Back to Journals » Infection and Drug Resistance » Volume 15
Characterization of Genetic Variants Associated with Rifampicin Resistance Level in Mycobacterium tuberculosis Clinical Isolates Collected in Guangzhou Chest Hospital, China
Authors Hameed HMA , Fang C , Liu Z, Ju Y, Han X, Gao Y, Wang S, Chiwala G , Tan Y, Guan P, Hu J, Xiong X, Peng J, Lin Y, Hussain M, Zhong N, Maslov DA, Cook GM, Liu J, Zhang T
Received 24 May 2022
Accepted for publication 11 September 2022
Published 27 September 2022 Volume 2022:15 Pages 5655—5666
DOI https://doi.org/10.2147/IDR.S375869
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
HM Adnan Hameed1– 4*, Cuiting Fang1– 4*, Zhiyong Liu,1– 3 Yanan Ju,1– 3 Xingli Han,1– 4 Yamin Gao,1– 4 Shuai Wang,1– 3,5 Gift Chiwala,1– 4 Yaoju Tan,6 Ping Guan,6 Jinxing Hu,6 Xiaoli Xiong,1– 3 Jiacong Peng,3,7 Yongping Lin,3,7 Muzammal Hussain,4 Nanshan Zhong,3,7,8 Dmitry A Maslov,9 Gregory M Cook,10,11 Jianxiong Liu,6 Tianyu Zhang1– 4
1State Key Laboratory of Respiratory Disease, Guangzhou Institutes of Biomedicine and Health (GIBH), Chinese Academy of Sciences (CAS), Guangzhou, People’s Republic of China; 2China-New Zealand Joint Laboratory of Biomedicine and Health, Guangzhou Institutes of Biomedicine and Health (GIBH), Chinese Academy of Sciences (CAS), Guangzhou, People’s Republic of China; 3Guangdong-Hong Kong-Macau Joint Laboratory of Respiratory Infectious Diseases, Guangzhou, People’s Republic of China; 4University of Chinese Academy of Sciences (UCAS), Beijing, People’s Republic of China; 5National Clinical Research Center for Infectious Diseases, Guangdong Provincial Clinical Research Center for Tuberculosis, Shenzhen Third People’s Hospital, Shenzhen, People’s Republic of China; 6State Key Laboratory of Respiratory Disease, Guangzhou Chest Hospital, Guangzhou, People’s Republic of China; 7State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China; 8Guangzhou National Laboratory, Guangzhou, People’s Republic of China; 9Laboratory of Bacterial Genetics, Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia; 10Department of Microbiology and Immunology, School of Biomedical Sciences, University of Otago, Dunedin, New Zealand; 11Maurice Wilkins Centre for Molecular Biodiscovery, The University of Auckland, Auckland, New Zealand
*These authors contributed equally to this work
Correspondence: Tianyu Zhang, Guangzhou Institutes of Biomedicine and Health (GIBH), Chinese Academy of Sciences (CAS), Room A207, 190 Kaiyuan Ave, Science Park, Huangpu District, Guangzhou, 510530, People’s Republic of China, Tel +86-2032015270, Email [email protected] Jianxiong Liu, Guangzhou Chest Hospital, 62 Hengzhigang Road, Yuexiu District, Guangzhou, People’s Republic of China, Tel +86-2083595977, Email [email protected]
Objective: Rifampicin (RIF)-resistance, a surrogate marker for multidrug-resistant tuberculosis (TB), is mediated by mutations in the rpoB gene. We aimed to investigate the prevalence of mutations pattern in the entire rpoB gene of Mycobacterium tuberculosis clinical isolates and their association with resistance level to RIF.
Methods: Among 465 clinical isolates collected from the Guangzhou Chest Hospital, drug-susceptibility of 175 confirmed Mtb strains was performed via the proportion method and Bactec MGIT 960 system. GeneXpert MTB/RIF and sanger sequencing facilitated in genetic characterization, whereas the MICs of RIF were determined by Alamar blue assay.
Results: We found 150/175 (85.71%) RIF-resistant strains (MIC: 4 to > 64 μg/mL) of which 57 were MDR and 81 pre-XDR TB. Genetic analysis identified 17 types of mutations 146/150 (97.33%) within RRDR (codons 426– 452) of rpoB, mainly at L430 (P), D435 (V, E, G, N), H445 (N, D, Y, R, L), S450 (L, F) and L452 (P). D435V 12/146 (8.2%), H445N 16/146 (10.9%), and S450L 70/146 (47.94%) were the most frequently encountered mutations. Mutations Q432K, M434V, and N437D are rarely identified in RRDR. Deletions at (1284– 1289 CCAGCT), (1295– 1303 AATTCATGG), and insertion at (1300– 1302 TTC) were detected within RRDR of three RIFR strains for the first time. We detected 47 types of mutations and insertions/deletions (indels) outside the RRDR. Four RIFR strains were detected with only novel mutations/indels outside the RRDR. Two of the four had (K274Q + C897 del + I491M) and (A286V + L494P), respectively. The other two had (G1687del + P454L) and (TT1835-6 ins + I491L) individually. Compared with phenotypic characterization, diagnostic sensitivities of GeneXpert MTB/RIF and sequencing analysis were 95.33% (143/150), and 100% (150/150) respectively.
Conclusion: Our findings underscore the key role of RRDR mutations and the contribution of non-RRDR mutations in rapid molecular diagnosis of RIFR clinical isolates. Such insights will support early detection of disease and recommend the appropriate anti-TB regimens in high-burden settings.
Keywords: rifampicin-resistance, Mycobacterium tuberculosis, rpoB, GeneXpert MTB/RIF, resistance-determining region
Introduction
Rifampicin (RIF), a semisynthetic derivative of rifamycin B, discovered in 1965 has a strong bactericidal activity against both active and latent bacilli.1 Resistance to RIF generally co-occurs with isoniazid (INH) resistance and Mycobacterium tuberculosis (Mtb) strains resistant to both of these drugs are classified as multidrug-resistant (MDR) TB. RIF-resistant (RIFR) TB is a proxy of MDR-TB and in 2018, WHO reported around 0.5 million RIFR cases, of which 78% were MDR-TB.2 After India, China has the second-highest burden of RIFR TB and accounts for 14% of the global RIFR TB cases.2,3 About 68,200 RIFR-TB cases were reported during 2015–2019, of which 48.1% were new cases.4 The number and detection rate of RIFR-TB incidences are increasing with each passing year, from (10,019 and 14.3%) in 2015 to (18,623 and 28.7%) in 2019.3,4
Among the confirmed TB cases reported in 2019, 81.9% of them were identified as resistant to RIF which was a considerable increase from 29.5% in 2015.4 The epidemic situation of drug-resistant TB in Guangdong province is still a serious challenge. Recently, of the 30,362 strains, the total drug-resistant rate and the mono-RIFR rate were 26.75% (8121/30,362) and 6.22% (1887/30,362) respectively.5 Taking this into consideration, it has been observed that RIFR/MDR-TB is one of the leading causes of morbidity and mortality among infectious diseases around the globe.2
RIF targets the DNA-dependent RNA polymerase (RNAP), impeding translocation followed by first phosphodiester bond formation, preventing RNA elongation, and thus inhibiting transcription.6 RIFR is mainly associated with mutations in the rpoB gene which encodes the β-subunit of RNA polymerase that is required for RNA transcription.6 About 90–95% of the RIFR strains harbor mutations within the 81-bp region of rpoB, known as RIF-resistance-determining region (RRDR) from codons 426 to 452 in Mtb or 507–533 (consensus numbering scheme of RNAP from Escherichia coli).7,8 The remaining ~5–10% RIFR strains contain mutations in the N-terminal or cluster II region of the rpoB or may have unexplored resistance mechanisms.9
Phenotypic susceptibility testing requires 2–8 weeks to collect the Mtb colonies, which not only causes a diagnostic delay but also hinders TB treatment.2 WHO endorsed the Mycobacteria Growth Indicator Tube (MGIT) and Xpert MTB/RIF assay for rapid phenotypic and genetic characterizations, respectively.10 The development of Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA, USA) is a breakthrough in molecular diagnosis as it is a fully automated and hemi-nested real-time nucleic acid amplification assay that provides rapid (2-hour) and reliable results using five overlapping molecular beacon probes (A–E) covering the RRDR to identify Mtb complex (MTBC) together with the mutations associated with resistance to RIF.7,11 Xpert MTB/RIF Ultra is a newer version of the Xpert assay.10 Commercial molecular assays are upgrading the landscape of diagnostic approaches, but these methods detect mutations only within the RRDR.7,11 A recent survey in Eswatini (formerly known as Swaziland) revealed that up to 30% of MDR-TB had RIF-resistance-related mutation outside the RRDR.12,13
Precise assessment of drug susceptibility based on the detection of resistance-conferring mutations in Mtb is essential for reliable early diagnosis and to determine effective treatment regimen(s). Yet there are limited data about the prevalence and the obscure role of various rpoB mutations leading to RIF resistance in MDR-TB patients in southern China. Using comprehensive phenotypic and genetic characterization, we investigated the distribution and frequency of RIFR-associated mutations in- and outside the RRDR in clinical isolates collected from Guangzhou, the central city in southern China. This prospective study will be useful in molecular diagnosis to identify the potential role of the newly discovered rpoB mutations causing RIF resistance in Mtb.
Materials and Methods
Collection of Mtb Clinical Isolates
A total of 465 clinical isolates were collected randomly from the patients (age range, 15 to 89 years) registered from September 2019 to November 2021 at Guangzhou Chest Hospital, the biggest specialized TB hospital in southern China. Medical records were further reviewed to categorize the TB patients who experienced poor clinical response based on previous treatment. Usually, the suspected cases are tested by sputum smear microscopy and culture testing, QuantiFERON–TB Gold In-Tube test (QFT–GIT), Tuberculin Skin Test (TST), and imaging evaluation. If the etiology is not in line with the clinical examination, the patients are given medications to treat common inflammation or lung infections. However, if the general anti-inflammatory drugs are found ineffective and the etiological tests support the diagnosis of TB, the patients are treated with anti-TB drugs while observing the treatment effects. The previously treated drug-resistant TB patients and the new cases with severe TB symptoms and positive etiological tests are admitted to this hospital. All admitted patients get TB culture testing and then drug susceptibility testing (DST) is performed only for those patients which are confirmed by positive TB cultures. Mtb species were also confirmed by Ziehl–Neelsen staining and commercial MPB64 monoclonal antibody assay (Genesis, Hangzhou, China).14 175 confirmed Mtb strains were selected to proceed this study.
Drug Susceptibility Testing of Mtb Strains
Phenotypic DST was determined using the proportion method on Löwenstein–Jensen (LJ) medium following previously recommended WHO-approved guidelines.15–19 The following critical concentrations (CC: μg/mL) for anti-TB drugs were used: INH (0.2), RIF (40.0), ethambutol (2.0), streptomycin (4.0), levofloxacin (3.0), moxifloxacin (2.0), amikacin (40.0), rifabutin 20.0, and prothionamide (40.0). To carry out DST, diluted bacteria were cultured onto LJ medium with and without drugs and incubated at 37℃ for 42 days. The critical concentrations were selected as the lowest concentrations of anti-TB drugs that inhibited the 99% in vitro growth of phenotypically susceptible Mtb strains. The strain was also considered as resistant to the tested drug when Mtb growth rate was ≥1% compared with the drug-free medium.17–19 Resistance to RIF and pyrazinamide (at 1 and 100 µg/mL, respectively) was also confirmed by Bactec MGIT 960 liquid culture system (Becton Dickinson, Franklin Lake, NJ, USA) according to the manufacturer’s instructions and in line with approved guidelines.15,18 The wild-type Mtb H37Rv (ATCC27294T) reference strain was used as a control.
Determination of Minimum Inhibitory Concentration (MIC)
The MIC was determined by microplate-based Alamar blue assay.20 The bacterial strains were cultured in 7H9 medium supplemented with 10% oleic acid albumin dextrose catalase (OADC, Difco, VWR, Radnor, PA) up to 0.4–0.8 at OD600. The cultures were then diluted to OD600: 0.01 and 100 µL of culture was added per well. Twofold dilutions of the drug were prepared in 7H9 broth in 96-well plates and the gradient concentrations of RIF ranging from 0.5 to 64 µg/mL were used to access MIC. Seven days post-incubation at 37°C, Alamar blue solution (Alamar Bio-sciences/Accumed, Westlake, OH) 10% of the total volume of the well was added to each well, and the plates were incubated at 37°C for 24 h. The MIC was defined as the lowest concentration of drug that prevented the change in color.
GeneXpert MTB/RIF Assay
To detect RIFR Mtb due to mutations in RRDR, Xpert MTB/RIF assay was performed using the first/older version of GeneXpert MTB/RIF (Cepheid, Sunnyvale, CA, USA) and results were read following the manufacturer’s instructions.10,21 Briefly, the sample reagent was mixed according to 3:1 to 0.5 mL of decontaminated specimen. The tube was manually stirred twice during incubation for 15 min at room temperature and then transferred 2 mL of the reagent-sample mixture to the Xpert-test cartridge. The cartridges containing this mixture were placed in the Xpert machine and the results readouts were obtained after 90 min of the fully automated process. The probes in the Xpert system hybridize to the sequence of RRDR of rpoB: A (codons 507–511), B (codons 512–518), C (codons 518–523), D (codons 523–529) and E (codons 529–533).11 If at least two of the five probes exhibit a cycle threshold (Ct) ≤38, it indicates the presence of MTBC. Whereas the strain displaying no hybridization of one or more probes or when the variance between first and last Ct is >3.5 is recognized as RIFR Mtb.11,22
Genetic Analysis of rpoB Gene
Genomic DNA was extracted from Mtb cultures using MagMAX Total Nucleic Acid Isolation Kit (Ambion, Life Technologies, NY, USA). The purified DNA was quantified with the NanoDropND-1000 spectrophotometer (NanoDrop Technologies). The complete 3519-bp long rpoB gene was amplified in RIFS and RIFR Mtb strains using the two sets of primers designed for this study (rpoB F1: 5’–ACTTGACACCGTGGTCTTAG–3’, rpoB R1: 5’–CGAGACGTCCATGTAGTCCAC–3’), covering 1098-bp (including 180-bp of the upstream region) and (rpoB F2: 5’–ATCGAAACGCCGTACCGCAAG–3’, rpoB B R2: 5’–GACCGATGCGGAGTTCATCG– 3’) 2088-bp (including 213-bp of the first part of the gene and 84-bp of the downstream region to avoid the omission during sequencing analysis), to detect the presence of resistance-associated mutations in- and outside the RRDR. PCR products were examined on 1.0% agarose gels, purified by PCR purification kit (Qiagen, Hilden, Germany), and sequences were determined via standard Sanger DNA sequencing on a ABI3730XL genetic analyzer (BGI, Guangzhou, China). Sequencing data were aligned with the reference sequence of wild-type Mtb H37Rv strain (GenBank accession number: NC_000962.3) using the software BioEdit version 7.2.6.1. The codon numbers were reported according to the Mtb rpoB gene numbering system (TubercuList: http://genolist.pasteur.fr/TubercuList/).
Statistical Analysis
The sensitivity, specificity and their 95% confidence intervals (CIs) were calculated, and the data were treated statistically by using MEDCALC statistical software (https:/www.medcalc.org/calc/diagnostic_test.php).
Results
Demographic Features
Among the 175 Mtb clinical isolates in this study, the proportion of TB cases was higher in male patients 115/175 (65.71%), while 60/175 (34.28%) were in female patients. The maximum number of positive Mtb cases 87/175 (49.71%) were from the age group of 45–65-year-old patients2,7,23 with the highest number 74/150 (49.33%) of RIFR TB patients. Moreover, most of the Mtb isolates 105/175 (60.00%) analyzed in this study were from retreated patients of TB, whereas 70/175 (40.00%) were listed as new TB cases (Table 1).
 |
Table 1 Demographic Features of 175 Mtb Clinical Isolates |
Drug Susceptibility Profile of Mtb Isolates
Of the 175 confirmed TB strains, 150/175 (85.71%) were identified as resistant and 25/175 (14.28%) were susceptible to RIF. Among them, 57 MDR and 81 pre-XDR Mtb strains were detected cumulatively covering 138/175 (78.85%) of the isolates. Four strains were mono-resistant to RIF, while the remaining 8/150 (5.33%) RIFR strains possessed different patterns of drug resistance. In this study, the number of previously treated RIFR Mtb patients 91/150 (60.66%) was considerably higher compared to the treatment-naive cases 59/150 (39.33%).
Genetic Characterization of rpoB in Mtb
The genetic testing of Mtb strains was determined by the detection of resistance-conferring mutations in rpoB gene. Besides the mutations in RRDR, we also identified the nonsynonymous mutations outside the RRDR which remain undetected by GeneXpert MTB/RIF assay. 150/150 (100%) RIFR strains contained resistance-conferring mutations or indels in rpoB gene. Sixty-seven different types of mutations were detected in the entire region of rpoB, comprising 46 nonsynonymous mutations, 13 synonymous mutations, 4 deletions, and 4 insertions.
RRDR of rpoB
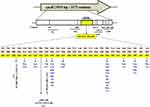
Interestingly, 146/150 (97.33%) RIFR isolates harbored single or multiple mutations/indels at codons from 426 to 452 of the rpoB in RIFR strains, of which 143/150 (95.33%) were covered by Xpert probes while 3/150 (2%) were only detected by sequencing of whole rpoB gene. Seventeen different types of nonsynonymous mutations were identified within the RRDR in RIFR Mtb strains (Figure 1). Most of the mutations were detected at codons L430 (P), D435 (V, E, G, N), N437 (D), H445 (N, D, Y, R, L), and S450 (L, F). The most frequently encountered mutations were D435V 12/146 (8.2%), H445N 16/146 (10.9%), and S450L 70/146 (47.94%) respectively. Compared to these substitutions, other RIFR-conferring rpoB mutations were present at lower frequencies. Amino acid substitutions Q432K (3/146; 2.05%), M434V (2/146; 1.36%), N437D (5/146; 3.42%), R448L (3/146; 2.05%) and L452P (3/146; 2.05%) are very rarely detected in RRDR but found in our study in 16 RIFR strains. Whereas 9/150 (6%) strains were identified with indels along with the mutations at codons 435, 445, or 450. By sequencing of whole rpoB gene, the insertion of an amino acid TTC at 1300–1302 (F434) without any other amino acid substitution is identified the first time in this study. Similarly, two phenotypically RIFR strains which also remained obscure by Xpert probes were found with the deletion of CCAGCT at 1284–1289 and AATTCATGG at 1295–1303 nucleotide positions for the first time in RRDR of rpoB.
Non-RRDR of rpoB
The sequencing results of the complete rpoB gene also showed 47 different types of non-RRDR mutations/indels (including 13 synonymous mutations) in rpoB (Figure 2). Among them, 27 nonsynonymous mutations/indels were observed only in resistant strains, two (R1162H, A1166T) were present in both resistant and susceptible strains, and five in only susceptible strains. Whereas I491 (L, M), P541 (T, K, Q), N673 (S), and A1166 (T) were frequently encountered non-RRDR mutations. Nonsynonymous mutations outside the RRDR of RIFR isolates were mostly found with mutations inside the RRDR which might be the reason for the increased level of RIF resistance in these strains.
It was interesting that the association of four Mtb strains with RIF resistance was not established when evaluated by Xpert MTB/RIF assay, though these strains were phenotypically resistant to RIF, but sequencing analysis of the complete region of rpoB revealed that missense mutations/indels were located only outside the RRDR which cannot be covered by Xpert probes (Figure 2). Two of them were detected with the novel nonsynonymous mutations (K274Q + C897 del + I491M + A1075A) and (A286V + L494P + A1075A), respectively. Of the remaining two strains, one showed deletion of G nucleotide at position 1687 along with P454L mutation and another showed the insertion of TT nucleotides at 1835–6 position + I491L + A1075A. The synonymous mutations A1075A and L1160L were present in most of the RIFR and RIFS strains. Besides, 25/175 (14.28%) RIFS strains presented synonymous mutations, ineffectual substitutions or wild-type rpoB gene.
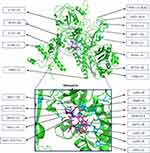 |
Figure 3 Locations of mutations and binding site residues in RpoB. |
Association Between rpoB Mutations and RIF Resistance Level
Minimum inhibitory concentration (MIC) testing indicated that mutations/indels in the RRDR of rpoB were mainly associated with resistance to RIF (CC > 1 µg/mL), with MICs ranging from 4 to >64 µg/mL. Interestingly, RIFR strains having double RRDR mutations or both RRDR and non-RRDR mutations together showed higher MIC levels compared to the strains with only a single mutation in RRDR (Table S1). Mainly, RIFR strains having mutations at codons 445 and S450L mutation developed a higher level of resistance. All RIFS isolates exhibited MICs of <0.5 µg/mL. Generally, there is a direct connotation between the distance of mutated amino acids at the drug-binding site and the level of drug resistance. The mutations mainly responsible for higher levels of RIF resistance in Mtb are found at exactly or near to RIF-binding pocket (RBP) (Figure 3). However, some frequently detected non-RRDR mutations are also marked in Figure 3. The presence of these mutations in variant distant regions facilitates to understand their effect on RIF binding and the resistance level.
Assessment of Xpert and DNA Sequencing Results
To evaluate the performance of molecular methods in the prediction of RIF resistance, the phenotypic and genotypic data for 175 Mtb isolates were compared statistically. Considering the phenotypic outcomes as a reference, detection of RIF resistance by Xpert assay showed a sensitivity of 95.33% (95% CI: 90.62–98.10%) and a specificity of 100% (95% CI: 86.28–100.00%) with the accuracy of 96.0% (95% CI: 91.93–98.38%). Whereas the sequencing analysis of the entire region of rpoB to detect resistance-conferring mutations exhibited a sensitivity of 100% (95% CI: 97.57–100%) and the specificity of 100% (95% CI: 86.28–100.00%) showing the accuracy of 100% (95% CI: 97.91–100%) (Table 2). Our analysis showed that covering the RRDR of rpoB by the Xpert system provides sensitivity (95.33%) almost similar to the entire rpoB gene sequencing (100%) with maximum accuracy of the tests regarding phenotypic resistance. However, Xpert system remained unresponsive to detect the RIF resistance of seven Mtb strains which later were clarified by sequencing of the whole rpoB gene. Of these, 3/7 were found with indels within RRDR and 4/7 had only non-RRDR mutations.
 |
Table 2 Evaluation of Molecular Susceptibility Testing Methods |
Discussion
RIF resistance is a surrogate marker for MDR-TB2,7,23 and the emergence of RIFR/MDR-TB is a serious public health problem. The lack of rapid TB diagnostic facilities and inappropriate treatment regimens for TB therapy are the key reasons for the emergence of RIFR/ MDR-TB. The exploration of molecular-level performance is essential to identify the RIFR-related rpoB mutations for better interpretation of phenotypic results and to continue the development of improved diagnostic assays. To investigate the factors associated with resistance to RIF among sputum smear and culture-positive new and previously treated drug-resistant TB patients have been scrutinized in various studies through genetic testing by commercially available molecular diagnostic approaches and sequencing analysis.24–27 Based on phenotypic and genetic characterization, the identification of 85.71% RIFR strains in the current study is higher than the study from Zhejiang (77.7%),25 and Vietnam (44.57%)26 but lower than that from Wuhan (96.71%)27 and South Korea (98.15%).20 The early diagnosis of TB and the rapid detection of resistance to anti-TB drugs are indispensable for the effective treatment and constrain the emergence of MDR-TB. Herein, we provide the detailed insights to comprehend the role of nonsynonymous mutations in- and outside of the RRDR of rpoB on RIF susceptibility.
In our study, 71/146 (48.63%) of RIFR RRDR mutants harbored mutations at codon 450 alone and with other mutations within the rpoB gene, whereas mutation S450L ranked first (70/146; 47.94%) in the data associated with high RIF resistance level. This endorsed an earlier report that codon 450 (531 in the E. coli numbering system) is the most frequently mutated in the rpoB gene.28 However, the frequency of S450L mutation can be different in various settings around the world, such as, Kazakhstan (80.9%),29 Bangladesh (69.4%),22 Taiwan (66.7%),30 China (60.9%),31 India (57.81%),32 South Africa (56%),33 South Korea (53.1%),24 Brazil (52.4%),34 Russia (47%)35 and Vietnam (37.8%).26
Besides, the next most abundant RRDR mutations occurred in the present study at codons positions 430 (13/146; 8.90%), 435 (31/146; 21.23%), and 445 (47/146; 32.19%) respectively in RIFR strains. The mutation frequencies at these codons have been reported within the ranges of 1.1–20.4%, 6.8–32%, and 0–8%, respectively.24,29,36–38 Our study elucidated that S450L, followed by H445N and D435V were the most common mutations for both RIF mono-resistant and MDR strains. The variances in the rpoB codons substitution and the frequency of rpoB mutations can have adverse effects on RNAP functions and DNA transcription. Therefore, the mutations at codons 430, 435, 445, and 452 have been associated with relapse or treatment failure in clinical settings.37
Genetic analysis in our study revealed 17 different types of mutations within RRDR in 146/150 (97.33%) RIFR Mtb strains which have been shown to confer resistance to RIF.24,37,39 Interestingly, the rarely detected RRDR mutations Q432K, M434V, and N437D in RIFR strains can also act as potential RIF resistance determinants consistent with an earlier report where mutations at codon 432 occurred exclusively in high-level RIFR strains.40 Moreover, the deletions at (1284–1289 CCAGCT), (1295–1303 AATTCATGG), and insertion at (1300–1302 TTC) were also detected within RRDR in three different RIFR strains first time in this study. These deletions and insertion were possibly involved in the impairment of RBP and therefore played a key role in the development of RIF resistance, as no other mutation was located in these resistant strains. Forty-seven different types of mutations were observed outside the RRDR, 13 of these mutations were synonymous, two of which D103D and A1075A were commonly documented in prior studies.38 Synonymous mutation A1075A has been associated with the Beijing genotype,41 and earlier reports confirmed the abundance of Beijing genotype in this region15,42; therefore, A1075A was observed in both RIFS and RIFR isolates.
Four RIFR strains remained obscure by Xpert MTB/RIF assay as it covers only RRDR of the rpoB. However, sequencing analysis of the entire rpoB gene revealed new mutations outside the RRDR. Among these four strains, two resistant strains conserved the nonsynonymous mutations and the other two strains, one had G1687 del along with (P454L + F248F + I271I + F294F) mutations, and the second was found with TT ins at 1835–6 position + I491L + A1075A. The role of rpoB mutations at codon 491 in developing RIF resistance has been investigated,12,40 and we found two different types of amino acid substitutions at this position, I491L in four and I491M in two MDR strains respectively. These four RIFR strains without RRDR mutations were initially classified as RIFR at CC (1 µg/mL) which were later confirmed by MIC determination assay where they were still able to grow at 16 µg/mL of RIF. Though, similar to earlier reports,40 the other nonsynonymous mutations outside the RRDR in RIFR strains co-existed with RRDR mutations; however, an increased level of RIF MIC was observed in these strains in comparison with those RIFR strains that possessed mutations only at codons 435, 445 and 450. Thus, the presence of these non-RRDR mutations only in RIFR strains and their influence on RIF MIC throws light on the potential role of non-RRDR mutations in conferring RIF resistance in Mtb strains.
Compared to other studies, we have detected a wide range of different types of mutations associated with the diverse range of resistance. In particular, mutations at codons 445 and 450 have consistently been associated with high levels of RIF resistance.30 Also, the MIC values for other RRDR mutants were somewhat lower than those having S450L mutations, but, higher than the previous reports of low levels of RIF resistance due to mutations at codons 430 and 435, particularly D435Y.43 The type and frequency of RIFR-related rpoB mutations may vary by settings and generally H445N and L430P mutations are associated with low-level RIF resistance. The mutations detected alone have been linked with low RIF MICs but showed higher levels of RIF resistance when multiple types of mutations were detected.44 Likewise, H445N and L430P mutants in the current study showed lower level of RIF resistance compared to those strains containing these mutations accompanied by high RIFR associated mutation (eg, S450L). A similar phenomenon was observed in another study where L430P RIFR mutant showed the MIC of 4 µg/mL but the MIC of a RIFR strain increased up to 32 µg/mL containing L430P mutation along with D435G.40 This indicates that H445N and L430P mutations alone are involved in low-level RIF resistance and only influence the MIC when co-exist with additional mutations responsible for high RIF resistance. Low-level RIF resistance causes diagnostic and treatment challenges and the data related to the low-level RIFR TB incidences are still lacking, so more comprehensive studies with a wider pool of isolates will facilitate to overcome such challenges.
Considering the structural relatedness and similar modes of action, it is assumed that all rifamycins hold common resistance mechanisms, causing a cross-resistance among all drugs of this group. This cross-resistance occurs, but strains resistant to RIF and susceptible to other rifamycins have been previously explained.45,46 In the current study, ~74.66% of RIFR strains were cross-resistant to rifabutin comparable to the other reports, where cross-resistance between the two drugs was observed from 72.2% to 85.4% of RIFR strains.45,46 Notably, the pattern of RIFR/rifabutin-susceptible has been associated with certain rpoB mutations, including M434I, D435Y, D435V, or H445L45,46 which was consistent with our findings, thus the presence of these mutations in clinical isolates may confirm not only RIF resistance but also indicates the susceptibility to rifabutin.
Resistance to RIF is generally caused by mutations in the RBP of RNAP leading to loss in drug affinity, and there is a strong association between the resistance level of RIF and the distance of the mutated residues to the drug-binding site in the RpoB.47 Therefore, majority (97.33%) of our detected mutations in the RRDR of rpoB have an obvious effect on RIF resistance, because most of the affected residues are located in the RBP site. Interestingly, similar to previous studies40,48 some mutations (R1162H, A1166T) were observed in both RIFS and RIFR isolates and R1162N, N1163R, N1163S, E1164G, S1167R substitutions only in RIFS isolates. These mutations were considered ineffectual on the interaction between RIF and RBP, therefore not involved in the development of RIF resistance. Considering previous observations, it was anticipated that such mutations may act as compensatory mutations to alleviate fitness impairment acquired by other mutations directly associated with drug resistance.40,48 However, the number of isolates harboring certain types of rpoB mutations was limited. Further investigation through mechanistic approaches will clarify the effects of non-RRDR mutations on the interaction between RpoB and RIF in susceptible and resistant strains, particularly when the patient’s diagnostic or treatment outcomes are not as expected. The RIF resistance level is important as some drugs may convert the low-level RIF-resistance into RIF-susceptible and the patients could be cured more easily.
Conclusions
In conclusion, this study revealed comprehensive phenotypic and genetic profiles of RIFR Mtb clinical isolates. However, evaluation of non-RRDR of rpoB reveals the unnoticed resistance-associated nonsynonymous mutations for the detection of false-susceptible strains and the patients with false-negative results most likely receive the treatment therapy for drug-susceptible TB and tend to have poor treatment outcome. However, we observed that assessing mutations outside the RDRR contributed only ~5% increase in the diagnostic sensitivity because Xpert probes covering the RRDR showed ~95% diagnostic sensitivity. Hence, improving the access of WHO-endorsed GeneXpert MTB/RIF could be useful for rapid molecular diagnosis of RIF resistance in Mtb. Lastly, resistance-associated mutations in- and outside the RRDR of rpoB highlight their role as potential diagnostic resistance markers and possible target sites for RIF and other rifamycins in Mtb. These findings may facilitate in designing the new probes for various alleles associated with RIF resistance to increase the sensitivity of molecular diagnostic methods for Mtb isolates in different geographic settings and also to understand RIF resistance for developing precise TB therapy for MDR-TB patients.
Ethical Approval
This study followed WHO guidelines and was approved by the ethics committee of Guangzhou Chest Hospital (GZXK-2019–21).
Funding
This work was supported by the National Key R&D Program of China (2021YFA1300904), partially by the National Natural Science Foundation of China (NSFC 81973372), Joint Research the Russian Science Foundation (RSF)-NSFC Collaboration grant numbers 21-45-00018 (to D.M.), 82061138019 (to T.Z.), the Joint Research Health Research Council of New Zealand (HRC)-NSFC Collaboration grant numbers 20/1211 (to G.C.), 82061128001 (to T.Z.), supported by China-New Zealand Joint Laboratory on Biomedicine and Health, Department of Science and Technology of Guangdong Province (2019B110233003), the Chinese Academy of Sciences (154144KYSB20190005, YJKYYQ20210026), State Key Laboratory of Respiratory Disease (SKLRD-OP-202324 to Tianyu Zhang), and China Postdoctoral Fellowship, Guangdong province, Huangpu, GIBH-University of Chinese Academy of Sciences, President’s International Fellowship Initiative-CAS and State Key Laboratory of Respiratory Disease(SKLRD-Z-202301 to H.M. Adnan Hameed). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
Dr Dmitry A Maslov reports grants from Russian Science Foundation, during the conduct of the study. All authors approved the study to publish in your esteemed journal and declare no competing interests.
References
1. Rastogi N, Labrousse V, Goh KS. In vitro activities of fourteen antimicrobial agents against drug susceptible and resistant clinical isolates of Mycobacterium tuberculosis and comparative intracellular activities against the virulent H37Rv strain in human macrophages. Curr Microbiol. 1996;33(3):167–175. doi:10.1007/s002849900095
2. World Health Organization. Global Tuberculosis Report. Geneva, Switzerland: World Health Organization; 2019.
3. Zhang H, Liu X, Xu C, et al. Guiding tuberculosis control through the healthy China initiative 2019–2030. China CDC Wkly. 2020;2(49):948–950. doi:10.46234/ccdcw2020.236
4. Su W, Ruan Y, Li T, Du X, Jiang J, Li R. Characteristics of rifampicin-resistant tuberculosis detection in China, 2015–2019. Infect Dis Pov. 2021;10(1):99. doi:10.1186/s40249-021-00883-8
5. Yan-mei CHEN, Hui-zhong WU, Liu-yue XU, Ke-hao PENG, Mei-ling YU. Analysis of monitoring results of tuberculosis drug-resistance in Guangdong province from 2016 to 2020. Chinese J of Antituber. 2022;44(7):685–689. doi:10.19982/j.issn.1000-6621.20220028
6. Loots DT. New insights into the survival mechanisms of rifampicin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother. 2015;71(3):655–660. doi:10.1093/jac/dkv406
7. Andre E, Goeminne L, Cabibbe A, et al. Consensus numbering system for the rifampicin resistance-associated rpoB gene mutations in pathogenic mycobacteria. Clin Microbiol Infect. 2017;23(3):167–172. doi:10.1016/j.cmi.2016.09.006
8. Hameed HMA, Islam MM, Chhotaray C, et al. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis strains. Front Cell Infect Microbiol. 2018;8:114. doi:10.3389/fcimb.2018.00114
9. Ahmad S, Mokaddas E. Recent advances in the diagnosis and treatment of multidrug-resistant tuberculosis. Respir Med. 2009;103(12):1777–1790. doi:10.1016/j.rmed.2009.07.010
10. Dorman SE, Schumacher SG, Alland D, et al. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis. 2018;18(1):76–84. doi:10.1016/S1473-3099(17)30691-6
11. Lawn SD, Nicol MP. Xpert MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Future Microbiol. 2011;6(9):1067–1082. doi:10.2217/fmb.11.84
12. André E, Goeminne L, Colmant A, Beckert P, Niemann S, Delmée M. Novel rapid PCR for the detection of Ile491Phe rpoB mutation of Mycobacterium tuberculosis, a rifampicin-resistance-conferring mutation undetected by commercial assays. Clin Microbiol Infect. 2017;23(4):267. e5–267. e7. doi:10.1016/j.cmi.2016.12.009
13. Sanchez-Padilla E, Merker M, Beckert P, et al. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med. 2015;372(12):1181–1182. doi:10.1056/NEJMc1413930
14. Pang Y, Dong H, Tan Y, et al. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci Rep. 2016;6:25330. doi:10.1038/srep25330
15. Hameed HA, Tan Y, Islam MM, et al. Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates in southern China. Infect Drug Resist. 2020;13:217. doi:10.2147/IDR.S230774
16. Islam MM, Tan Y, Hameed HMA, et al. Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect. 2018;25:1041.e1–1041.e7. doi:10.1016/j.cmi.2018.12.008
17. World Health organization. Policy Guidance on Drug-Susceptibility Testing (DST) of Second-Line Antituberculosis Drugs. Geneva: World Health Organization; 2008.
18. World Health organization. Technical Manual for Drug Susceptibility Testing of Medicines Used in the Treatment of Tuberculosis. World Health Organization; 2018.
19. Islam MM, Tan Y, Hameed HMA, et al. Prevalence and molecular characterization of amikacin resistance among Mycobacterium tuberculosis clinical isolates from southern China. J Glob Antimicrob. 2020;22:290–295. doi:10.1016/j.jgar.2020.02.019
20. Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41(5):1004–1009. doi:10.1128/AAC.41.5.1004
21. Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48(1):229–237. doi:10.1128/jcm.01463-09
22. Uddin MKM, Rahman A, Ather MF, et al. Distribution and frequency of rpoB mutations detected by Xpert MTB/RIF assay among Beijing and Non-Beijing rifampicin resistant Mycobacterium tuberculosis isolates in Bangladesh. Infect Drug Resist. 2020;13:789–797. doi:10.2147/IDR.S240408
23. Ahmad S, Mokaddas E. Current status and future trends in the diagnosis and treatment of drug-susceptible and multidrug-resistant tuberculosis. J Infect Public Health. 2014;7(2):75–91. doi:10.1016/j.jiph.2013.09.001
24. Jnawali HN, Hwang SC, Park YK, et al. Characterization of mutations in multi- and extensive drug resistance among strains of Mycobacterium tuberculosis clinical isolates in Republic of Korea. Diagn Microbiol Infect Dis. 2013;76(2):187–196. doi:10.1016/j.diagmicrobio.2013.02.035
25. Liu Z, Zhang M, Wang J, et al. Longitudinal analysis of prevalence and risk factors of rifampicin-resistant tuberculosis in Zhejiang, China. Biomed Res Int. 2020:3159482. doi:10.1155/2020/3159482
26. Minh NN, Van BN, Son NT, et al. Molecular characteristics of rifampin-and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Vietnam. J Clin Microbiol. 2012;50(3):598–601. doi:10.1128/JCM.05171-11
27. Huang H, Zhang Y, Li S, et al. Rifampicin resistance and multidrug-resistant tuberculosis detection using Xpert MTB/RIF in Wuhan, China: a retrospective study. Microb Drug Resist. 2017;24(5):675–679. doi:10.1089/mdr.2017.0114
28. Farhat MR, Sultana R, Iartchouk O, et al. Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am J Respir Crit Care Med. 2016;194(5):621–630. doi:10.1164/rccm.201510-2091OC
29. Kozhamkulov U, Akhmetova A, Rakhimova S, et al. Molecular Characterization of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Kazakhstan. Jpn J Infect Dis. 2011;64(3):253–255. doi:10.7883/yoken.64.253
30. Lin YH, Tai CH, Li CR, Lin CF, Shi ZY. Resistance profiles and rpoB gene mutations of Mycobacterium tuberculosis isolates in Taiwan. J Microbiol Immunol Infect. 2013;46(4):266–270. doi:10.1016/j.jmii.2012.06.008
31. Jing W, Pang Y, Zong Z, et al. Rifabutin resistance associated with double mutations in rpoB gene in Mycobacterium tuberculosis isolates. Front Microbiol. 2017;8(1768). doi:10.3389/fmicb.2017.01768
32. Prasad PG, Jasmine MS, Deepthi K, Allam US, Allam US. Analysis of drug resistance mutations in pulmonary Mycobacterium tuberculosis isolates in the Southern coastal region of Andhra Pradesh, India. Braz J Infect Dis. 2019;23:281–290. doi:10.1016/j.bjid.2019.07.002
33. Rukasha I, Said HM, Omar SV, et al. Correlation of rpoB Mutations with minimal inhibitory concentration of rifampin and rRifabutin in Mycobacterium tuberculosis in an HIV/AIDS endemic setting, South Africa. Front Microbiol. 2016;7(1947). doi:10.3389/fmicb.2016.01947
34. Valim ARM, Rossetti MLR, Ribeiro MO, Zaha A. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J Clin Microbiol. 2000;38(8):3119–3122. doi:10.1128/JCM.38.8.3119-3122.2000
35. Casali N, Nikolayevskyy V, Balabanova Y, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46(3):279–286. doi:10.1038/ng.2878
36. Poudel A, Nakajima C, Fukushima Y, et al. Molecular characterization of multidrug-resistant Mycobacterium tuberculosis isolated in Nepal. Antimicrob Agents Chemother. 2012;56(6):2831–2836. doi:10.1128/AAC.06418-11
37. Rahim Z, Nakajima C, Raqib R, et al. Molecular mechanism of rifampicin and isoniazid resistance in Mycobacterium tuberculosis from Bangladesh. Tuberculosis. 2012;92(6):529–534. doi:10.1016/j.tube.2012.07.005
38. Tang K, Sun H, Zhao Y, et al. Characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Sichuan in China. Tuberculosis. 2013;93(1):89–95. doi:10.1016/j.tube.2012.10.009
39. Luo T, Zhao M, Li X, et al. Selection of mutations to detect multidrug-resistant Mycobacterium tuberculosis strains in Shanghai, China. Antimicrob Agents Chemother. 2010;54(3):1075–1081. doi:10.1128/AAC.00964-09
40. M-c L, Lu J, Lu Y, et al. rpoB mutations and effects on rifampin resistance in Mycobacterium tuberculosis. Infect Drug Resist. 2021;14:4119. doi:10.2147/IDR.S333433
41. Wan L, Liu H, Li M, et al. Genomic analysis identifies mutations concerning drug-resistance and Beijing genotype in multidrug-resistant Mycobacterium tuberculosis isolated from China. Front Microbiol. 2020;11:1444. doi:10.3389/fmicb.2020.01444
42. Hameed HMA, Tan Y, Islam MM, et al. Phenotypic and genotypic characterization of levofloxacin- and moxifloxacin-resistant Mycobacterium tuberculosis clinical isolates in southern China. J Thorac Dis. 2019;11(11):4613–4625. doi:10.21037/jtd.2019.11.03
43. Feuerriegel S, Oberhauser B, George AG, et al. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol. 2012;12(1):90. doi:10.1186/1471-2180-12-90
44. Shea J, Halse TA, Kohlerschmidt D, et al. Low-level rifampin resistance and rpoB mutations in Mycobacterium tuberculosis: an analysis of whole-genome sequencing and drug susceptibility test data in New York. J Clin Microbiol. 2021;59(4):e01885–20. doi:10.1128/JCM.01885-20
45. Cavusoglu C, Karaca‐Derici Y, Bilgic A. In‐vitro activity of rifabutin against rifampicin‐resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Clin Microbiol Infect. 2004;10(7):662–665. doi:10.1111/j.1469-0691.2004.00917.x
46. Jamieson F, Guthrie J, Neemuchwala A, Lastovetska O, Melano R, Mehaffy C. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol. 2014;52(6):2157–2162. doi:10.1128/JCM.00691-14
47. Phelan J, Coll F, McNerney R, et al. Mycobacterium tuberculosis whole genome sequencing and protein structure modelling provides insights into anti-tuberculosis drug resistance. BMC Med. 2016;14(1):31. doi:10.1186/s12916-016-0575-9
48. Jagielski T, Bakuła Z, Brzostek A, et al. Characterization of mutations conferring resistance to rifampin in Mycobacterium tuberculosis clinical strains. Antimicrob Agents Chemother. 2018;62(10):e01093–e0118. doi:10.1128/AAC.01093-18
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.