3p inhibited the cell damage by CSE via targeting TLR4.
Knockdown of Circ-HACE1 Regulated Cell Injury by Upregulating miR-485-3p to Decrease TLR4 Expression in CSE-Treated 16HBE Cells
The potential expression regulation between circ-HACE1 and TLR4 was further studied. TLR4 mRNA (Figure 7A) and protein (Figure 7B) levels were suppressed in CSE+si-circ-HACE1 group compared to CSE group in 16HBE cells, whereas TLR4 expression suppression by si-circ-HACE1 was subsequently alleviated following the anti-miR-485-3p transfection. The expression analysis suggested that knockdown of circ-HACE1 downregulated the level of TLR4 by increasing miR-485-3p expression. To further investigate whether the function of circ-HACE1 was associated with the miR-485-3p-mediated TLR4 expression change, we have performed the reverted transfection (si-NC, si-circ-HACE1, si-HACE1+pcDNA, si-circ-HACE1+TLR4) after 16HBE cells were treated with 2% CSE. The results have demonstrated that the introduction of TLR4 relived all those effects of si-circ-HACE1 on biological behaviors of CSE-treated 16HBE cells (Supplementary Figure 2). Overall, the circ-HACE1 function was ascribed to the regulation of miR-485-3p-mediated TLR4 as least in part.
Discussion
In this report, we affirmed that the dysregulation of circ-HACE1, miR-485-3p and TLR4 was associated with the CSE-induced COPD. Circ-HACE1 downregulation ameliorated the bronchial epithelial cell injury caused by CSE via targeting miR-485-3p to regulate TLR4 expression level.
Dysregulated circRNAs are usually related to the progression of human respiratory diseases.19 Blood circRNAs have been suggested as reliable targets for the diagnosis of active pulmonary tuberculosis.20 Li et al found 67 dysregulated circRNAs in idiopathic pulmonary fibrosis via a circRNA microarray and clarified the circRNA/miRNA/mRNA network.21 Lin et al exhibited that circHIPK3 contributed to proliferation and migration of airway smooth muscle cells in the development of asthma via the miR-326/STIM1 axis.22 The researches of COPD have revealed that circBbs9 enhanced lung inflammation via targeting the miR-30e-5p/Adar pathway,23 and hsa_circRNA_0008672 promoted the pathogenesis of COPD via acting on the miR-1265/MAPK1 axis.24
Herein, circ-HACE1 expression was significantly increased in smokers or smokers with COPD compared to non-smokers and 16HBE cells treated with CSE contrasted with the untreated cells. Thus, circ-HACE1 upregulation might be one of the molecular manifestations of COPD occurrence. Circ-HACE1 could be used as a biomarker to improve the clinical diagnosis of COPD. Cellular experiments demonstrated that CSE stimulation triggered the inhibitory effects on cell viability and cell cycle progression but promotive effects on apoptosis and inflammation. Interestingly, knockdown of circ-HACE1 has reverted these effects in 16HBE cells. In addition to primary inflammation, oxidative stress can induce mitochondrial dysfunction to further drive lung inflammation in COPD.25 Through the detection of MDA and SOD, we found the downregulation of circ-HACE1 relieved the oxidative stress evoked by CSE. These results identified that downregulating circ-HACE1 ameliorated the CSE-induced cell damages, indicating that the knockdown of circ-HACE1 could be used a therapeutic strategy to inhibit the progression of COPD in clinic. This molecular therapy may be beneficial for the treatment of COPD, and further research in clinical presentation needs to be performed.
To explore the potential circRNA/miRNA/mRNA axis underpinning circ-HACE1, Circinteractome software was used to seek the miRNA target of circ-HACE1. MiR-485-3p was shown to interact with circ-HACE1 and circ-HACE1 acted as a sponge to negatively regulate the miR-485-3p expression. The level of miR-485-3p was lower in smokers and smokers with COPD than that in non-smokers, and its expression was also reduced by CSE in 16HBE cells. Furthermore, the alleviation of si-circ-HACE1 on the CSE-stimulated cell injury was blocked by miR-485-3p inhibition. Circ-HACE1 was firstly identified as a miRNA sponge, and circ-HACE1/miR-485-3p axis was confirmed to be implicated in the pathological development of COPD in vitro. Regulating target gene expression is the most common functional mechanism of miRNAs. In COPD, miR-206 facilitated cell apoptosis in human pulmonary microvascular endothelial cells by the direct regulation of Notch3 and VEGFA;26 miR-664a-3p overexpression was accountable for CSE-induced COPD through inhibiting FHL1.27 Tang et al declared that miR-29b downregulation participated in the airway inflammation via upregulating BRD4.28 In this chapter, miR-485-3p was discovered to have target relation with TLR4. TLR4 was highly expressed in smokers and smokers with COPD relative to non-smokers, as well as in CSE-treated 16HBE cells relative to control cells. MiR-485-3p played a protective role for 16HBE cells against CSE via downregulating the expression of TLR4, which also suggested that TLR4 acted as a pathogenic gene in CSE-induced COPD. The miR-485-3p/TLR4 signal axis was also disclosed in the COPD research for the first time. More importantly, we found that circ-HACE1 knockdown reduced the expression of TLR4 via miR-485-3p. Meanwhile, the overexpression of TLR4 eliminated the alleviation of circ-HACE1 siRNA on CSE-induced cell damages. TLR4 was again affirmed to promote the CSE-induced COPD progression, and the promoting effect of circ-HACE1 on CSE-induced COPD was also attributed to the TLR4 upregulation. Our results have clearly clarified how circ-HACE1 acted in the developing process of CSE-induced COPD. The discovery of circ-HACE1/miR-485-3p/TLR4 axis could contribute to the understanding of molecular mechanism in COPD progression.
TLR4 is associated with the inflammatory response in some diseases. Downregulation of circ_0068087 impeded the high glucose-induced cell dysfunction and inflammation in diabetes mellitus via targeting miR-197 to regulate TLR4.29 CircLRP6 regulated inflammation and oxidative stress in diabetic nephropathy via the miR-205/TLR4 axis.30 CircTUBD1 acted as a pro-inflammatory factor in radiation-induced liver disease by modulating TLR4 as the miR-146a-5p sponge.31 Our findings exhibited that the miR-485-3p/TLR4 axis was partly responsible for the function of circ-HACE1 in CSE-induced COPD. Overall, we have provided the first information for the function of mechanism of circ-HACE1 in COPD.
In conclusion, knockdown of circ-HACE1 suppressed CSE-triggered apoptosis, inflammation and oxidative stress in 16HBE cells via the miR-485-3p/TLR4 axis. The diagnostic efficiency of COPD can be enhanced using circ-HACE1 as a molecular target, and the therapeutic management of COPD may also be improved through the expression inhibition of circ-HACE1.
Abbreviations
COPD, chronic obstructive pulmonary disease; Circ-HACE1, CircRNA HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; TLR4, toll-like receptor 4; CSE, cigarette smoke extract; HBE, human bronchial epithelial cells; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde-phosphate dehydrogenase; CCK-8, cell counting kit-8; FITC, fluorescein isothiocyanate; PI, propidium iodide; ELISA, enzyme-linked immunosorbent assay; IL-1β, interleukin-1beta; TNF-α, tumor necrosis factor-alpha; MDA, malondialdehyde; SOD, superoxide dismutase; 3ʹ-UTR, 3ʹ-untranslated region.
Data Sharing Statement
Please contact the correspondence author for the data request.
Ethics Approval and Consent Participate
Written informed consent was obtained from patients with approval by the Institutional Review Board in 4th Affiliated Hospital of Anhui Medical University.
Funding
This work was supported by key research projects of natural science in colleges and universities of Anhui province (KJ2018A0858).
Disclosure
The authors declare that they have no financial or other conflicts of interest.
References
1. Moon J-Y, Leitao Filho FS, Shahangian K, et al. Blood and sputum protein biomarkers for chronic obstructive pulmonary disease (COPD). Expert Rev Proteomics. 2018;15(11):923–935. doi:10.1080/14789450.2018.1539670
2. Eapen MS, Myers S, Walters EH, et al. Airway inflammation in chronic obstructive pulmonary disease (COPD): a true paradox. Expert Rev Respir Med. 2017;11:827–839. doi:10.1080/17476348.2017.1360769
3. Hikichi M, Mizumura K, Maruoka S, et al. Pathogenesis of chronic obstructive pulmonary disease (COPD) induced by cigarette smoke. J Thorac Dis. 2019;11:S2129–S2140. doi:10.21037/jtd.2019.10.43
4. Sun Y, Zhou J. New insights into early intervention of chronic obstructive pulmonary disease with mild airflow limitation. Int J Chron Obstruct Pulmon Dis. 2019;14:1119–1125. doi:10.2147/COPD.S205382
5. Mei D, Tan WSD, Tay Y, et al. Therapeutic RNA strategies for chronic obstructive pulmonary disease. Trends Pharmacol Sci. 2020;41:475–486. doi:10.1016/j.tips.2020.04.007
6. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi:10.1016/j.pharmthera.2018.01.010
7. Zhang C, Huo ST, Wu Z, et al. Rapid development of targeting circRNAs in cardiovascular diseases. Mol Ther Nucleic Acids. 2020;21:568–576. doi:10.1016/j.omtn.2020.06.022
8. Azari H, Mousavi P, Karimi E, et al. The expanding role of CDR1-AS in the regulation and development of cancer and human diseases. J Cell Physiol. 2020. doi:10.1002/jcp.29950
9. Zeng N, Wang T, Chen M, et al. Cigarette smoke extract alters genome-wide profiles of circular RNAs and mRNAs in primary human small airway epithelial cells. J Cell Mol Med. 2019;23:5532–5541. doi:10.1111/jcmm.14436
10. Huang X, Zhu Z, Guo X, et al. The roles of microRNAs in the pathogenesis of chronic obstructive pulmonary disease. Int Immunopharmacol. 2019;67:335–347. doi:10.1016/j.intimp.2018.12.013
11. Mohamed A, Kunda NK, Ross K, et al. Polymeric nanoparticles for the delivery of miRNA to treat Chronic Obstructive Pulmonary Disease (COPD). Eur J Pharm Biopharm. 2019;136:1–8. doi:10.1016/j.ejpb.2019.01.002
12. Li X, Yu X, He Y, et al. Integrated analysis of MicroRNA (miRNA) and mRNA profiles reveals reduced correlation between microRNA and target gene in cancer. Biomed Res Int. 2018;2018:1972606. doi:10.1155/2018/1972606
13. Musri MM, Coll-Bonfill N, Maron BA, et al. MicroRNA dysregulation in pulmonary arteries from chronic obstructive pulmonary disease. relationships with vascular remodeling. Am J Respir Cell Mol Biol. 2018;59:490–499. doi:10.1165/rcmb.2017-0040OC
14. Shen W, Liu J, Zhao G, et al. Repression of Toll-like receptor-4 by microRNA-149-3p is associated with smoking-related COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:705–715. doi:10.2147/COPD.S128031
15. Jia R, Zhao XF. MicroRNA-497 functions as an inflammatory suppressor via targeting DDX3Y and modulating toll-like receptor 4/NF-kappaB in cigarette smoke extract-stimulated human bronchial epithelial cells. J Gene Med. 2020;22:e3137. doi:10.1002/jgm.3137
16. Song J, Wang Q, Zong L. LncRNA MIR155HG contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-128-5p/BRD4 axis. Biosci Rep. 2020;40:BSR20192567. doi:10.1042/BSR20192567
17. Maurya PK, Noto C, Rizzo LB, et al. The role of oxidative and nitrosative stress in accelerated aging and major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2016;65:134–144. doi:10.1016/j.pnpbp.2015.08.016
18. Zhou Q, Zhu L, Zhang D, et al. Oxidative stress-related biomarkers in postmenopausal osteoporosis: a systematic review and meta-analyses. Dis Markers. 2016;2016:7067984. doi:10.1155/2016/7067984
19. Wang J, Zhu M, Pan J, et al. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20:3. doi:10.1186/s12931-018-0962-1
20. Qian Z, Liu H, Li M, et al. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine. 2018;27:18–26. doi:10.1016/j.ebiom.2017.12.007
21. Li R, Wang Y, Song X, et al. Potential regulatory role of circular RNA in idiopathic pulmonary fibrosis. Int J Mol Med. 2018;42:3256–3268. doi:10.3892/ijmm.2018.3892
22. Lin J, Feng X, Zhang J. Circular RNA circHIPK3 modulates the proliferation of airway smooth muscle cells by miR-326/STIM1 axis. Life Sci. 2020;255:117835. doi:10.1016/j.lfs.2020.117835
23. Li M, Hua Q, Shao Y, et al. Circular RNA circBbs9 promotes PM2.5-induced lung inflammation in mice via NLRP3 inflammasome activation. Environ Int. 2020;143:105976. doi:10.1016/j.envint.2020.105976
24. Duan R, Niu H, Yu T, et al. Identification and bioinformatic analysis of circular RNA expression in peripheral blood mononuclear cells from patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:1391–1401. doi:10.2147/COPD.S252896
25. Wiegman CH, Michaeloudes C, Haji G, et al. Oxidative stress-induced mitochondrial dysfunction drives inflammation and airway smooth muscle remodeling in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2015;136:769–780. doi:10.1016/j.jaci.2015.01.046
26. Sun Y, An N, Li J, et al. miRNA-206 regulates human pulmonary microvascular endothelial cell apoptosis via targeting in chronic obstructive pulmonary disease. J Cell Biochem. 2019;120:6223–6236. doi:10.1002/jcb.27910
27. Zhong S, Chen C, Liu N, et al. Overexpression of hsa-miR-664a-3p is associated with cigarette smoke-induced chronic obstructive pulmonary disease via targeting FHL1. Int J Chron Obstruct Pulmon Dis. 2019;14:2319–2329. doi:10.2147/COPD.S224763
28. Tang K, Zhao J, Xie J, et al. Decreased miR-29b expression is associated with airway inflammation in chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 2019;316:L621–L629. doi:10.1152/ajplung.00436.2018
29. Cheng J, Liu Q, Hu N, et al. Downregulation of hsa_circ_0068087 ameliorates TLR4/NF-kappaB/NLRP3 inflammasome-mediated inflammation and endothelial cell dysfunction in high glucose conditioned by sponging miR-197. Gene. 2019;709:1–7. doi:10.1016/j.gene.2019.05.012
30. Chen B, Li Y, Liu Y, et al. circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J Cell Physiol. 2019;234:21249–21259. doi:10.1002/jcp.28730
31. Niu H, Zhang L, Chen YH, et al. Circular RNA TUBD1 acts as the miR-146a-5p sponge to affect the viability and pro-inflammatory cytokine production of LX-2 cells through the TLR4 pathway. Radiat Res. 2020;193:383–393. doi:10.1667/RR15550.1
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.