Back to Archived Journals » Oncolytic Virotherapy » Volume 4
Characterization of an oncolytic adenovirus vector constructed to target the cMet receptor
Authors Sakr H , Coleman D, Cardelli J, Mathis M
Received 26 April 2015
Accepted for publication 26 June 2015
Published 4 September 2015 Volume 2015:4 Pages 119—132
DOI https://doi.org/10.2147/OV.S87369
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Hany I Sakr,1–3 David T Coleman,3,4 James A Cardelli,3,4 J Michael Mathis,2,3,5
1Department of Cellular Biology and Anatomy, 2Gene Therapy Program, 3Feist-Weiller Cancer Center, 4Department of Microbiology and Immunology, LSU Health Shreveport, Shreveport, 5Department of Comparative Biomedical Sciences, School of Veterinary Medicine, Louisiana State University, Baton Rouge, LA, USA
Abstract: The cMet receptor is a homodimer with tyrosine kinase activity. Upon stimulation with its ligand, hepatocyte growth factor (HGF), the receptor mediates wide physiologic actions. The HGF-cMet signaling pathway is dysregulated in many cancers, which makes cMet an important target for novel therapeutic interventions. Oncolytic adenoviruses (Ads) have been used for the past three decades as a promising therapeutic approach for a wide array of neoplastic diseases. To date, achieving cancer-specific replication of oncolytic Ads has been accomplished by either viral genome deletions or by incorporating tumor selective promoters. To achieve novel specificity of oncolytic Ad infection of cancer cells that overexpress cMet, we inserted the HGF NK2 sequence, corresponding to a competitive antagonist of HGF binding to the cMet receptor, into the Ad serotype 5 (Ad5) fiber gene. The resulting vector, Ad5-pIX-RFP-FF/NK2, was rescued, amplified in HEK293 cells, and characterized. Binding specificity and viral infectivity were tested in various cancer cell lines that express varying levels of cMet and hCAR (the Ad5 receptor). We found that Ad5-pIX-RFP-FF/NK2 demonstrated binding specificity to the cMet receptor. In addition, there was enhanced viral infectivity and virus replication compared with a non-targeted Ad vector. Although NK2 weakly induces cMet receptor activation, our results showed no receptor phosphorylation in the context of an oncolytic Ad virus. In summary, these results suggest that an oncolytic Ad retargeted to the cMet receptor is a promising vector for developing a novel cancer therapeutic agent.
Keywords: bacteriophage T4 fibritin, fiber protein, gene therapy, hCAR, hepatocyte growth factor, NK2 isoform, oncolytic virotherapy, pIX protein, red fluorescent protein
Introduction
Adenovirus-based gene therapy represents a rational approach to developing novel cancer treatment regimens to improve cancer survival rates in patients. Oncolytic adenoviruses (Ads) have emerged as a promising therapeutic tool to enable the virus-mediated lysis of infected tumor cells.1 After initial binding to its native receptor, hCAR (the human coxsackievirus and adenovirus receptor), an Arg-Gly-Asp (RGD) motif on the Ad penton capsid protein interacts with integrins on the cell surface to trigger internalization.2 Ad virions are then disassembled in the endosome, followed by endosomal escape and nuclear translocation, where viral gene expression and DNA replication take place.3 Subsequently, the release of the viral progeny during infection results in further propagation in surrounding tumor cells. Despite being attractive delivery vehicles, applying Ad vectors in clinical trials has been limited by their poor infection efficiency in cells that express low levels of the native hCAR.4 Since hCAR is downregulated in many cancers,5 development of novel strategies to enhance viral affinity toward tumor cells over normal cells is a key approach to overcome a major hurdle in widespread cancer therapy application.
The cMet oncogene is a homodimer cell surface receptor with tyrosine kinase activity. Activation of the cMet pathway in cancer has multiple effects on mitogenesis, motogenesis, morphogenesis, and anoikis/survival of tumor cells, as well as angiogenesis.6–8 The cMet oncogene is dysregulated and activated in the majority of malignancies, including lung, breast, and hepatocellular carcinomas (HCCs),9 and is associated with poor outcome. Activation of the cMet receptor initiates signaling through a number of downstream pathways and can crosstalk with multiple other cell surface receptors.10,11 The widespread involvement in cancer makes cMet a good target for novel therapeutic interventions.
The ligand for cMet, hepatocyte growth factor (HGF) or scatter factor, is a heterodimeric molecule composed of two chains, a 69 kDa α-chain and a smaller 34 kDa β-chain. The HGF α-chain has an N-terminal hairpin domain and four kringle domains, while the β-chain has serine protease-like activity. The N-terminal hairpin loop and kringle-1 domain of the α-chain are essential for specific binding to cMet.12,13 In addition, HGF has multiple isoforms,14 and extensive characterization of these isoforms has identified an antagonist of HGF/cMet binding. It is reported that the NK1 isoform has partial agonist/antagonist activity,15 and the NK2 and NK3 isoforms exert some motogenic activity, while the NK4 isoform fails to exert any observable biological activities through cMet.12,16–19
Many modifications have been incorporated into Ad vectors to increase their overall infection potential, proliferation efficiency, tropism, and ability to penetrate solid tumors. Among the most popular strategies is the genetic modification of the fiber protein.20 In the present study, we sought to test and validate a fiber-modified oncolytic Ad serotype 5 (Ad5) vector to retarget infection specificity to cancer cells that overexpress the cMet receptor. We hypothesized that retargeting Ad5 using the HGF NK2 isoform would allow selective infection and killing of cMet expressing cancer cells.
Material and Methods
Cell lines and culture conditions
Use of human cell lines (Biosafety Level 2) was approved by the Institutional Biosafety Committees at LSU Health Shreveport and at Louisiana State University. The human HCC cell lines (Hep 3B2.1-7 and SNU-449), the human breast cancer cell lines (MDA-MB-231, SK-BR-3, ZR-75-1, BT-549, and MCF-7), the human non-small cell lung cancer cell line NCI-H1993, and the human embryonic kidney cell line HEK293 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The HEK293/F28 helper cell line has been previously described21 and was provided by David T Curiel (Washington University School of Medicine at St Louis, St Louis, MO, USA). The Hep 3B2.1-7 and MCF-7 cell lines were maintained in Eagle’s minimum essential medium (Mediatech, Inc., Manassas, VA, USA), and the SNU-449 and NCI-H1993 cell lines were maintained in RPMI-1640 (Mediatech, Inc.). All other breast cancer cell lines, as well as the HEK293 cell line, were maintained in Dulbecco’s minimum essential medium (Mediatech, Inc.). The HEK293/F28 cell line was grown under the selection of zeocin (400 μg/mL) in DMEM. All cell line media were supplemented with 10% heat-inactivated fetal bovine serum and 1% antibiotic solution (Thermo Fisher Scientific, Waltham, MA, USA). All the cell lines were maintained in a humidified atmosphere of 5% CO2 at 37°C.
Construction and characterization of the recombinant adenovirus vector
Initially, we sought to incorporate NK4 isoform (447 amino acids) into Ad5/fiber. However, Ad infection depends on a critical fiber shaft length,22 and the NK2 isoform at 257 amino acids provides a more optimal length for incorporation. We replaced the native Ad5 fiber with a chimeric fiber by fusion of the N-terminus tail of the Ad5 fiber with the T4-phage fibritin protein linked with the HGF/NK2 isoform at the carboxyl (C) terminus. In brief, an XbaI-SwaI fragment encompassing the chimeric Ad fiber gene was synthesized (GenScript, Piscataway, NJ, USA) and used to replace an XbaI-SwaI fragment of the wild-type Ad sequence within the pAdEasy-1 plasmid (Agilent Technologies, Santa Clara, CA, USA), introducing the T4-phage rod-like trimeric fibritin molecule. Ampicillin-resistant colonies were selected following transformation; DNA was extracted, and identities of positive clones were confirmed by restriction digestion and polymerase chain reaction (PCR). A pIX-RFP reporter gene was introduced into the AdEasy-1 by homologous recombination with a modified pShuttle vector containing a wild-type Ad5 E1A gene and the mCherry coding sequence inserted downstream of the Ad5 minor capsid pIX gene to generate a C-terminal pIX fusion protein (pShuttle-E1A-pIX-RFP), a kind gift from Anton V Borovjagin (University of Alabama at Birmingham, Birmingham, AL, USA). Recombinants were selected on kanamycin agar plates and confirmed by restriction digestion and PCR analysis. DNA sequencing was performed to confirm the identity of the inserted fragments.
Rescue, propagation, and purification of Ad virions
As described previously,23 the genome of the fiber-modified virus was used to transfect HEK293/F28 cells that stably express the Ad5 wild-type fiber, by using CaPO4 co-precipitation kit (Stratagene). To obtain a homogenous population of Ad5-pIX-RFP-FF/NK2 virions, the rescued virus was used to reinfect HEK293 cells. The recombinant Ad virus was then purified by equilibrium ultracentrifugation on CsCl gradients. The virus titer of each Ad preparation was determined by spectrophotometry using a conversion factor of 1.1×1012 viral particles (VP) per absorbance unit at 260 nm.
Virus binding assay
Cells were infected with Ad5-pIX-RFP-FF/NK2 or the Ad5-pIX-RFP-wt/Fiber control. When indicated, cells were pretreated with cMet-blocking polyclonal antibody (R&D Systems, Inc., Minneapolis, MN, USA) for 30 minutes at 4°C. The incubation temperature was 4°C unless otherwise specified. In all, 1×105 cells were washed once with ice-cold phosphate-buffered saline (PBS). The virus was added to wells or microcentrifuge tubes at the indicated multiplicity of infection (MOI) and incubated for 30 minutes or 1 hour (specified in Figures 2–9). Following incubation, cells were washed three times with ice-cold PBS, collected, and resuspended in 0.2 mL PBS. Total genomic DNA was extracted using a DNA-mini kit, (Qiagen NV, Venlo, the Netherlands), according to the manufacturer’s instructions. Aliquots of the extracted DNA (2.0 μL) were used for real-time PCR to measure E4 copy number.
Virus infectivity assay
Cell lines were infected with Ad5-pIX-RFP-FF//NK2 or with Ad5-pIX-RFP-wt/Fiber control at an increasing MOI of 1.0 VP/cell, 10 VP/cell, 100 VP/cell, and 1,000 VP/cell. When indicated, cells were pretreated with human HGF (Lonza, Walkersville, MD, USA) at increasing concentrations of 0.5 ng, 5.0 ng, and 50 ng. The cells were incubated for 2 hours at 37°C. The cells were then washed with PBS twice, and complete growth medium was added. After 48 hours, cells were washed twice with PBS, collected, and resuspended in 400 μL PBS. RFP expression was measured by flow cytometry.
Virus replication assay
Cell lines were infected with Ad5-pIX-RFP-FF//NK2 or Ad5-pIX-RFP-wt/Fiber (control) at an MOI of 100 VP/cell. The infected cells were incubated for 4 days at 37°C. At each time point, cells were collected and resuspended in 0.2 mL PBS. Total genomic DNA was extracted using a Qiagen DNA-mini kit and used for real-time PCR to measure E4 copy number as an indicator of replication.
Virus oncolysis assay
Each cell line was seeded in replicates into 96-well plates (3×103 cells/well) and infected with Ad5-pIX-RFP-FF/NK2, Ad5-pIX-RFP-wt/Fiber, or a non-replicating control virus (Ad5-CMV-GFP/Luc) at an MOI of 1,000 VP/cell. The infected cells were incubated for 7 days at 37°C. Uninfected cells were included as controls. At each day after infection, cell survival was determined using a colorimetric assay based on the CellTiter-Blue reagent (Promega Corporation, Fitchburg, WI, USA) that quantifies the number of living cells based on mitochondrial activity. At each time point, 20 μL of the dye was added to each well and the plate was shaken for 10 seconds and then incubated for 20 hours at 37°C to obtain maximal fluorescence emission. After incubation, 50 μL of 3% SDS solution was added, the plate was shaken again, and the fluorescence emission was read at 590 nm.
Flow cytometry
A fluorescein isothiocyanate-labeled anti-cMet antibody and its isotype control (R&D Systems, Inc.) and a phycoerythrin-labeled anti-hCAR antibody (Santa Cruz Biotechnology Inc., Dallas, TX, USA) and its isotype control (Abcam, Cambridge, MA, USA) were used for this assay. For each cell line, 1×105 cells were incubated with the antibodies for 45 minutes at 4°C in the dark. After incubation, the cells were washed twice with PBS, pelleted, resuspended in 400 μL PBS, and analyzed using a FACSCalibur cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) using CellQuest software.
cMet knockdown in the Hep 3B2.1-7 cell line
The Hep3B2.1 cells stably producing shRNAs targeting cMet expression (Hep3B2.1-cMet/KD) or non-target shRNA (Hep3B2.1-NT/Ctrl) were generated using MISSION lentiviral transduction particles (Sigma-Aldrich Co, St Louis, MO, USA). Five individual clones for each were established by selection in the medium containing 10 μg/mL puromycin.
PCR and qPCR
A pair of oligonucleotide primers specific for the human NK2 isoform was synthesized (Thermo Fisher Scientific) and used to confirm the Ad vector construction:
- forward: 5′-GAATTCAAAAAATCAGCAAAGACTACCC-3″ and
- reverse: 5′-GGATATCTTTCAGGCAAGAATTTG-3′.
PCR reactions were performed using a PfuUltra II Hotstart PCR Master Mix (Agilent Technologies) for 36 cycles at 94°C for 1 min, 55°C for 30 seconds, and 72°C for 30 seconds. The final PCR products were analyzed by electrophoresis on 1% agarose gels containing 1 μg/mL ethidium bromide and visualized on a UV transilluminator.
For real-time qPCR of DNA, the following sets of primers and probes were synthesized (Thermo Fisher Scientific) and used:
- human Ad5 E4 gene
- forward: 5′-GGGTCGCCACTTAATCTACCT-3′;
- reverse: 5′-GCAAGGCGCTGTATCCAA-3′; and
- probe: 5′FAM-CGCTTGTGGTATGATGGCC ACGT-TAMRA3′.
- human GAPDH gene
- forward: 5′-dACCAGGTGGTCTCCTCTGAC-3′;
- reverse: 5′-dTTGCTGTAGCCAAATTCGTT-3′; and
- probe: 5′FAM-dTTTGTCAAGCTCATTTCC TGGTATGAC-TAMARA3′.
In this assay, the extracted DNAs were amplified for 40 cycles at 94°C for 5 seconds and 60°C for 30 seconds, using a TaqMan Real-Time PCR Master Mix (Thermo Fisher Scientific) on an Applied Biosystems 7900HT system (Thermo Fisher Scientific).
For the human cyclophilin mRNA quantification, the primers used were as follows:
- sense: 5′-CTTGGGCCGCGTCTCC-3′ and
- antisense: 5′-GCAGGAACCCTTATAACCAAA TCC-3′.
In this assay, the extracted DNAs were amplified for 40 cycles of 94°C for 5 seconds and 60°C for 30 seconds using an iQ SYBR Green Supermix (Bio-Rad Laboratories Inc., Hercules, CA, USA) on an Applied Biosystems 7900HT system (Thermo Fisher Scientific).
Western blot analysis
Western blot analysis was performed as previously described.24 In brief, cells were grown in 24-well plates to approximately 70% confluency. Following treatment, the protein was harvested with reducing Laemmli buffer followed by 5 minutes of boiling. Primary antibodies used were as follows: anti-HGFα (Santa Cruz Biotechnology Inc.), anti-Ad-fiber [4D2] (Abcam), anti-Met (Y1234/35), anti-pAkt (S476) (Cell Signaling Technology, Danvers, MA, USA), anti-cMet (Thermo Fisher Scientific), GAPDH (EMD Millipore, Billerica, MA, USA), anti-β-actin (Abcam), and anti-α-tubulin (Neomarkers Inc., Fremont, CA, USA). Blots were probed with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology Inc.) and visualized using an enhanced chemiluminescence system (GE Healthcare Biosciences; Pittsburgh, PA) for protein detection.
Statistical analysis
All data are expressed as means ± SEM unless otherwise specified. Statistical analyses were performed using Student’s t-test (two tailed). A P-value <0.05 was considered to be statistically significant.
Results
Production of recombinant Ad5-pIX-RFP-FF/NK2 virus
To construct the recombinant Ad vector, we created a recombinant fiber gene containing 80 amino acids from the N-terminus of the Ad fiber (corresponding to the tail domain), 257 amino acids from the bacteriophage T4 fibritin protein (containing the fibritin shaft and foldon trimerization domains), a 15 amino acid spacer (GGGGSGGGGSGGGGS), and 254 amino acids of the naturally occurring human HGF isoform sequence (corresponding to NK2). This Ad5 fiber–T4 fibritin–HGF/NK2 construct was inserted into the pAdEasy-1 plasmid as shown in Figure 1. In addition, the minor capsid protein pIX gene was modified by fusion with the monomeric red fluorescent protein 1 (mRFP1) sequence as previously described.25
The identity of HGF/NK2 within our Ad5 vector construct was confirmed by performing a PCR analysis with NK2-specific oligonucleotides and by DNA sequencing (data not shown). The Ad vector was rescued by transfecting HEK293/F28 cells with the linearized recombinant plasmid. The purified Ad5-pIX-RFP-FF/NK2 was further propagated in the parental HEK293 cell line. This step was essential to eliminate contamination of wild-type fibers in the virus stock and ensure homogenous virus preparations. The rescued viral clone was amplified and purified and subjected to selective DNA sequence analysis to confirm the appropriate location and reading frame for the NK2 insert.
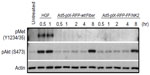
To confirm the rescue of viable virions with the proper Ad5 fiber–T4 fibritin–HGF/NK2 chimera incorporation, a Western blot analysis was performed on HEK293 cells infected with Ad5-pIX-RFP-FF/NK2 at an MOI of 50 VP/cell. Uninfected cells or cells infected with Ad5-pIX-RFP-wt/Fiber were used as controls. At 2 days after infection, 70%–80% of the cells expressed the pIX-RFP reporter gene. Cell lysates were subjected to Western blot analysis using either an antibody specific for the Ad fiber protein (N-terminus) or an antibody specific for human HGF. These results revealed a band with a molecular weight of approximately 68 kDa, representing the chimeric Ad5 fiber–T4 fibritin–HGF/NK2 protein (Figure 2).
Analysis of Ad5-pIX-RFP-FF/NK2 infection in cancer cells that overexpress cMet
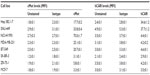
To validate that the FF/NK2 modification in Ad5-pIX-RFP-FF/NK2 could bind to cancer cells overexpressing the cMet receptor, we first analyzed surface expression of the cMet and hCAR receptors in different cell lines by flow cytometry. We examined a panel of cell lines, including two HCC cell lines (Hep 3B2.1-7 and SNU-449), one lung cancer cell line (NCI-H1993), and four breast cancer cell lines (MDA-MB-231, BT-549, SK-BR-3, ZR-75-1, and MCF-7). We measured both percentages of cells expressing the appropriate receptor as well as the mean level of expression. Analysis of cMet receptor expression showed that the NCI-H1993 lung cancer cell line had the highest level followed by the HCC cell lines Hep 3B2.1-7 and SNU-449. The MDA-MB-231 and BT-549 breast cancer cell lines demonstrated moderate cMet levels, while the other breast cancer cells showed cell lines that had minimal cMet levels (Figure 3A). Analysis of hCAR receptor expression showed that the two HCC cell lines (Hep 3B2.1-7 and SNU-449) had moderate levels of the receptor. All other cancer cells demonstrated minimal hCAR levels (Figure 3B). These results are summarized as the mean fluorescence intensity (MFI) of each peak in Table 1.
To correlate the infection efficiency with receptor expression levels, Ad infectivity assays were performed on the panel of cancer cell lines, using an increasing range of MOI (1,000 VP/cell, 100 VP/cell, 10 VP/cell, and 1.0 VP/cell). Uninfected cells were included as a negative control. In addition, the Ad5-pIX-RFP-FF/NK2 infectivity was compared with Ad5-pIX-RFP-wt/Fiber, which contains a wild-type fiber gene and uses hCAR for binding. The percentages of cells positive for the pIX-RFP reporter gene expression (an indication of an expression of viral genes) were determined by flow cytometry at 48 hours after infection. In the HCC cell lines (Hep 3B2.1-7 and SNU-449), there was a marked enhancement of infectivity using Ad5-pIX-RFP-FF/NK2 compared with using the control Ad5-pIX-RFP-wt/Fiber (Figure 4A and B). This increase in Ad5-pIX-RFP-FF/NK2 infectivity was statistically significant starting as low as 10:1 MOI in Hep 3B2.1-7 cells. The BT-549 breast cancer cell line showed moderate levels of enhanced infectivity using Ad5-pIX-RFP-FF/NK2 compared with using the control Ad5-pIX-RFP-wt/Fiber (Figure 4C). The NCI-H1993, MDA-MB-231, and SK-BR-3 cells also demonstrated modest levels of enhanced infectivity using Ad5-pIX-RFP-FF/NK2 (Figure 4D–F). In contrast, the ZR-75-1 and MCF-7 cells exhibited the least levels enhanced infectivity using Ad5-pIX-RFP-FF/NK2 compared with using the control Ad5-pIX-RFP-wt/Fiber (Figure 4G and H). In summary, these data show that the Ad5-pIX-RFP-FF/NK2 infection efficiency was higher in almost all cell lines screened compared with the Ad5-pIX-RFP-wt/Fiber control virus. However, we did not observe a perfect correlation between infectivity and Ad5-pIX-RFP-FF/NK2 and cMet receptor levels. For instance, infectivity was relatively low in NCI-H1993 cells, which express high levels of cMet.
Analysis of Ad5-pIX-RFP-FF/NK2 binding specificity to cMet
To examine the binding specificity of Ad5-pIX-RFP-FF/NK2, HGF was used to investigate competition for virus binding. Hep 3B2.1-7 cells were used for the binding specificity, and binding of Ad5-pIX-RFP-FF/NK2 was compared with the Ad5-pIX-RFP-wt/Fiber control. As shown in Figure 5A, HGF pretreatment resulted in a concentration-dependent reduction in Ad5-pIX-RFP-FF/NK2 infectivity. This competitive effect was statistically significant at each concentration of HGF used. In contrast, Ad5-pIX-RFP-wt/Fiber exhibited weak infection that was not affected by HGF pretreatment of the cells.
As an additional approach to examining the specificity of Ad5-pIX-RFP-FF/NK2 binding to cMet, we used a polyclonal anti-cMet antibody to compete for virus binding. In this experiment, Hep 3B2.1-7 cells were preincubated with increasing concentrations of blocking antibody for 30 minutes at 4°C and then infected with Ad5-pIX-RFP-FF/NK2 or with Ad5-pIX-RFP-wt/Fiber (at an MOI of 10 VP/cell) for 30 minutes at 4°C. Real-time PCR analysis revealed a decrease in the number of Ad5-pIX-RFP-FF/NK2 virus particles bound to the cells at all concentrations of blocking antibody used. In contrast, Ad5-pIX-RFP-wt/Fiber binding was not affected by lower concentrations of anti-cMet antibody. However, there was an apparent reduction in virus particles bound at the highest concentration of antibody used (50 ng), which could be a non-specific (off-target) effect of the antibody (Figure 5B).
Effect of cMet knockdown on Ad5-pIX-RFP-FF/NK2 infection
To more directly examine the binding of Ad5-pIX-RFP-FF/NK2 to cells expressing cMet, we used lentivirus delivered shRNA to knock down endogenous cMet expression in Hep 3B2.1-7 cells. As a control, we delivered a non-specific shRNA to Hep 3B2.1-7 cells. In Hep3B2.1-7 cMet/KD cells expressing the cMet shRNA, Western blot analysis demonstrated a protein level knockdown of >90% compared with parental Hep 3B2.1-7 cells (Figure 6A inset). In contrast, control Hep 3B2.1-7 NT cells expressing a non-specific shRNA showed no change in cMet expression compared with parental Hep 3B2.1-7 cells. We also analyzed surface expression of cMet in the different cell lines by flow cytometry. As shown in Figure 6A, a reduction in cMet staining from an MFI of 17.3±0.4 in parental Hep 3B2.1-7 cells to 8.7±0.1 in Hep3B2.1-7 cMet/KD cells was observed.
To test the effect of cMet knockdown on infectivity of Ad5-pIX-RFP-FF/NK2, 1×105 Hep 3B2.1-7 NT and Hep 3B2.1-7 cMet/KD were treated for 1 hour at 4°C (Figure 6B). Following virus treatment, the cells were washed with PBS, DNA was extracted, and relative Ad5 E4 levels were measured by real-time PCR. Real-time PCR analysis revealed a significant reduction in Ad5-pIX-RFP-FF/NK2 VP bound to Hep3B2.1-7 cMet/KD cells at an MOI of 1,000 VP/cell and 2,500 VP/cell.
As a control, the effect of cMet knock down on infectivity of Ad5-pIX-RFP-wt/Fiber in Hep 3B2.1-7 NT and Hep 3B2.1-7 cMet/KD was also determined. As shown in Figure 6C, the Ad5-pIX-RFP-wt/Fiber control showed no difference in binding between the two cell lines.
Analysis of Ad5-pIX-RFP-FF/NK2 replication in cancer cells overexpressing cMet
To investigate the efficacy of virus replication, we quantified the increase in Ad5-pIX-RFP-FF/NK2 or Ad5-pIX-RFP-wt/Fiber after infection of Hep 3B2.1-7, SNU-449, and MCF-7 cell lines. As shown in Figure 7, the Ad5 E4 copy number in cells infected with Ad5-pIX-RFP-FF/NK2 was compared with cells infected using the Ad5-pIX-RFP-wt/Fiber virus. Real-time PCR analysis revealed an increase in Ad5 E4 copy number in Hep 3B2.1-7 cells (Figure 7A) and SNU-449 cells (Figure 7B) infected with Ad5-pIX-RFP-FF/NK2 cell lines, which was 6–16-fold higher than cells infected with Ad5-pIX-RFP-wt/Fiber. Interestingly, infection of Hep 3B2.1-7 cells with Ad5-pIX-RFP-FF/NK2 showed a decrease in Ad5 E4 copy number at day 4 compared with that at day 3 detection. This result is likely due to induction of an oncolytic effect in the Hep 3B2.1-7 cells after day 3. Levels of Ad5-pIX-RFP-FF/NK2 E4 copy number were negligible in MCF-7 cells (Figure 7C), which express low levels of cMet and hCAR. Importantly, levels of Ad5-pIX-RFP-wt/FiberE4 copy number were negligible in all three analyzed cancer cell lines.
Analysis of Ad5-pIX-RFP-FF/NK2-mediated cancer cell killing
To test the oncolytic activity of Ad5-pIX-RFP-FF/NK2, we used cell viability assays to test cell killing in Hep 3B2.1-7, SNU-449, and MCF-7 cell lines. The Ad5-pIX-RFP-FF/NK2 oncolytic activity was compared with Ad5-pIX-RFP-wt/Fiber, as well as with a non-replicating Ad5-CMV-GFP/Luc control. In this experiment, the cells were plated in 96-well dishes, treated with the viruses, and incubated over 7 days to examine an oncolytic effect with time. These results demonstrate a strong oncolytic effect of Ad5-pIX-RFP-FF/NK2 in Hep 3B2.1-7 cells (Figure 8A) and SNU-449 cells (Figure 8B), resulting in the lysis of 50%–80% of cells at 2 days post infection. By day 7, almost 97% of Hep 3B2.1-7 cells were killed. Importantly, the oncolytic activity of Ad5-pIX-RFP-wt/Fiber was less than the oncolytic activity of Ad5-pIX-RFP-FF/NK2 in both the tested HCC cells. In MCF-7 cells, both viruses did not show any significant cell lysis except at day 5, in which the Ad5-pIX-RFP-FF/NK2 showed some killing effect (Figure 8C). This oncolytic activity differed among the cell lines examined and was consistent with the differences in Ad infection and replication efficiency.
Analysis of Ad5-pIX-RFP-FF/NK2 binding on cMet activation
To investigate the effect of incorporating the chimeric Ad5 fiber–T4 fibritin–HGF/NK2 into an Ad vector on cMet receptor activation, Western blot analysis was performed using Hep 3B2.1-7 cells infected with Ad5-pIX-RFP-FF/NK2 or with Ad5-pIX-RFP-wt/Fiber. HGF treatment (33 ng/mL) was also used as a positive control for cMet activation. As shown in Figure 9, Western blot analysis demonstrated that HGF induced cMet phosphorylation of cMet, as early as 30 minutes post treatment. However, infection with neither Ad5-pIX-RFP-FF/NK2 nor Ad5-pIX-RFP-wt/Fiber induced detectable cMet autophosphorylation at an MOI of 500 VP/cell. In addition to cMet autophosphorylation, activation of the Akt pathway was examined. In this experiment, HGF induced Akt phosphorylation, as early as 30 minutes post treatment. However, neither Ad5-pIX-RFP-wt/Fiber nor Ad5-pIX-RFP-FF/NK2 induced Akt phosphorylation until 8 hours post treatment. This late effect on Akt phosphorylation Hep 3B2.1-7 cells is not likely due to initial cMet binding, but rather a non-specific effect of cell manipulation.
Discussion
The HGF/cMet pathway plays a pivotal role in the process of carcinogenesis. Specific monoclonal antibodies against HGF or cMet, decoy receptors, dominant negative receptors, and small molecule inhibitors of cMet have all been investigated as targeting modalities.26,27 Oncolytic Ads have been used as an adjuvant therapeutic for cancer treatment with promising effects. There are other oncolytic viruses that have also been investigated,28,29 yet Ad vectors remain popular due to their well-characterized structure and biology, efficient methods for generating recombinant virus, systemic stability, and ability to infect dividing as well as quiescent cells.30
The success of Ad vectors as a therapeutic intervention infection depends on infection efficiency, infection potential, and achieving cancer cell specificity. Modification of the Ad5 genome,31,32 use of tumor selective promoters, different Ad fiber replacements, and fiber knob modifications have all been investigated. Ad infection depends on a critical fiber shaft length with a correct spatial arrangement of the knob and penton RGD motifs. To achieve infectivity comparable to the wild-type Ad5, we designed a truncated fragment of the HGF/NK2 peptide inserted downstream of eight-coiled coils of the bacteriophage T4 fibritin, as a proof-of-principle oncolytic Ad construct that would bind cMet.
We first analyzed cMet and hCAR surface levels on panels of cancer cells. We then performed infectivity assays to find a correlation with Met surface expression levels and compare infectivity to a wild-type fiber-containing virus. Our data showed that substituting an HGF/NK2 isoform for the Ad5 fiber knob domain in Ad5-pIX-RFP-FF/NK2 resulted in better infectivity of cancer cells than Ad5-pIX-RFP-wt/Fiber. These data corroborated the approach adopted by Noureddini et al33 and Krasnykh et al34 that the bacteriophage T4 fibritin motif successfully maintained proper fiber trimerization. Unexpectedly, the infection levels did not show perfect correlation with the cMet expression levels. This was observed in the results of similar infection levels of Ad5-pIX-RFP-FF/NK2 between SK-BR-3 and MDA-MB-231 cells (MDA-MB-231 cells express higher cMet than SK-BR-3 cells). Likewise, lower infection of Ad5-pIX-RFP-FF/NK2 was observed in NCI-H1993 cells compared with the HCC cell lines (NCI-H1993, SNU-449, and Hep 3B2.1-7 cells show similar cMet).
To investigate if the modified virus bound to cMet, we performed competition assays using HGF (the cMet ligand) as well as using an anti-cMet-blocking antibody. The cMet-blocking experiments demonstrated that the NK2 targeted virus can bind cMet, unlike the wild-type fiber-containing virus Ad5-pIX-RFP-wt/Fiber. However, cMet may not be the only site for binding of Ad5-pIX-RFP-FF/NK2, as binding was reduced but not completely abolished. Nonetheless, the cMet antibody reduced Ad5-pIX-RFP-FF/NK2 binding to Hep 3B2.1-7 cells at all concentrations tested. In a similar fashion, HGF blocked Ad5-pIX-RFP-FF/NK2 infectivity in a concentration-dependent manner.
To investigate replication potential of Ad5-pIX-RFP-FF/NK2, we examined replication in three different cancer cell lines by measuring Ad5 E4 DNA using real-time PCR. The data we obtained demonstrated efficient replication potential of Ad5-pIX-RFP-FF/NK2 in Hep 3B2.1-7 and SNU-449 cells, which also showed enhanced infectivity. These data confirmed that the fiber modification did not compromise the translocation of the virions to the nucleus, since viral replication starts only at a certain threshold concentration of intranuclear viral genomes.21 The intended outcome of oncolytic Ad design is cell killing, a feature of Ads that can be utilized for cancer therapy. To investigate Ad5-pIX-RFP-FF/NK2 oncolytic activity, different cancer cell lines were infected with replicating or non-replicating viruses and followed in a 7-day cell viability assay. The Ad5-pIX-RFP-FF/NK2 was shown to enhance oncolysis of HCC cells over Ad5-pIX-RFP-wt/Fiber.
Studies of the NK2 isoform have suggested that this domain alone can cause cMet dimerization and phosphorylation, although some controversy remains.35–37 To address if NK2, the fragment in the context of an oncolytic Ad, would active cMet, Hep3B2.1 cells were infected with Ad5-pIX-RFP-FF/NK2 and the protein lysates were analyzed by Western blot for cMet activation. We demonstrated that Ad5-pIX-RFP-FF/NK2 did not activate cMet, since we did not observe any autophosphorylation of cMet by Ad5-pIX-RFP-FF/NK2. Nonetheless, there was some Akt phosphorylation observed at 8 hours after infection, which may simply be a cellular response to the stress of infection. One potential explanation for the lack of cMet activation is that the steric hindrance in the context of an Ad may block cMet dimerization. Heparin and related glycosaminoglycans have been shown to stabilize the interaction of NK2 with cMet.38 It may be possible to test the effect of heparin to enhance Ad5-pIX-RFP-FF/NK2 binding to cMet in future studies. Nonetheless, the inability of Ad5-pIX-RFP-FF/NK2 to activate cMet encourages its use without the concern of the virus functioning as a potential cMet agonist.
Ad5 virus infection is a complex process involving many primary receptors for initial binding. The initially accepted concept of the two-step entry pathway, which involves hCAR and integrins, described for Ad5 infection has increased dramatically in complexity. The human Ad serotypes bind to the MHC-Iα2,39 integrins,40–42 lactoferrin, sialic acid, and blood coagulation factors (eg, FX, FIX, FVII, and protein C43,44) and use them as primary binding receptors.45,46 The SK-BR-3 cells express moderate levels of integrins (data not shown). It is possible that integrins on SK-BR-3 cells compensated for the low cMet levels; this hypothesis is supported by data from Takayama et al.47
The NCI-H1993 cells have an aggressive malignant phenotype and express very high levels of cMet. It has been demonstrated that tumor cells overexpressing cMet also tend to shed more of the cMet extracellular domain.48 It is possible that the soluble shed forms of cMet could have an antagonistic effect in reducing Ad5-pIX-RFP-FF/NK2 binding. In addition, the naturally occurring NK2 isoform binds heparan sulfate proteoglycans (HSPGs) weakly via the N-terminal domain,49 which may add to the complexity of Ad5-pIX-RFP-FF/NK2 interaction with the cells.44 Differences in HSPG expression among the cell lines may play a role accounting for the differences in the observed infectivity levels.
In summary, using oncolytic Ads, designed to divide in and specifically kill tumor cells, is a promising virotherapy approach for cancer treatment. The safety, as well as the cancer cell specificity of our virus, could be further enhanced by additional modifications of the Ad5 genome to further restrict its infectivity and limit replication only in tumor cells. Using appropriate in vivo cancer models will further validate infectivity, toxicity, as well as the therapeutic efficacy of our retargeted Ad5 agent. Biodistribution experiments are also required to examine whether cMet targeting in vivo is correlated to the tumor targeting observed in vitro.
Acknowledgments
The authors thank Dr Wayne Orr (LSU Health Shreveport) for his helpful comments in writing this paper. This work was supported, in part, by the Louisiana Gene Therapy Research Consortium and a grant from the National Institutes of Health – National Cancer Institute (5R01CA154697). These funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the paper.
Disclosure
The authors report no conflicts of interest in this work.
References
Stoff-Khalili MA, Rivera AA, Nedeljkovic-Kurepa A, et al. Cancer-specific targeting of a conditionally replicative adenovirus using mRNA translational control. Breast Cancer Res Treat. 2008;108(1):43–55. | |
Nemerowa GR, Pachea L, Reddya V, Stewartb PL. Insights into adenovirus host cell interactions from structural studies. J Virol. 2009;384(2):380–388. | |
Leopold PL, Ferris B, Grinberg I, Worgall S, Hackett NR, Crystal RG. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther. 1998;9:367–378. | |
Kim M, Zinn KR, Barnett BG, et al. The therapeutic efficacy of adenoviral vectors for cancer gene therapy is limited by a low level of primary adenovirus receptors on tumor cells. Eur J Cancer. 2002;38(14):1917–1926. | |
Kuster K, Koschel A, Rohwer N, Fischer A, Wiedenmann B, Anders M. Down regulation of the coxsackie and adenovirus receptor in cancer cells by hypoxia depends on HIF-1a. Cancer Gene Ther. 2010;17:141–146. | |
Furge KA, Zhang Y-W, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000;19:5582–5589. | |
Boros P, Miller CM. Hepatocyte growth factor: a multifunctional cytokine. Lancet. 1995;345:293–295. | |
Piscaglia AC, Shupe TD, Pani G, Tesori V, Gasbarrini A, Petersen BE. Establishment of cancer cell lines from rat hepatocholangiocarcinoma and assessment of the role of granulocyte-colony stimulating factor and hepatocyte growth factor in their growth, motility and survival. J Hepatol. 2009;51(1):77–92. | |
Christensena JG, Burrowsc J, Salgiab R. Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. | |
Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19(10):542–551. | |
Alam N, Goel HL, Zarif MJ, et al. The integrin – growth factor receptor duet. J Cell Physiol. 2007;213:649–653. | |
Matsumoto K, Kataoka H, Date K, Nakamura T. Cooperative interaction between α- and β-chains of hepatocyte growth factor on cMet receptor confers ligand-induced receptor tyrosine phosphorylation and multiple biological responses. J Biol Chem. 1998;273(36):22913–22920. | |
Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. Crystal structure of the HGF β-chain in complex with the sema domain of the Met receptor. EMBO J. 2004;23:2325–2335. | |
Lokker NA, Mark MR, Luis EA, et al. Structure-function analysis of hepatocyte growth factor: identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J. 1992;11(7):2503–2510. | |
Cioce V, Csaky KG, Chan AM-L, et al. Hepatocyte growth factor (HGF)/NK1 is a naturally occurring HGF/scatter factor variant with partial agonist/antagonist activity. J Biol Chem. 1996;271(22):13110–13115. | |
Date K, Matsumoto K, Shimura H, Tanaka M, Nakamura T. HGF/NK4 is a specific antagonist for pleiotrophic actions of hepatocyte growth factor. FEBS Lett. 1997;420(1):1–6. | |
Kuba K, Matsumoto K, Date K, Shimura H, Tanaka M, Nakamura T. HGF/NK4, a four-kringle antagonist of hepatocyte growth factor, is an angiogenesis inhibitor that suppresses tumor growth and metastasis in mice. Cancer Res. 2000;60:6737–6743. | |
Sakai K, Nakamura T, Matsumoto K, Nakamura T. Angioinhibitory action of NK4 involves impaired extracellular assembly of fibronectin mediated by perlecan-NK4 association. J Biol Chem. 2009;284(33):22491–22499. | |
Matsumoto K, Nakamura T. Mechanisms and significance of bifunctional NK4 in cancer treatment. Biochem Biophys Res Commun. 2005; 333(2):316–327. | |
Kanerva A, Hemminki A. Modified adenoviruses for cancer gene therapy. Int J Cancer. 2004;110:475–480. | |
Belousova N, Korokhov N, Krendelshchikova V, et al. Genetically targeted adenovirus vector directed to CD40-expressing cells. J Virol. 2003;77(21):11367–11377. | |
Shayakhmetov DM, Lieber A. Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol. 2000;74(22):10274–10286. | |
Krasnykh V, Dmitriev I, Mikheeva G, Miller CR, Belousova N, Curiel DT. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72(3):1844–1852. | |
Bigelow RLH, Cardelli JA. The green tea catechins, (-)-epigallocatechin-3-gallate (EGCG) and (-)-epicatechin-3-gallate (ECG), inhibit HGF/Met signaling in immortalized and tumorigenic breast epithelial cells. Oncogene. 2006;25(13):1922–1930. | |
Le LP, Le HN, Dmitriev IP, et al. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006;98(3):203–214. | |
Bellusci S, Moens G, Gaudino G, et al. Creation of a hepatocyte growth factor/scatter factor autocrine loop in carcinoma cells induces invasive properties associated with increased tumorigenicity. Oncogene. 1994;9(4):1091–1099. | |
You W-K, McDonald DM. The hepatocyte growth factor/cMet signaling pathway as a therapeutic target to inhibit angiogenesis. BMB Rep. 2008;41(12):833–839. | |
Liu T-C, Kirn D. Gene therapy progress and prospects cancer: oncolytic viruses. Gene Ther. 2008;15:877–884. | |
Nemunaitis J, Edelman J. Selectively replicating viral vectors. Cancer Gene Ther. 2002;9:987–1000. | |
Rein DT. Current developments in adenovirus-based cancer gene therapy. Future Oncol. 2006;2(1):137–143. | |
Green NK, Seymour LW. Adenoviral vectors: systemic delivery and tumor targeting. Cancer Gene Ther. 2002;9:1036–1042. | |
Yu W, Fang H. Clinical trials with oncolytic adenovirus in China. Curr Cancer Drug Targets. 2007;7:141–148. | |
Noureddini SC, Krendelshchikov A, Simonenko V, et al. Generation and selection of targeted adenoviruses embodying optimized vector properties. Virus Res. 2006;116(1–2):185–195. | |
Krasnykh V, Belousova N, Korokhov N, Mikheeva G, Curiel DT. Genetic Targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J Virol. 2001;75(9):4176–4183. | |
Otsuka T, Jakubczak J, Vieira W, et al. Disassociation of Met-mediated biological responses in vivo: the natural hepatocyte growth factor/scatter factor splice variant NK2 antagonizes growth but facilitates metastasis. Mol Cell Biol. 2000;20(6):2055–2065. | |
Hartmann G, Naldinit L, Weidner KM, Sachs M, Vigna E, Comoglio PM. A functional domain in the heavy chain of scatter factor/hepatocyte growth factor binds the cMet receptor and induces cell dissociation but not mitogenesis. Proc Natl Acad Sci U S A. 1992;89:11574–11578. | |
Guerin C, Luddy C, Abounader R. Glioma inhibition by HGF/NK2, an antagonist of scatter factor/hepatocyte growth factor. Biochem Biophys Res Commun. 2000;273(1):287–293. | |
Schwall RH, Chang LY, Godowski PJ, et al. Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists NK1 and NK2. J Cell Biol. 1996;133(3):709–718. | |
Hong SS, Karayan L, Tournier J, Curiel DT, Boulanger PA. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16(9):2294–2306. | |
Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use αv integrins for infection. J Virol. 1994;68(10):6811–6814. | |
Nemerow GR, Stewart PL. Role of alpha (v) integrins in adenovirus cell entry and gene delivery. Microbiol Mol Biol Rev. 1999;63(3):725–734. | |
Lyle C, McCormick F. Integrin αvβ5 is a primary receptor for adenovirus in CAR-negative cells. J Virol. 2010;7:148. | |
Waddington SN, McVey JH, Bhella D, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. | |
Lenaerts L, van Dam W, Persoons L, Naesens L. Interaction between mouse adenovirus type 1 and cell surface heparan sulfate proteoglycans. PLoS One. 2012;7(2):e31454. | |
Bergelson JM. Receptors mediating adenovirus attachment and internalization. Biochem Pharmacol. 1999;57(9):975–979. | |
Henaff D, Salinas S, Kremer EJ. An adenovirus traffic update: from receptor engagement to the nuclear pore. Future Microbiol. 2011;6(2):179–192. | |
Takayama K, Ueno H, Pei XH, Nakanishi Y, Yatsunami J, Hara N. The levels of integrin alpha v beta 5 may predict the susceptibility to adenovirus-mediated gene transfer in human lung cancer cells. Gene Ther. 1998;5(3):361–368. | |
Athauda G, Giubellino A, Coleman JA, et al. cMet ectodomain shedding rate correlates with malignant potential. Clin Cancer Res. 2006; 12(14 pt 1):4154–4162. | |
Lyon M, Deakin JA, Lietha D, Gherardi E, Gallagher JT. The interactions of hepatocyte growth factor/scatter factor and its NK1 and NK2 variants with glycosaminoglycans using a modified gel mobility shift assay. J Biol Chem. 2004;279(42):43560–43567. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.