Back to Journals » Infection and Drug Resistance » Volume 12
Characterization of a small plasmid carrying the carbapenem resistance gene blaOXA-72 from community-acquired Acinetobacter baumannii sequence type 880 in China
Authors Jia H, Sun Q, Ruan Z , Xie X
Received 29 January 2019
Accepted for publication 22 May 2019
Published 6 June 2019 Volume 2019:12 Pages 1545—1553
DOI https://doi.org/10.2147/IDR.S202803
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Joachim Wink
Huiqiong Jia,1 Qingyang Sun,2 Zhi Ruan,1 Xinyou Xie1
1Department of Clinical Laboratory, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, People’s Republic of China; 2Department of Clinical Laboratory, The 117th hospital of People’s Liberation Army, Hangzhou, People’s Republic of China
Background: Acinetobacter baumannii has emerged as an important pathogen associated with hospital- and community-acquired infections. Community-acquired A. baumannii pneumonia is characterized by a fulminant course and high mortality rates. In this study, we report the identification of a community-acquired carbapenem-resistant A. baumannii strain carrying the blaOXA-72 gene.
Methods: This A. baumannii isolate was recovered from a male patient diagnosed with community-acquired pneumonia, septic shock, and respiratory failure. Antimicrobial susceptibility testing were performed and the minimum inhibitory concentrations were determined by the broth microdilution method. Whole-genome sequencing was performed using both long-read MinION and short-read Illumina platforms to fully characterize the blaOXA-72-carrying plasmid of the A. baumannii A52. The in silico multilocus sequence typing and genomic epidemiological analysis of the closely related isolates were further elucidated by our recently updated BacWGSTdb server.
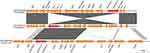
Results: The isolate was resistant to meropenem and remained susceptible to several other antimicrobial agents. Whole-genome sequencing and bioinformatics analysis indicated that this A. baumannii isolate belonged to the rare sporadic clone sequence type 880 and the blaOXA-72 gene was located on the 8,493-bp plasmid pA52-OXA-72. This plasmid exhibited only partial similarity to different OXA-72-encoding plasmids (size range: 8,771–12,056 bp) in various Acinetobacter spp. recovered from patients and other reservoirs in different countries.
Conclusion: This study described the first case of fulminant carbapenem-resistant community-acquired A. baumannii pneumonia caused by a rare sporadic clone in China. Adequate surveillance is warranted to monitor the emergence of A. baumannii as a community pathogen.
Keywords: Acinetobacter baumannii, community-acquired pneumonia, blaOXA-72, carbapenem resistance, plasmid
Introduction
Over the past decades, Acinetobacter baumannii has become a common causative agent of multidrug-resistant, hospital-acquired infections worldwide. In addition, it has been shown to be an important cause of community-acquired infection.1,2 Unlike hospital-acquired A. baumannii, community-acquired pneumonia caused by A. baumannii is characterized by a fulminant course and high mortality rates.3 Carbapenem-resistant A. baumannii is a major threat for public health. In 2017, the World Health Organization classified this pathogen as the top priority for the development of additional antibiotics.4 The most important mechanism of carbapenem resistance in A. baumannii is associated with the production of carbapenem-hydrolyzing class D OXA-type β-lactamases. Of note, five groups of OXA-type carbapenemases (ie, OXA-23-, OXA-24/40-, OXA-51-, OXA-58-, and OXA-143-like) are frequently encountered.5 The gene blaOXA-72 – one of the most important allelic variants within the blaOXA-24/40 group – was initially identified in an A. baumannii strain isolated in 2004 in Thailand (GenBank accession no. AY739646). Subsequently, blaOXA-72-carrying A. baumannii strains from human and animal origins have been widely identified. However, thus far, few blaOXA-72-positive A. baumannii strains have been reported in China and the genetic context of blaOXA-72 is largely unknown.6 In the present study, we reported the first identification of a carbapenem-resistant A. baumannii isolate associated with fulminant community-acquired pneumonia. This isolate belonged to a rare sporadic clone, harboring both the blaOXA-72 and the intrinsic blaOXA-259 genes. Whole-genome sequencing and microbiological analysis were performed to elucidate its mechanism of resistance to carbapenems.
Materials and methods
A 66-year-old male patient was hospitalized with symptoms indicative of community-acquired pneumonia (CAP), septic shock, respiratory failure, and fever. A. baumannii isolate A52 was cultured from the sputum sample of the patient within 24 hrs after admission. This patient resided in the countryside, without a history of recent travel or hospitalization. The presence of CAP was considered if pneumonia was not acquired in a hospital, and the interval between the onset of symptoms and previous discharge from hospital was >30 days. Community-associated A. baumannii was defined as isolates cultured from sputum and/or a blood specimen obtained from CAP patients and collected within 48 hrs after admission.3,7 The quality of the sputum specimen for culture was determined through microscopy. The patient was successfully treated with a one-week administration of imipenem and linked to a good prognosis. The initial identification of species was performed using Vitek 2 (bioMérieux, Marcy-l’Étoile, France) and Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Corp., Billerica, MA, USA). A. baumannii A52 was subjected to antimicrobial susceptibility testing by the microdilution broth method for the following antimicrobial agents: amikacin, ceftazidime, cefotaxime, cefepime, ciprofloxacin, colistin, gentamicin, imipenem, meropenem, minocycline, piperacillin, and tigecycline, which were purchased from Sigma-Aldrich (St. Louis, MO, USA). The results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) 2018 guidelines, except for tigecycline.8 Although officially there are no clinical breakpoints for tigecycline against A. baumannii given by either CLSI or European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines, the US FDA breakpoints (MIC ≤2 mg/L for susceptibility and MIC ≥8 mg/L for resistance) or EUCAST breakpoints version 8.0 (MIC ≤1 mg/L for susceptibility and MIC >2 mg/L for resistance) for Enterobacteriaceae are commonly applied (
Whole-genome sequencing (WGS) of A. baumannii A52 was performed using both the HiSeq X10 (Illumina, San Diego, CA, USA), with the 150 bp paired-end protocol, and the MinION (Nanopore, Oxford, UK) platforms. Hybrid assembly of both short Illumina reads and long Nanopore reads was constructed using Unicycler v 0.4.7 with the Pilon v1.23 option on for the modification of the assembled reads.9 The genome annotation was performed using NCBI Prokaryotic Genome Annotation Pipeline. Antibiotic resistance genes was queried using the ResFinder database at the Center for Genomic Epidemiology (
Results and discussion
Antimicrobial susceptibility testing showed that A. baumannii A52 was resistant to meropenem. However, it remained susceptible to several other antimicrobial agents, including imipenem (Table S1). In particular, for the broth microdilution method, the minimum inhibitory concentrations of meropenem and imipenem were 8 mg/L and 2 mg/L, respectively. In addition to carbapenemases, the permeability defects, such as overexpression of efflux system, may also contribute to carbapenem resistance in A. baumannii. The sequences of efflux pump regulatory genes were analyzed, which are known to be involved in antibiotic resistance, namely, the AdeR/S two-component system and AdeN and AdeB, which regulate the AdeIJK and AdeAB efflux pumps, respectively. The regulatory efflux pumps AdeR and AdeS of A. baumannii A52 showed 99% (1 substitution) and 100% amino acid sequence identities, respectively, with those of A. baumannii ATCC 17978, whereas AdeN and AdeB of A. baumannii A52 showed 100% amino acid sequence identities with those of A. baumannii ATCC 17978. These findings highlight the efflux pump regulatory genes in this strain may have a negative influence in resistance to carbapenems. WGS to assess the sequence type (ST) identified A. baumannii A52 as ST880 and ST77, according to the Oxford and Pasteur MLST schemes, respectively. It contained three genes (blaOXA-72, blaOXA-259, and blaADC-26) conferring resistance to β-lactams, which were not preceded by an insertion element. This finding was consistent with the phenotypic data. In addition, we found that this strain lacked the A. baumannii antibiotic resistance island, which confers resistance to multiple antibiotics (eg, aminoglycosides, beta-lactams, sulfonamides, and tetracyclines). The absence of an antibiotic resistance island in A. baumannii may explain the high susceptibility of the community-acquired strain to antibiotics.
The blaOXA-72 gene was located on the 8,493-bp plasmid pA52-OXA-72. The backbone of the pA52-OXA-72 plasmid is almost identical to that of pAbIHIT32296 – a blaOXA-72-carrying plasmid in A. baumannii recovered from a captive grey parrot in Germany. The shared sequence of these plasmids contains the blaOXA-72 gene. However, in pA52-OXA-72, this gene is in a different location and orientation. Furthermore, the plasmid pA52-OXA-72 exhibited only partial similarity to other OXA-72-encoding plasmids (size range: 8,771–12,056 bp) in various Acinetobacter spp. recovered from patients in different countries (Figure 1, Table S2). Plasmid pA52-OXA-72 contained 11 open reading frames and encoded a replication protein belonging to the replication protein 3 superfamily1.13 In addition, it carried genes encoding a putative type II toxin–antitoxin system (VapC2-VapB2). These genes were also present in other OXA-72-encoding plasmids (eg, pIEC338SCOX) and may be involved in plasmid maintenance (Figure 2).14 The blaOXA-72 gene was flanked by XerC/XerD recombination sites, which were identical to those of previously reported plasmids. This finding indicated the importance of mobilization through the site-specific recombination mechanism. As previously suggested, the XerC/XerD-sites may be involved in a site-specific recombination system responsible for the mobilization of the blaOXA-72 gene among Acinetobacter spp. Moreover, a conserved region (ie, ydaF2-vapB2-vapC2-orf1) in pA52-OXA-72 also bracketed with the XerC/XerD-sites and showed high sequence identity to the regions in plasmid pIEC338SCOX. This observation suggests that different plasmids may exchange carbapenem-hydrolyzing class D OXA-type β-lactamases genes and other components within a restricted region between the two closest XerC/XerD binding sites.
 | Figure 2 Plasmid sequence alignment of blaOXA-72-carrying plasmids that revealed partial sequence identity to pA52-OXA-72. |
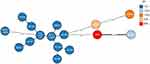
The origin of A. baumannii A52 remains unknown. We analyzed the phylogenetic relationship between A. baumannii A52 and a total of 3,417 A. baumannii strains currently deposited in the NCBI GenBank database. This analysis was performed using the following two bacterial source tracking strategies: SNP and cgMLST. Both strategies suggested that A. baumannii A52 belongs to a rare sporadic clone. Notably, the closest relative was an ST1325 (single-locus variant of ST880) isolate known as HC9436, which was recovered from the endobronchial intubation tube of a patient in Honduras in 2015 (Table S3, Figure S1). However, the present patient had no history of recent overseas travel or direct contact with foreigners. Hence, there was no epidemiological link identified between this Chinese patient and the previously reported case. A. baumannii CAP exhibits seasonal variation, with a higher incidence reported in the warmer and more humid months of the year.3 Consistent with previously reported cases, the present patient was admitted in August (ie, the hot and rainy season in China).15–17
The OXA-72-producing A. baumannii isolates have been mainly obtained from hospitals and other reservoirs (ie, livestock/companion animals, and environment) in South America, Southern Asia, and Eastern Europe.18–20 More recently, a cross-sectional study reported a high prevalence and clonal dissemination of OXA-72-producing A. baumannii in a Chinese hospital, corresponding to the predominant international clone 2. However, noninternational clone 2 A. baumannii strains (ie, ST78 in Germany, ST1 in Serbia, and ST79 in Brazil) carrying blaOXA-72 have also been reported.20–22 Therefore, based on the genetic flexibility mediated by the elements for site-specific recombination, the blaOXA-72-carrying plasmids of A. baumannii may represent powerful vehicles for the acquisition, horizontal-transfer, and evolution of carbapenem resistance.
Conclusion
We reported the identification of a carbapenem-resistant community-acquired A. baumannii isolate in China. This isolate belongs to a rare sporadic clone harboring a unique blaOXA-72-carrying plasmid. Our study highlights the transmission potential of OXA-72-producing A. baumannii in the community. Continuous surveillance is necessary to monitor the transmission dynamics of the blaOXA-72 gene in the community.
Abbreviation list
A. baumannii, Acinetobacter baumannii; CAP, community-acquired pneumonia; MIC, minimum inhibitory concentration; CLSI, Clinical and Laboratory Standards Institute; EUCAST, European Committee on Antimicrobial Susceptibility Testing; WGS, whole-genome sequencing; MLST, multilocus sequence typing; ST, sequence type; cgMLST, core genome multilocus sequence typing; SNP, single nucleotide polymorphism.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81401698, 81871696) and Zhejiang Province Public Welfare Technology Application Research Project (LGF18H190001).
Author contributions
ZR and XYX conceived and designed the study. HQJ and QYS collected samples and performed experiments, ZR and HQJ performed data analysis, ZR wrote the paper. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Clinical SB and Pathophysiological overview of acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30(1):409–447. doi:10.1128/CMR.00058-16
2. Ruan Z, Chen Y, Jiang Y, et al. Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int J Antimicrob Agents. 2013;42(4):322–328. doi:10.1016/j.ijantimicag.2013.06.019
3. Dexter C, Murray GL, Paulsen IT, Peleg AY Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther. 2015;13(5):567–573. doi:10.1586/14787210.2015.1025055
4.
5. Evans BA, Amyes SG OXA beta-lactamases. Clin Microbiol Rev. 2014;27(2):241–263. doi:10.1128/CMR.00117-13
6. Chen Y, Yang Y, Liu L, et al. High prevalence and clonal dissemination of OXA-72-producing Acinetobacter baumannii in a Chinese hospital: a cross sectional study. BMC Infect Dis. 2018;18(1):491. doi:10.1186/s12879-018-3109-6
7. Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44 Suppl 2:S27–S72. doi:10.1086/511159
8.
9. Wick RR, Judd LM, Gorrie CL, Holt KE Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005731
10. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
11. Ruan Z, Feng Y BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44(D1):D682–687. doi:10.1093/nar/gkv1004
12. Sullivan MJ, Petty NK, Beatson SA Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi:10.1093/bioinformatics/btr039
13. Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(10):4168–4177. doi:10.1128/AAC.00542-10
14. Jurenaite M, Markuckas A, Suziedeliene E Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J Bacteriol. 2013;195(14):3165–3172. doi:10.1128/JB.00237-13
15. Peng C, Zong Z, Fan H Acinetobacter baumannii isolates associated with community-acquired pneumonia in West China. Clin Microbiol Infect. 2012;18(12):E491–E493. doi:10.1111/1469-0691.12017
16. Ong CW, Lye DC, Khoo KL, et al. Severe community-acquired Acinetobacter baumannii pneumonia: an emerging highly lethal infectious disease in the Asia-Pacific. Respirology. 2009;14(8):1200–1205. doi:10.1111/j.1440-1843.2009.01630.x
17. Leung WS, Chu CM, Tsang KY, Lo FH, Lo KF, Ho PL Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest. 2006;129(1):102–109. doi:10.1378/chest.129.1.102
18. Klotz P, Jacobmeyer L, Stamm I, et al. Carbapenem-resistant Acinetobacter baumannii ST294 harbouring the OXA-72 carbapenemase from a captive grey parrot. J Antimicrob Chemother. 2018;73(4):1098–1100. doi:10.1093/jac/dkx490
19. Kuo HY, Hsu PJ, Chen JY, et al. Clonal spread of blaOXA-72-carrying Acinetobacter baumannii sequence type 512 in Taiwan. Int J Antimicrob Agents. 2016;48(1):111–113. doi:10.1016/j.ijantimicag.2016.04.020
20. Dortet L, Bonnin RA, Bernabeu S, et al. First occurrence of OXA-72-producing Acinetobacter baumannii in Serbia. Antimicrob Agents Chemother. 2016;60(10):5724–5730. doi:10.1128/AAC.01016-16
21. Pfeifer Y, Hunfeld KP, Borgmann S, et al. Carbapenem-resistant Acinetobacter baumannii ST78 with OXA-72 carbapenemase and ESBL gene blaCTX-M-115. J Antimicrob Chemother. 2016;71(5):1426–1428. doi:10.1093/jac/dkv462
22. Pagano M, Rocha L, Sampaio JL, Martins AF, Barth AL Emergence of OXA-72-producing Acinetobacter baumannii belonging to high-risk clones (CC15 and CC79) in different Brazilian States. Infect Control Hosp Epidemiol. 2017;38(2):252–254. doi:10.1017/ice.2016.287
Supplementary materials
 | Table S1 Antibiotic resistance profile of A. baumannii isolate A52 |
 | Table S2 Characterisation of Acinetobacter spp. plasmids containing blaOXA-72 gene recovered from different countries |
 | Table S3 Information of closely related strains to A. baumannii A52 |
References
1. Klotz P, Jacobmeyer L, Stamm I, et al. Carbapenem-resistant Acinetobacter baumannii ST294 harbouring the OXA-72 carbapenemase from a captive grey parrot. J Antimicrob Chemother. 2018;73(4):1098–1100. doi:10.1093/jac/dkx490
2. Chagas TPG, Tavares EOTR, D’Alincourt Carvalho-Assef AP, Albano RM, Asensi MD Carbapenem-resistant Acinetobacter pittii strain harboring blaOXA-72 from Brazil. Diagn Microbiol Infect Dis. 2017;88(1):93–94. doi:10.1016/j.diagmicrobio.2017.01.022
3. Kuo HY, Hsu PJ, Chen JY, et al. Clonal spread of blaOXA-72-carrying Acinetobacter baumannii sequence type 512 in Taiwan. Int J Antimicrob Agents. 2016;48(1):111–113. doi:10.1016/j.ijantimicag.2016.04.020
4. Merino M, Acosta J, Poza M, et al. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob Agents Chemother. 2010;54(6):2724–2727. doi:10.1128/AAC.01674-09
5. Tada T, Miyoshi-Akiyama T, Shimada K, Shimojima M, Kirikae T Dissemination of 16S rRNA methylase ArmA-producing acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob Agents Chemother. 2014;58(5):2916–2920. doi:10.1128/AAC.01212-13
6. Povilonis J, Seputiene V, Krasauskas R, et al. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J Antimicrob Chemother. 2013;68(5):1000–1006. doi:10.1093/jac/dks499
7. D’Andrea MM, Giani T, D’Arezzo S, et al. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(8):3528–3533. doi:10.1128/AAC.00178-09
8. Dortet L, Bonnin RA, Bernabeu S, et al. First occurrence of OXA-72-producing Acinetobacter baumannii in Serbia. Antimicrob Agents Chemother. 2016;60(10):5724–5730. doi:10.1128/AAC.01016-16
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.