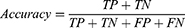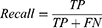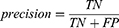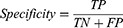Back to Journals » Medical Devices: Evidence and Research » Volume 15
Cervix Type and Cervical Cancer Classification System Using Deep Learning Techniques
Authors Habtemariam LW, Zewde ET , Simegn GL
Received 13 March 2022
Accepted for publication 8 June 2022
Published 16 June 2022 Volume 2022:15 Pages 163—176
DOI https://doi.org/10.2147/MDER.S366303
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Lidiya Wubshet Habtemariam,1 Elbetel Taye Zewde,1,2 Gizeaddis Lamesgin Simegn1,2
1Biomedical Imaging Unit, School of Biomedical Engineering, Jimma Institute of Technology, Jimma University, Jimma, Ethiopia; 2AI and Biomedical Imaging Research unit, Jimma Institute of Technology, Jimma University, Jimma, Ethiopia
Correspondence: Gizeaddis Lamesgin Simegn, Tel +251913925481, Email [email protected]
Purpose: Cervical cancer is the 4th most common cancer among women, worldwide. Incidence and mortality rates are consistently increasing, especially in developing countries, due to the shortage of screening facilities, limited skilled professionals, and lack of awareness. Cervical cancer is screened using visual inspection after application of acetic acid (VIA), papanicolaou (Pap) test, human papillomavirus (HPV) test and histopathology test. Inter- and intra-observer variability may occur during the manual diagnosis procedure, resulting in misdiagnosis. The purpose of this study was to develop an integrated and robust system for automatic cervix type and cervical cancer classification using deep learning techniques.
Methods: 4005 colposcopy images and 915 histopathology images were collected from different local health facilities and online public datasets. Different pre-trained models were trained and compared for cervix type classification. Prior to classification, the region of interest (ROI) was extracted from cervix images by training and validating a lightweight MobileNetv2-YOLOv3 model to detect the transformation region. The extracted cervix images were then fed to the EffecientNetb0 model for cervix type classification. For cervical cancer classification, an EffecientNetB0 pre-trained model was trained and validated using histogram matched histopathological images.
Results: Mean average precision (mAP) of 99.88% for the region of interest (ROI) extraction, and test accuracies of 96.84% and 94.5% were achieved for the cervix type and cervical cancer classification, respectively.
Conclusion: The experimental results demonstrate that the proposed system can be used as a decision support tool in the diagnosis of cervical cancer, especially in low resources settings, where the expertise and the means are limited.
Keywords: cervical cancer, cervix type, classification, deep learning, detection, histopathology image, transformation zone
Introduction
The cervix is part of the female reproductive organ structurally found at the lower fibromuscular portion of the uterus. It contains two kinds of cells: rectangular columnar cells and flat scale-like squamous cells. Abnormal cells are developed in the transformation zone, which is an area where columnar cells are constantly changing into squamous cells.1 The location of the transformation zone varies among women. Based on transformation zone location, cervix is classified as cervix type 1, type 2, and type 3.2,3 The cells in the transformation zone develop gradually to abnormal cells and could change into cervical cancer through time.
Cervical cancer is one of the leading causes of cancer-related deaths in women, worldwide, with 80% of the cases occurring in developing countries.4,5 According to the human papillomavirus (HPV) information center report, about 6294 new cervical cancer cases are diagnosed annually in Ethiopia.6 In remote areas, which have poor medical conditions with insufficient healthcare accessibility and unqualified medical staff, cervical cancer incidence and mortality are estimated to be higher.
Cervical cancer screening techniques include visual inspection after application of acetic acid (VIA),7 papanicolaou test (Pap),3,8 and human papillomavirus (HPV) test.9 The type of cervical cancer is confirmed by visual examination of histopathology images under a microscope. These manual techniques are usually tedious, time-taking, and prone to error due to inter- and intra-observer variability. Computer-aided diagnosis (CAD) techniques have the potential to automate the manual screening and diagnosis procedures and help medical professionals in a wide range of decision-making, including disease detection, localization of cervix, cancer grading, and treatment planning.
Cervix type classification has been proposed in the literature using the transfer learning approach.10–12 Inception v3, ResNet 50, and ResNet 34 pre-trained models were employed in these literatures to classify cervix into type 1, type 2 and type 3. However, the accuracies reported were generally low. Similarly, a two-stage solution which performs classification of cervix type following region of interest (ROI) extraction were proposed in different literatures.13–19 Cruz et al13 proposed a color assembly method for ROI extraction and used it to train an artificial neural network (ANN), and an accuracy of 64% were reported. A similar approach was proposed by Lei et al14 using Inception v3 and an accuracy of 74% were reported. The highest accuracy (96.77%) was reported by Gorantla et al18 using Mask RCNN for ROI extraction and Hierarchical Convolutional Mixture of Experts (HCME) neural network for classification.
Deep learning models have also been proposed for diagnosis of cervical cancer by different literatures.20–24 Guo et al20 proposed a localized fusion-based approach to classify squamous epithelium into four classes, training a support vector machine (SVM) and linear discriminant analysis (LDA) using 61 digitized histopathologic hematoxylin and eosin-stained images. Similarly, Almubarak et al21 used 65 digitized histology images to train a CNN for classification of cervical cancer and an accuracy of 77.27% was claimed. Likewise, Sornapudi et al23 proposed an epithelium detection module using 150 whole-slide images (WSI) and reported 85% accuracy, but this was limited to classifying only WSIs. Wei et al24 proposed a method to classify the histopathology image as normal, cervical intraepithelial neoplasia (CIN) lesion, and cancer using 90 histology images based on texture and lesion feature, yielding an accuracy of 90%. Better cervical cancer classification accuracies were reported by training a deep learning model with a relatively large dataset.25,26 However, these approaches were magnification power-dependent and also limited to classifying a few classes of cancer.
Most of the studies proposed in the literature were designed to classify either the cervix type or few types of cervical cancers. Moreover, the generalization abilities of the models are low, and the classifications were limited only to a few classes and applicable for images acquired using specific magnification powers. In this paper, a full-fledged, integrated, magnification power independent and accurate system is proposed for cervical cancer screening and diagnosis by automating both the pre-screening (cervix type classification) and cervical cancer type classification procedures.
Methods
Cervix Type Classification
Figure 1 demonstrates the general steps of the proposed cervix type classification system. After data collection, annotation and labeling, all the cervix images were first fed to the object detection network for cervix localization (ROI extraction). Then, the ROI extracted images were used to train and validate the cervix type classification model.
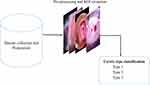 |
Figure 1 Block diagram of the cervix type classification system. |
Dataset
A total of 4005 images were collected for cervix type classification. 133 colposcope images were collected from Tercha General Hospital (south west Ethiopia) using a speculum (to access the cervix), and 13 MP and 18 MP Tecno smartphone cameras after application of 5% acetic acid on the cervix. The collected images were then labeled into three classes (type 1, type 2 and type 3) by three expert gynecologists: an integrated emergency surgical officer, and senior medical doctors. The other images were collected from the publicly available online Kaggle dataset.27 Ethical approvals from the institutional research review board of Jimma Institute of Health, Jimma University, and a written informed consent forms from all subjects, were obtained prior to data collection.
Detection of Cervical Region (ROI Extraction)
Before feeding images to the detection network, annotation was performed using the Dark Label annotation tool. The data format of annotation was Pascal Visual Object Class (VOC) and was set to detect only one object or cervix.
The MobileNetv2-YOLOv3 lightweight network was used for localizing the cervix (ROI extraction). YOLOv3 is the third version of you only look once (YOLO) which predicts at three different scales. It divides an image into grid cells, produces the cell’s probabilities, and predicts boxes. The detection layer makes detection at feature maps of three different sizes, having strides 32, 16, and 8 which are factors to down sampling image.28 MobileNetv2 is used as a backbone to facilitate migration to the mobile terminal and improve the generalization ability of the model and extract features. Furthermore, it combines the anti-residual module with the depth wise separable convolution. The depth wise separable convolution can effectively improve the computational efficiency and reduce the number of parameters. This enables the network to perform well with the lightweight character.24,29
Prior to training, the network was configured according to the cervix type dataset. The dataset was split into 80% for training, 10% for validation, and 10% for testing with respective annotation files. The annotation files and images were converted to TFRecord file format. Then, the input image, which was originally 416x416, was down-sampled using strides of 32, 16, and 8 resulting in 13×13, 26×26, and 52×52 output images. The detection network was then trained with these data using an Adam optimizer, 0.001 initial learning rate and batch size of 8.
Cervix Type Classification Model
For the cervix type classification, four pre-trained models were selected, trained, validated and compared. The selected models were VGGNet, ResNet, EfficientNet, and Resnet50 ensembled with MobilenetV2. The models were trained and evaluated twice using two types of datasets: (1) ROI extracted images and (2) raw (ROI unextracted) images. Adam and stochastic gradient descent optimizers with learning rates of 0.001–0.0001 and batch size of 32 were used for all models. A 10-fold cross-validation technique was used to partition the data. The dataset was split 10-folds and set to 90% training and 10% validation in each fold. Moreover, the L2 regularization technique30 was applied in each layer to reduce overfitting.
Cervical Cancer Classification
Figure 2 demonstrates the procedures used to train and validate the cervical cancer classification system. These include data collection, augmentation, preprocessing, model training, model validation and testing.
 |
Figure 2 Block diagram of the cervical cancer classification system. |
Dataset
A total of 915 hematoxylin and eosin (H&E)-stained histopathology images labeled with four cancer classes (normal, precancer, adenocarcinoma, and squamous cell carcinoma), were gathered from Jimma University Medical Center (JUMC) and St. Paul Hospital for the cervical cancer classification. A smartphone camera and digital camera placed on an OPTIKA light microscope, as well as a digital slide scanner, were used to acquire the images. Microscopic images were acquired by 4x, 10x, 40x and 100x magnification powers with a resolution as small as 419×407 and as large as 2048×1536.
Image Preprocessing
H&E stain is one of the primary tissue stains for histology. This is because H stain makes nuclei easily visible in blue against a pink background of cytoplasm and other tissue regions.31 This enables a pathologist to easily identify and evaluate the tissue; a highly manual process. For automated image analysis, these H&E stained images need to be normalized. This is because of the significant variation in image color arising from both sample preparation and imaging conditions. Therefore, normalizing H&E images is important for classification. Histogram matching technique was used as a color normalizer to reduce the color variation of the images.
Cervical Cancer Classification Model
For cervical cancer classification, the state-of-the-art pre-trained model called EffecientNetB0 was trained and validated. The SoftMax layer of the model was trained with raw histopathology mage (without applying histogram matching) and preprocessed image (histogram matched image) for comparison. Adam optimizer with a learning rate of 0.0001, 50 number of epochs and batch size 32 were used for training. The comparison was performed to evaluate the generalization ability of the model with color normalized and raw images. Finally, unseen (new) data were used to test the performance of the model.
Performance Evaluation Metrics
Accuracy, precision, recall, F1 score, kappa score, and ROC-AUC plot were used to evaluate the performance of the models. The metrics were calculated from the model’s confusion matrix based on TP (True Positive), TN (True Negative), FP (False Positive), and FN (False Negative) values as demonstrated in Equations (1–6). AUC-ROC plot was used to visualize how well the model can distinguish between classes.
Results
ROI Detection and Cervix Type Classification
ROI Detection
Figure 3 demonstrates the loss versus epoch graph of the cervix ROI detection model. Both the training and validation losses decreased inversely with epoch, yielding a training loss of 0.48 and validation loss of 0.39 at the 60th epoch. The network mean average precision (mAP) was 99.89%. Figure 4 show the sample images generated from the detection model. The results demonstrate that, the detection model was able to remove the irrelevant region from the images preserving the cervix region. Figure 5 shows the intersection over union (IoU) score for some selected test images generated by the model, demonstrating how accurately the model predicted the ROI with respect to the ground truth. The model performs well and the IoU score was greater than 0.5.
 |
Figure 3 Performance of ROI detection model (A) Training loss (B) Validation loss. |
 |
Figure 4 Sample results of the Cervix ROI extraction model (A) input images (B) ROI extracted images. |
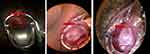 |
Figure 5 Sample test images and their respective IoU score of ROI detected images. |
Result of Cervix Type Classification
Table 1 demonstrates the training results of VGG16, Resnet50, EfficientNetB0 and the hybrid model (ResNet50 with MobilenetV2) for cervix type classification trained using raw data and ROI extracted data. As indicated, EfficientNetB0 model trained with ROI extracted image outperformed the other models with an accuracy of 92.19%, and it was selected for cervix type classification. Better results were obtained for all models trained with an ROI extracted dataset compared to models trained with a raw dataset. Figure 6 illustrates the selected EfficientNetB0 model accuracy and loss curves.
 |
Table 1 Results of Models Trained Using ROI Extracted Data and Raw Data |
 |
Figure 6 Training and validation performance of EfficientNetB0 model for cervix type classification (A) Accuracy (B) Loss. |
The selected model (EfficientNetB0) was further evaluated using a test dataset. Test results are demonstrated using the confusion matrix and ROC-AUC plot in Figure 7. The precision, recall, F1-score and Kappa score are calculated from the values of the confusion matrix. An overall test accuracy of 96.84% and kappa score of 0.95 were achieved using the EfficientNetB0 model for cervix type classification. Table 2 illustrates the summary of classification results.
 |
Table 2 Result of Cervix Type Classification with Different Metrics for the Final Model |
 |
Figure 7 Test results of EfficientNetB0 model for cervix type classification (A) Confusion matrix (B) ROC-AUC plot. |
Cervical Cancer Classification Results
Preprocessing Results
The first step of cervical cancer classification was preprocessing histopathologic images. Image resizing and color normalization using a histogram matching technique to all the dataset, and data augmentation to the training dataset, were applied prior to model training. Initially, the images were resized to 224×224. Then augmentation technique using image transformation (45°, 90°, 180°, and 270° rotation) was applied to all images. Figure 8 demonstrates sample image and augmented images using affine image transformation. The effect of histogram matching on sample images is demonstrated in Figure 9.
 |
Figure 8 Image augmentation using different degrees of rotation. |
 |
Figure 9 Effect of histogram matching on cervical histopathology sample images (A) Reference image (B) Source images (C) Histogram matched images. |
Cervical Cancer Classification
The training result of EffecientnetB0 model for classifying histopathological images into normal, precancer, adenocarcinoma, and squamous cell carcinoma is shown using the accuracy and loss curves in Figure 10. The highest validation accuracy, 97.8%, was achieved at the 50th epoch.
 |
Figure 10 Training and validation performance of EfficientNetB0 model for cervical cancer type classification (A) Accuracy and (B) Loss. |
The model was also further tested using a test dataset. The confusion matrix and ROC-AUC plot (Figure 11) demonstrates the test results of the model. The AUC scores for all classes were close to 1 confirming the model’s good measure of separability. Table 3 presents the summary of the model’s test performance. The model was found to be 94.5% accurate in classifying the test dataset.
 |
Table 3 Result of Cervical Cancer Classification with Different Metrics |
User Interface (UI)
A user interface has been developed for ease of use of the proposed system (sample snap shot demonstrated on Figure 12). The UI includes a “Browse” button to browse images from the local machine and the “Preprocess” button that allows the preprocessed images to be displayed. The “Result” button will return the models predicted cervix/cancer type.
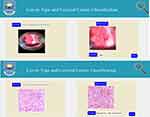 |
Figure 12 User interface for the proposed cervix type and cervical cancer classification system. |
Discussion
In this paper, a deep learning approach has been employed for cervix transformation zone detection, and for cervix type and cervical cancer classification. A total of 4005 cervix images were collected from local health facilities and online public datasets for training and validating deep learning models for the cervix type classification. For the cervical cancer classification, a total of 915 cervical tissue biopsy images were collected from Jimma University Medical Center and St. Paul Hospital.
The cervix type classification system was developed, initially by extracting the ROI from raw cervix images. MobileNetv2-YOLOv3 end-to-end object detection model was used for extracting the ROI. The model input size was 416×416 and the detection was conducted at three different scales by downsampling the image by 32, 16, and 8. Then the model crops the images preserving the required ROI using bounding box vertices as demonstrated in Figures 4 and 5. A mAP of 99.89% was obtained using the proposed detection model. Then, deep learning models including VGG16, ResNet 50, an ensemble of ResNet50 and Mobilenetv2, and EfficientNet families were trained, validated and compared for cervix type classification using raw and ROI extracted cervix images. The models were trained with an initial learning rate of 0.00001, and by applying L2 weight decay regularizes in each layer. A better result was obtained using the EfficientNetB0 model trained with ROI extracted cervix images, yielding a validation accuracy of 92.19% and test accuracy of 96.84%.
For the cervical cancer classification, the same EfficientNetB0 model was trained with an 0.0001 learning rate, an Adam optimizer and batch size of 32. A validation accuracy of 97.8%, and test accuracy of 94.5% were achieved for classifying histopathology images into four classes (normal, precancer, adenocarcinoma, and squamous cell carcinoma).
The current work presents an integrated cervical cancer screening system with better performance compared to previous literatures proposed for the same purpose,20–26 which were designed to classify either only the cervix type or a few types of cervical cancers. Moreover, the generalization abilities of the models are low, and the classifications were limited only for microscopic images acquired using specific magnification powers. In summary, in this paper, a full-fledged, integrated, magnification power independent and robust system is proposed for cervical cancer screening and diagnosis system by automating both the pre-screening (cervix type classification) and cervical cancer type classification tasks. We acknowledge that the staging of cervical cancer has not been included in this work. Moreover, adding more data may improve the accuracies of the proposed systems.
Conclusion
This paper presents two clinically related image classification systems: cervix type and cervical cancer classification. The cervix type classification model classifies cervix images into type 1, type 2, and type 3 after detecting a ROI, and the cervical cancer classification model classifies histopathology images into normal, precancer, adenocarcinoma, and squamous cell carcinoma types. A mean average precision (mAP) of 99.88% for the ROI detection and, average classification test accuracies of 96.84% and 94.5% were achieved for the cervix type and cervical cancer classification, respectively. The results obtained demonstrate that, the proposed system has the potential to be used as a decision support system for physicians, especially those in low resource setting, where both the experts and the means are limited.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This research has been approved by Jimma University’s research ethics institutional review board (IRB). This research has been performed in accordance with the ethical standards as laid down in the 1964\Declaration of Helsinki and its later amendments or comparable ethical standards.
Acknowledgment
The resources required for this research were provided by the school of Biomedical Engineering, Jimma Institute of Technology, Jimma University and Jimma University Medical Center (JUMC), St. Paul Millennium Hospital and Tercha General Hospital.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
Resources required for this study were provided by the School of Biomedical Engineering, Jimma Institute of Technology, Jimma University, Ethiopia.
Disclosure
The authors declare that they have no competing interests.
References
1. Grover A, Pandey D. Anatomy and physiology of cervix. In: Colposcopy of Female Genital Tract. Springer; 2017:3–16.
2. Payette J, Rachleff J, de Graaf C. Intel and mobileodt cervical cancer screening kaggle competition: cervix type classification using deep learning and image classification. Stanford University; 2017.
3. Prendiville W, Sankaranarayanan R. Colposcopy and treatment of cervical precancer. International Agency for Research on Cancer, World Health Organization; 2017.
4. Small W Jr, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis cancer. Cancer. 2017;123:2404–2412. doi:10.1002/cncr.30667
5. Jassim G, Obeid A, Al Nasheet HA. Knowledge, attitudes, and practices regarding cervical cancer and screening among women visiting primary health care centres in Bahrain. BMC Public Health. 2018;18(1):1–6. doi:10.1186/s12889-018-5023-7
6. Bruni L, Barrionuevo-Rosas L, Albero G, et al. Human papillomavirus and related diseases report. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre); 2019.
7. Natae SF, Nigatu DT, Negawo MK, Mengesha WW. Cervical cancer screening uptake and determinant factors among women in Ambo town, Western Oromia, Ethiopia: community-based cross-sectional study. Cancer Med. 2021;10(23):8651–8661. doi:10.1002/cam4.4369
8. World Health Organization. WHO technical guidance and specifications of medical devices for screening and treatment of precancerous lesions in the prevention of cervical cancer; 2020.
9. Sawaya GF, Huchko MJ. Cervical cancer screening. Med Clin North Am. 2017;101(4):743–753. doi:10.1016/j.mcna.2017.03.006
10. Arora M, Dhawan S, Singh K. Deep learning in health care: automatic cervix image classification using convolutional neural network. mobile radio communications and 5g networks. Springer; 2021:145–151.
11. Asawa C, Homma Y, Stuart S. Deep learning approaches for determining optimal cervical cancer treatment. Stanford University Report; 2017.
12. Arora M, Dhawan S, Singh K. Exploring deep convolution neural networks with transfer learning for transformation zone type prediction in cervical cancer. In: Soft Computing: Theories and Applications. Springer; 2020:1127–1138.
13. Cruz DA, Villar-Patiño C, Guevara E, Martinez-Alanis M, editors. Cervix type classification using convolutional neural networks.
14. Lei L, Xiong R, Zhong H. Identifying cervix types using deep convolutional networks. Stanford University Report; 2017.
15. Kaur N, Panigrahi N, Mittal A. Automated cervical cancer screening using transfer learning. Int J Adv Res Sci Eng. 2017;6:2110–2119.
16. Bijoy M, Ansal Muhammed A, Jayaraj P, editors. Segmentation based preprocessing techniques for predicting the cervix type using neural networks.
17. Zhang X, Zhao SG. Cervical image classification based on image segmentation preprocessing and a CapsNet network model. Int J Imaging Syst Technol. 2019;29(1):19–28. doi:10.1002/ima.22291
18. Gorantla R, Singh RK, Pandey R, Jain M, editors. Cervical cancer diagnosis using cervix net-a deep learning approach.
19. Alyafeai Z, Ghouti L. A fully-automated deep learning pipeline for cervical cancer classification. Expert Syst Appl. 2020;141:112951. doi:10.1016/j.eswa.2019.112951
20. Guo P, Banerjee K, Stanley RJ, et al. Nuclei-based features for uterine cervical cancer histology image analysis with fusion-based classification. IEEE j Biomed Health Inform. 2015;20(6):1595–1607. doi:10.1109/JBHI.2015.2483318
21. Almubarak HA, Stanley RJ, Long R, et al. Convolutional neural network based localized classification of uterine cervical cancer digital histology images. Procedia Comput Sci. 2017;114:281–287. doi:10.1016/j.procs.2017.09.044
22. AlMubarak HA, Stanley J, Guo P, et al. A hybrid deep learning and handcrafted feature approach for cervical cancer digital histology image classification. Int J Healthc Inf Syst Inform. 2019;14(2):66–87. doi:10.4018/IJHISI.2019040105
23. Sornapudi S, Addanki R, Stanley RJ, et al. Cervical whole slide histology image analysis toolbox. medRxiv. 2020. doi:10.1101/2020.07.22.20160366
24. Wei L, Gan Q, Ji T. Cervical cancer histology image identification method based on texture and lesion area features. Computer Assisted Surg. 2017;22(sup1):186–199. doi:10.1080/24699322.2017.1389397
25. Wu M, Yan C, Liu H, Liu Q, Yin Y. Automatic classification of cervical cancer from cytological images by using convolutional neural network. Biosci Rep. 2018;38(6):BSR20181769.
26. Cho B-J, Kim J-W, Park J, et al. Automated diagnosis of cervical intraepithelial neoplasia in histology images via deep learning. Diagnostics. 2022;12(2):548. doi:10.3390/diagnostics12020548
27. Competition K. Intel & MobileODT cervical cancer screening; 2017.
28. Zhao H, Zhou Y, Zhang L, et al. Mixed YOLOv3-LITE: a lightweight real-time object detection method. Sensors. 2020;20(7):1861. doi:10.3390/s20071861
29. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C, editors. Mobilenetv2: inverted residuals and linear bottlenecks.
30. Hoerl AE, Kennard RW. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 1970;12(1):55–67. doi:10.1080/00401706.1970.10488634
31. Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008:
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.