Back to Journals » Journal of Asthma and Allergy » Volume 14
Cell Adhesion Molecules are Upregulated and May Drive Inflammation in Chronic Rhinosinusitis with Nasal Polyposis
Authors Blight BJ, Gill AS, Sumsion JS, Pollard CE, Ashby S, Oakley GM, Alt JA, Pulsipher A
Received 19 February 2021
Accepted for publication 20 April 2021
Published 27 May 2021 Volume 2021:14 Pages 585—593
DOI https://doi.org/10.2147/JAA.S307197
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Luis Garcia-Marcos
Brennan J Blight, Amarbir S Gill, Jorgen S Sumsion, Chelsea E Pollard, Shaelene Ashby, Gretchen M Oakley, Jeremiah A Alt, Abigail Pulsipher
Division of Otolaryngology-Head and Neck Surgery, Rhinology-Sinus and Skull Base Surgery Program, Department of Surgery, University of Utah School of Medicine, Salt Lake City, UT, USA
Correspondence: Abigail Pulsipher
Division of Otolaryngology-Head and Neck Surgery, Rhinology-Sinus and Skull Base Surgery Program, Department of Surgery, University of Utah School of Medicine, 50 North Medical Drive, Room 3C120, Salt Lake City, UT, 84132, USA
Email [email protected]
Purpose: Chronic rhinosinusitis with nasal polyps (CRSwNP) is predominantly characterized by eosinophil- and T helper 2 cell (Th2)-biased inflammation. Integrins and intercellular adhesion molecules (ICAMs) are superfamilies of cell adhesion molecules (CAMs) that facilitate the recruitment and trafficking of immune cells and have been implicated in coordinating eosinophil and Th2 cell adhesion and signaling in asthma. The roles of CAMs in CRSwNP, however, remain poorly understood. The purpose of this study was to characterize the systemic and local expression of CAMs and identify which CAMs are potentially involved in CRSwNP pathology.
Materials and Methods: A prospective observational study was conducted using peripheral blood and anterior ethmoid tissues of patients with CRSwNP (n=32) and controls (n=15). Multiplex gene analysis and Pearson correlations were performed to identify associations between systemically and locally expressed CAMs. Based on the gene expression results, immunohistochemical evaluation and quantification of cells expressing integrins ITGAM, ITGAX, and ITGB2, as well as ICAM-3 in sinonasal tissues were conducted to compare local protein expression patterns.
Results: Integrin and ICAM genes were significantly elevated in the blood (p< 0.001 to p< 0.05) and sinuses (p< 0.0003 to p< 0.05) of patients with CRSwNP compared to controls. Strong positive correlations of genes expressed in the blood (p< 0.01 to p< 0.05) and sinuses (p< 0.01) were observed between ITGAM, ITGAX, ITGB2, and ICAM3. ITGAM-, ITGB2-, ICAM-3-, and ICAM-3/ITGB2-positive cell counts were significantly increased in CRSwNP compared to controls (p< 0.0001 to p< 0.04), and a positive correlation between ICAM-3/ITGB2- and ITGAM-positive cell counts was observed (p< 0.02).
Conclusion: The systemic and local expression of ITGAM, ITGB2, and ICAM-3 is significantly upregulated in CRSwNP, suggesting that integrin complex ITGAM/ITGB2 and ICAM-3 serve a potential role in inflammation-mediated signaling in CRSwNP.
Keywords: integrins, intercellular adhesion molecule 3, macrophage-1 antigen, Th2 pathophysiology, transcriptional analysis, allergy, asthma
Introduction
Chronic rhinosinusitis with nasal polyps (CRSwNP) is a heterogeneous inflammatory disease of the paranasal sinuses characterized by the recruitment and trafficking of leukocytes from the circulatory system into the sinonasal mucosa in response to local signaling. Cell adhesion molecules (CAMs), such as integrins, intercellular adhesion molecules (ICAMs), cadherins, and selectins, facilitate these processes as part of the innate immune response.1–3 Although CAMs have been implicated in numerous chronic inflammatory conditions, including inflammatory bowel disease, ulcerative colitis, Crohn’s disease, allergic respiratory diseases, and multiple sclerosis,4–13 studies investigating CAM contributions to inflammatory signaling in CRSwNP remain limited.6,14–16 With the emergence of approved immunotherapies targeting CAMs for inflammatory intestinal diseases, increased investigations into this area could be key for developing new CAM-targeting therapies for CRS.7,8
CAMs have been shown to be important in coordinating the adaptive immune response between T helper 2 (Th2) cells and eosinophils via adhesion and/or activation events in allergic asthma and allergic rhinitis.9,10 Allergic asthma and allergic rhinitis are generally eosinophil-predominant and share overlapping pathophysiology to that of eosinophilic CRSwNP.14,17,18 We therefore hypothesize that CAMs may serve similar roles in initiating and coordinating the immune cell signaling between Th2 cells and eosinophils in CRSwNP.4,9,14,17–21 Our objective was to identify specific CAMs and CAM associations to further our understanding of the complex interplay between the local and systemic immune response in CRSwNP.
Materials and Methods
Subjects and Data Collection
This study was conducted in accordance with the Declaration of Helsinki. Study participants were enrolled in the University of Utah Sinus and Skull Base Surgery Clinic using informed consent protocols with approval from the University of Utah Institutional Review Board (IRB 00074325). Inclusion criteria for the CRSwNP cohort consisted of a diagnosis of CRSwNP according to the Academy of Otolaryngology Adult sinusitis guidelines22 for patients who had elected to undergo endoscopic sinus surgery after being treated with the appropriate medical management. Inclusion criteria for control subjects consisted of patients who had elected to undergo non-CRS-related endoscopic sinus surgery.
A total of 47 surgical patients were enrolled in this study: 15 control subjects and 32 patients with CRSwNP. Each study participant underwent evaluation at the University of Utah Sinus and Skull Base Surgery Clinic with a complete demographic and medical history, head and neck physical exam, nasal endoscopy, and review of computed tomography scans. Patient health information was de-identified and stored in a secure electronic database. The following exclusion criteria were implemented to decrease the potential non-CRS-associated contributions to systemic inflammation: patients on oral steroid therapy within two weeks of surgery and patients with a diagnosis of allergic fungal rhinosinusitis, eosinophilic granulomatosis with polyangiitis, systemic lupus erythematosus, chronic obstructive pulmonary disease, multiple sclerosis, cystic fibrosis, cancer, and smoking or alcohol use.
Sinonasal Tissue Collection and Preparation for Gene Expression Analysis
Anterior ethmoid biopsies (4.0 mm2) were obtained intraoperatively per the standard of care and only if the study participant’s (CRSwNP and control subjects) endoscopic sinus surgical procedure involved the excising of tissues from the ethmoid sinuses that would have otherwise been discarded. Sinonasal tissues were immediately snap-frozen and stored at −80°C in 2.0-mL Eppendorf tubes (Fisher Scientific; Waltham, MA) until analyzed. Sinonasal tissues were then thawed on ice, transferred to 0.6 mL of lysis RLT buffer (Qiagen; Hilden, Germany) containing stainless-steel beads (5 mm), and homogenized using a Qiagen Tissuelyser LT. Nucleic acid was extracted using a Qiagen AllPrep RNA microRNA Universal Kit and a QiaCube, followed by subsequent quantification with a NanoDrop 8000 and a Qubit (Thermo Fisher Scientific; Pittsburg, PA), according to manufacturer’s instructions.
Peripheral Blood Collection and Preparation
Peripheral blood (1–3 mL) was drawn from each study participant into a K2-EDTA collection tube (Becton Dickinson; Franklin Lakes, NJ) prior to endoscopic sinus surgery per the standard of care, of which 0.5 mL of blood was added to 1 mL of RNAlater (AppliedBiosystems/Life Technologies Corp., Carlsbad, CA) and stored at −80°C until analyzed. The blood/RNAlater mixture was then thawed on ice, centrifuged at 8,000 rpm at 4°C (Labnet International; Edison, NJ), and the RNAlater was aspirated. RNA was isolated using a RiboPure Blood Kit (Invitrogen; Carlsbad, CA), followed by quantification with a NanoDrop 8000 and a Qubit (Thermo Fisher Scientific; Pittsburg, PA), according to manufacturer’s protocols.
Multiplex Gene Expression Analysis
Sinonasal and blood transcriptional analysis of a total of 594 genes associated with innate and adaptive immunity, allergy, autoimmunity, and infectious diseases was performed by the Molecular Diagnostics Core at the Huntsman Cancer Institute (Salt Lake City, UT) using Human Immunology v2 Panels and the nCounter® MAX Analysis System (NanoString Technologies, Inc.; Seattle, WA). From this large list of genes, the most noticeable transcriptional differences between mRNA transcript copy number ratios of CAM-related genes achieving and approaching statistical significance in CRSwNP:controls were selected for further analysis. After nucleic acid extraction, the sample concentrations were normalized and added (5 µL) to hybridization buffer (70 µL) containing reporter (8 µL) and capture (2 µL) probes. The mixtures were then subjected to hybridization via incubation for 16 hours at 65°C, followed by mRNA transcript copy quantification with a Digital Analyzer. A background subtraction was performed, and the raw transcript copy counts were normalized using the geometric mean of 15 internal reference genes (ABCF1, ALAS1, EEF1G, G6PD, GAPDH, GUSB, HPRT1, OAZ1, POLRIB, POLR2A, PPIA, RPL19, SDHA, TBP, and TUBB) for each lane and the arithmetic mean of all geometric means to create a normalization ratio of the arithmetic mean:geometric mean per lane using nSolver version 3.0 software.23,24 Data are reported as individual values ± standard deviation of the mean mRNA transcript copy number for each patient group.
Sinonasal Tissue Collection and Preparation for Immunohistochemical Analysis
Anterior ethmoid biopsies (4.0 mm2) were obtained as outlined above and immediately placed into 2.0-mL Eppendorf tubes (Fisher Scientific; Waltham, MA) containing 4% neutral formalin (Ted Pella; Redding, CA). Tissues were fixed for 48 hours, trimmed, and sectioned under an Olympus FSX100 stereoscope (Olympus Inc.; Center Valley, PA), followed by paraffin embedding, cutting (4–6-μm thick), and slide mounting by the Research Histology Core at the Huntsman Cancer Institute (Salt Lake City, UT).
Sinonasal Tissue Immunohistochemistry (IHC)
Slide-mounted anterior ethmoid tissues were deparaffinized in xylene (3 x 5 min) and rehydrated in ethanol (100% ethanol, 2×5 min; 95% ethanol, 5 min; 70% ethanol, 5 min) and Nanopure ddH2O (ddH2O, 2×5 min) (Thermo Fisher Scientific; Pittsburg, PA). After staining of each respective antigen described below, tissues were counterstained with hematoxylin (IHC World; Woodstock, MD), dehydrated following the reverse protocol described above, and slide mounted with Permount (ThermoFisher Scientific; Pittsburg, PA). Tissues were evaluated under an Olympus BX43 upright microscope (Olympus Inc.; Pittsburg, PA), imaged with an EOS Rebel T2i digital SLR camera (Canon Inc.; Melville, NY), and processed with Digital Photo Professional version 4.0 software (Canon Inc.; Melville, NY).
Tissue Co-Staining Procedure for Integrin Subunit Alpha x (ITGAX) and Integrin Subunit Alpha M (ITGAM)
After following the hydration protocol described above, the tissues were subjected to heated antigen retrieval in Tris/EDTA buffer (pH 9.0), followed by blocking in BLOXALL (30 min) and 10% goat serum in PBS (20 min) obtained from Vector Labs (Burlingame, CA) and detection of rabbit anti-ITGAX (1:100, 4°C, overnight) (Abcam, ab52632; Cambridge, MA) using ImmPRESSTMHRP anti-rabbit IgG and ImmPACT DAB Peroxidase kits from Vector Labs. The process was repeated for the detection of mouse anti-ITGAM (1:500, 4°C, overnight) (R&D Systems, MAB16991; Minneapolis, MN), employing ImmPRESSTMHRP anti-mouse IgG (clone #238446) and ImmPACT SG Peroxidase kits from Vector Labs. Tissues were also stained with and without primary and secondary antibodies for both antigens to serve as staining controls.
Tissue Co-Staining Procedure for Integrin Subunit Beta 2 (ITGB2) and Intercellular Adhesion Molecule 3 (ICAM-3)
After following the hydration protocol described above, the tissues were subjected to heated antigen retrieval in citrate buffer (pH 6.0), blocking in BLOXALL (30 min) and 10% goat serum in PBS (20 min) obtained from Vector Labs (Burlingame, CA), and detection of rabbit anti-ITGB2 (1:100, 4°C, overnight) (Abcam, ab131044; Cambridge, MA) using ImmPRESSTMHRP anti-rabbit IgG and ImmPACT DAB Peroxidase kits from Vector Labs. The process was repeated for the detection of mouse anti-ICAM-3 (1:1000, 4°C, overnight) (Abcam, ab10804; Cambridge, MA), employing ImmPRESSTMHRP anti-mouse IgG (clone ab10804) and ImmPACT SG Peroxidase kits from Vector Labs. Tissues were also stained with and without primary and secondary antibodies for both antigens to serve as staining controls.
Integrin- and ICAM-Positive Cell Quantification
Integrin- and ICAM-expressing cell counts were enumerated after performing IHC staining of ITGAM, ITGAX, ITGB2, and ICAM-3 in anterior ethmoid tissue samples obtained from controls and patients with CRSwNP. These four CAMs were selected due to the results of the gene expression analysis in ethmoid tissue and blood, which demonstrated the greatest upregulation of these particular molecules in the CRSwNP cohort compared to the control population. Tissue slides were randomized and blinded, and the number of cells that were stained positive for ITGAM, ITGAX, ITGB2, ICAM-3, and co-stained for ITGB2/ICAM-3 were counted and averaged from five randomized high-power (40X) fields with similar histopathologic features per patient. Data are reported as the individual value with upper and lower quartiles ± standard deviation of the mean cell count of respective CAM-positive cells for each group.
Statistical Analyses
Data analysis was conducted using R statistical software (R Core Team, Vienna, Austria, Version 3.6.1, 2019) with figures produced using the ggplot2 package.25 Simple descriptive statistics were calculated to analyze patient characteristics (ie, age, gender, and race) and comorbidities (ie, asthma, allergy, diabetes, gastroesophageal reflux disease (GERD), and aspirin-exacerbated respiratory disease (AERD)) between groups. Gene expression values were compared across groups using Welch’s t-tests due to differences in distribution variance. Two-tailed Pearson’s correlations were used to determine the association between gene expression (ie, mRNA transcript copy counts) in sinonasal and blood tissues. Cell counts were similarly compared across groups using Welch’s t-tests, and associations were revealed using two-tailed Pearson’s correlations. To assess the potential confounding of comorbid asthma and allergy with mRNA transcript copy values and CAM-positive cell counts for both CRSwNP and control cohorts, we performed a series of subgroup analyses utilizing Welch’s t-tests. Significance was evaluated at p<0.05 for all tests.
Results
Cohort Demographics
Forty-seven participants were prospectively enrolled into this study and classified into the following diagnostic groups: control (n=15) or CRSwNP (n=32). Demographic and comorbid information is presented in Table 1. Asthma (p<0.023) and allergy (p<0.029) were significantly different between control and CRSwNP groups.
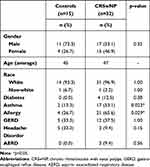 |
Table 1 Demographic Comparisons of the Study Cohort with Respect to Diagnostic Group |
CAM Transcriptional Analysis
From the NanoString gene database, CRSwNP:control expression ratios of CAM-related genes that achieved (p<0.05) or approached statistical significance were selected for further analysis and included genes from the ICAM and integrin superfamilies. The remaining non-CAM genes that were significantly upregulated in the ethmoid sinus and peripheral blood in patients with CRSwNP compared to controls are listed in Supplementary Data Table S1.
The expression of ICAM3 (p<0.02), ITGAM (p<0.05), and ITGB2 (p<0.02) was significantly upregulated in the anterior ethmoid tissues of patients with CRSwNP compared to controls (Figure 1A). Notably, the transcriptional changes in ICAM3, ITGAM, and ITGAX were significantly increased by more than 3-fold in the sinuses of CRSwNP compared to controls. In the peripheral blood of patients with CRSwNP, ICAM3 (p<0.02) and ITGB2 (p<0.001) were significantly upregulated compared to controls (Figure 1B).
Correlation Analysis of Local and Systemic Gene Expression of CAMs
Pearson correlations were conducted to identify potential associations between key identified CAM genes expressed in the sinonasal tissues (Table 2) and peripheral blood (Table 3) in patients with CRSwNP. The expression values of ICAM3, ITGAM, ITGAX, and ITGB2 were strongly correlated to one another both locally in sinonasal tissues (p<0.01 to p<0.05) and systemically in the blood (p<0.05), whereas few gene associations were found between the combinations of other cell adhesion molecules (ICAM1, ICAM2, ITGAE, and ITGB1) expressed in the sinuses and blood.
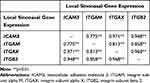 |
Table 2 Pearson Correlations Between the Local Gene Expression of Cell Adhesion Molecules in the Sinonasal Tissues of Patients with CRSwNP |
 |
Table 3 Pearson Correlations Between the Systemic Gene Expression of Cell Adhesion Molecules in the Peripheral Blood of Patients with CRSwNP |
ITGAM, ITGAX, ITGB2, and ICAM-3 Expression in Sinonasal Tissues
Immunohistochemical analysis of cells expressing ICAM-3, ITGB2, ITGAM, and ITGAX in anterior ethmoid tissues was performed to compare protein expression patterns between CRSwNP and controls. These proteins were selected for analysis based on the strong, positive gene correlations observed in CRSwNP tissues and previous studies that report important roles for these CAMs in inflammatory conditions with overlapping pathophysiology.9,13,26
Increased expression of ITGAM, ITGB2, and ICAM-3 was observed in the sinonasal tissues of patients with CRSwNP compared to controls (Figure 2A). The number of cells expressing ITGAM, ICAM-3, and ITGB2, as well as cells co-expressing ICAM-3/ITGB2 was significantly increased in CRSwNP compared to controls: ITGAM, p<0.0004 (Figure 2B); ICAM-3, p<0.0001 (Figure 2D); ITGB2, p<0.04 (Figure 2E); and ICAM-3/ITGB2, p<0.002 (Figure 2F). The number of cells expressing ITGAX was also increased in CRSwNP compared to controls but was not significant (Figure 2C, p<0.34). Moreover, Pearson correlation analysis demonstrated a significant positive correlation between the counts of ITGAM- and ICAM-3/ITGB2-expressing cells in CRSwNP (Figure 2G, R=0.55, p<0.02), suggesting that these CAMs may be interacting with one another to mediate cell migration and adhesion signaling events in CRSwNP, similar to that reported after stimulation with allergic-related molecules in prior studies.9,10,26
Subgroup Analyses
Subgroup analyses were performed to understand the potential confounding effect of comorbid asthma and allergy on CAM expression. No evidence of confounding was found for all subgroup comparison tests of CAM gene expression values and CAM-positive cell counts. For example, the number of cells expressing ITGAM (p<0.4), ICAM-3 (p<0.7), and ITGB2 (p<1.0), as well as cells co-expressing ICAM-3 and ITGB2 (p<0.2), was not significantly different in CRSwNP with asthma compared to CRSwNP without asthma. Similarly, the number of cells expressing ITGAM (p<0.3), ICAM-3 (p<0.2), and ITGB2 (p<0.5), as well as cells co-expressing ICAM-3 and ITGB2 (p<0.3), was not significantly different in CRSwNP with allergy compared to CRSwNP without allergy. Regarding the control cohort, the number of cells expressing ICAM-3 (p<0.7), ITGB2 (p<0.4), and co-expressing ICAM-3 and ITGB2 (p<0.7) was not significantly different in controls with allergy compared to controls without allergy. Similarly, the number of cells expressing ITGB2 (p<0.3) and co-expressing ICAM-3 and ITGB2 (p<0.06) was not significantly different in controls with asthma compared to controls without asthma. The number of cells expressing ICAM-3 (p<0.01) was significantly different between these two groups; however, only 2 of 15 control subjects had comorbid asthma. Due to the increased variability within statistical tests with small sample sizes, this sample size was too small to unequivocally determine that asthma was a confounding variable.
Discussion
This study focused on identifying specific CAMs as potential contributors to local and systemic inflammatory responses in CRSwNP based on gene and protein expression in sinonasal and blood tissues. The upregulation of gene and protein expression, as well as the significant positive correlations between integrins ITGAM, ITGAX, and ITGB2 and ICAM3 in blood and sinonasal tissues and sinonasal tissue co-staining protein patterns in CRSwNP, suggest that these CAMs may serve key systemic and local roles in regulating the pathogenesis of CRSwNP in a coordinated manner.
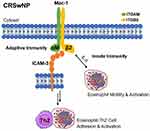
ITGAM and ITGB2 were found to be significantly elevated in the sinonasal mucosa of patients with CRSwNP compared to controls. These two integrins heterodimerize to form integrin αMβ2, termed macrophage-1 antigen (Mac-1).27 Mac-1 is expressed in many types of leukocytes, particularly in eosinophils.26 Mac-1 serves a key role in both innate and adaptive immune responses28 and mediates eosinophil motility upon IL-5 stimulation.26 Becker and colleagues demonstrated that blocking Mac-1 on peripheral blood mononuclear cells isolated from patients with allergic bronchopulmonary aspergillosis resulted in the reduction of cytokines IL-5 and IL-13, which are known to promote eosinophilia and inflammation in CRSwNP.17,19 Acknowledging that Mac-1 is upregulated on immune cells upon allergic stimulation, Mac-1 is poised to be a critical player involved in the cyclic recruitment and trafficking of eosinophils other and leukocytes to locally inflamed sinonasal sites in CRSwNP.
Several mechanistic studies have also linked ICAM-3 to Mac-1.9,10,29 In a rabbit model of allergic pulmonary inflammation, investigators posited that lymphocyte recruitment was mediated by a combination of Mac-1 and lymphocyte-associated antigen 1 (LFA-1), which is comprised of integrin αLβ2 subunits, and is a receptor for ICAM-3.10 A separate study showed that blocking ICAM-3 results in the downregulation and reduced activity of Mac-1 in neutrophils.29 Moreover, Douglas et al demonstrated that ICAM-3 mediates eosinophil adhesion to activated CD4+ lymphocytes and that blocking ICAM-3 inhibited the expression of Mac-1 in the context of allergic asthma.9 CD4+ lymphocytes and eosinophils secrete Th2-associated cytokines IL-4 and IL-13 and are key effector cells involved in allergic airway inflammatory responses, supporting the potential contribution of ICAM-3 in the pathophysiology of CRSwNP. As evidenced by strong positive correlations in systemic and local gene expression and local protein co-localization, our data loosely suggest that Mac-1 may potentially coordinate with ICAM-3 to mediate immune cell adhesion and inflammatory signaling in CRSwNP (Figure 3).9,10,29,30 Future mechanistic studies investigating ICAM-3 interactions with Mac-1 and their involvement in immune cell migration, adhesion, and activation in the context of allergic airway inflammation in CRSwNP are currently underway.
ITGAX also heterodimerizes with ITGB2, forming integrin αXβ2 known as complement receptor 4 (CR4).27 CR4 is not as well studied as Mac-1, but this complex also interacts with numerous ligands and is expressed on eosinophils, basophils, and other leukocytes.31 Our data demonstrate that ITGAX is significantly increased by 3.5-fold in the sinonasal tissues of patients with CRSwNP compared to controls and is positively correlated to ITGB2 expression in blood and the sinuses. ITGAX- and ITGB2-positive immune cells were increased in the sinonasal tissues in patients with CRSwNP compared to controls. Altogether, our data suggest a new finding of CR4 involvement in CRSwNP. Future studies are necessary to further understand the mechanistic implications of ITGAX and ITGB2 interactions and CR4-mediated outcomes in CRSwNP pathology.
There are several limitations to consider while interpreting these findings. Although the sample size is not as robust as desired, we believe the cohort size is ample based on the criteria for performing genome-wide association studies.6 Comorbid allergy and/or asthma were present in both controls and patients with CRSwNP. Although we found no evidence of confounding in subgroup analysis, allergy and asthma have been linked to chronic inflammation and elevated systemic inflammatory biomarker levels.
Conclusion
We report new findings that suggest Mac-1 and ICAM-3 may be involved in coordinating inflammatory cell migration, adhesion, and activation in CRSwNP (Figure 3), as previously shown in allergic asthma and allergic rhinitis.6,9,26 We also demonstrate several significant associations between locally and systemically expressed integrins and ICAMs in CRSwNP, serving as a key investigation to propel further mechanism-driven research in the pursuit of identifying new potential disease biomarkers. For example, several monoclonal antibodies based on integrins (ie, integrin α4: natalizumab, integrin α4β7: vedolizumab and integrin αEβ7: etrolizumab) are approved to treat inflammatory intestinal diseases, and future mechanistic investigations on CAMs could be key for developing new CAM-targeting therapies for CRSwNP.7,8 Phenotyping studies to identify which specific CAMs are upregulated with respect to peripheral blood leukocytes and granulocytes in CRSwNP, as well as mechanistic investigations to explore the interactions of these molecules and further clarify the pathophysiologic outcomes of CAM-mediated signaling in CRSwNP are currently underway.
Acknowledgments
The authors thank the Research Histology and Molecular Diagnostics divisions of the Biorepository and Molecular Pathology Shared Resource at the Huntsman Cancer Institute (Salt Lake City, UT) for assistance with tissue processing, nucleic acid isolation, and gene expression analyses.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was funded by grants from the National Institute of Allergy and Infectious Diseases (R44AI126987 to A. P. and J. A. A.) and Flight Attendant Medical Research Institute (CIA160008 to J.A.A.).
Disclosure
A. P. and J. A. A. have a financial interest in GlycoMira Therapeutics (Salt Lake City, UT). J. A. A. reports personal fees from Optinose and Medtronic, outside the submitted work, is a consultant for OptiNose (Pennsylvania, PA), and is involved in clinical research for OptiNose and AstraZeneca (Wilmington, DE). None of these potential conflicts of interest are affiliated with this research. The authors report no other potential conflicts of interest for this work.
References
1. Mitroulis I, Alexaki VI, Kourtzelis I, Ziogas A, Hajishengallis G, Chavakis T. Leukocyte integrins: role in leukocyte recruitment and as therapeutic targets in inflammatory disease. Pharmacol Ther. 2015;147:123–135.
2. Fagerholm SC, Guenther C, Llort Asens M, Savinko T, Uotila LM. Beta2-integrins and interacting proteins in leukocyte trafficking, immune suppression, and immunodeficiency disease. Front Immunol. 2019;10:254. doi:10.3389/fimmu.2019.00254
3. Yee NK, Hamerman JA. beta(2) integrins inhibit TLR responses by regulating NF-kappaB pathway and p38 MAPK activation. Eur J Immunol. 2013;43:779–792. doi:10.1002/eji.201242550
4. Chen X, Corry DB, Li E. Mechanisms of allergy and adult asthma. Curr Opin Allergy Clin Immunol. 2020;20:36–42. doi:10.1097/ACI.0000000000000601
5. Lees KR, Diener HC, Asplund K, Krams M. UK-279,276, a neutrophil inhibitory glycoprotein, in acute stroke: tolerability and pharmacokinetics. Stroke. 2003;34:1704–1709. doi:10.1161/01.STR.0000078563.72650.61
6. Liu M, Guo P, An J, Guo C, Lu F, Lei Y. Genomewide profiling of lncRNA and mRNA expression in CRSwNP. Mol Med Rep. 2019;19:3855–3863. doi:10.3892/mmr.2019.10005
7. McLean LP, Cross RK. Integrin antagonists as potential therapeutic options for the treatment of Crohn’s disease. Expert Opin Investig Drugs. 2016;25:263–273. doi:10.1517/13543784.2016.1148137
8. Singh H, Grewal N, Arora E, Kumar H, Kakkar AK. Vedolizumab: a novel anti-integrin drug for treatment of inflammatory bowel disease. J Nat Sci Biol Med. 2016;7:4–9. doi:10.4103/0976-9668.175016
9. Douglas IS, Leff AR, Sperling AI. CD4+ T cell and eosinophil adhesion is mediated by specific ICAM-3 ligation and results in eosinophil activation. J Immunol. 2000;164:3385–3391. doi:10.4049/jimmunol.164.6.3385
10. Gascoigne MH, Holland K, Page CP, et al. The effect of anti-integrin monoclonal antibodies on antigen-induced pulmonary inflammation in allergic rabbits. Pulm Pharmacol Ther. 2003;16:279–285. doi:10.1016/S1094-5539(03)00069-5
11. Gu P, Theiss A, Han J, Feagins LA. Increased cell adhesion molecules, PECAM-1, ICAM-3, or VCAM-1, predict increased risk for flare in patients with quiescent inflammatory bowel disease. J Clin Gastroenterol. 2017;51:522–527. doi:10.1097/MCG.0000000000000618
12. Karta MR, Rosenthal PS, Beppu A, et al. β2 integrins rather than β1 integrins mediate Alternaria-induced group 2 innate lymphoid cell trafficking to the lung. J Allergy Clin Immunol. 2018;141:329–338 e312. doi:10.1016/j.jaci.2017.03.010
13. Vainer B, Nielsen OH, Horn T. Comparative studies of the colonic in situ expression of intercellular adhesion molecules (ICAM-1, −2, and −3), beta2 integrins (LFA-1, Mac-1, and p150,95), and PECAM-1 in ulcerative colitis and Crohn’s disease. Am J Surg Pathol. 2000;24:1115–1124. doi:10.1097/00000478-200008000-00009
14. Koennecke M, Klimek L, Mullol J, Gevaert P, Wollenberg B. Subtyping of polyposis nasi: phenotypes, endotypes and comorbidities. Allergo J Int. 2018;27:56–65. doi:10.1007/s40629-017-0048-5
15. Shin HW, Kim DK, Park MH, et al. IL-25 as a novel therapeutic target in nasal polyps of patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2015;135:1476–1485 e1477. doi:10.1016/j.jaci.2015.01.003
16. Oyer SL, Nagel W, Mulligan JK. Differential expression of adhesion molecules by sinonasal fibroblasts among control and chronic rhinosinusitis patients. Am J Rhinol Allergy. 2013;27:381–386. doi:10.2500/ajra.2013.27.3934
17. Cao PP, Wang ZC, Schleimer RP, Liu Z. Pathophysiologic mechanisms of chronic rhinosinusitis and their roles in emerging disease endotypes. Ann Allergy Asthma Immunol. 2019;122:33–40. doi:10.1016/j.anai.2018.10.014
18. Rosati MG, Peters AT. Relationships among allergic rhinitis, asthma, and chronic rhinosinusitis. Am J Rhinol Allergy. 2016;30:44–47. doi:10.2500/ajra.2016.30.4252
19. Becker KL, Gresnigt MS, Smeekens SP, et al. Pattern recognition pathways leading to a Th2 cytokine bias in allergic bronchopulmonary aspergillosis patients. Clin Exp Allergy. 2015;45:423–437. doi:10.1111/cea.12354
20. Cheung PF, Wong CK, Ip WK, Lam CW. IL-25 regulates the expression of adhesion molecules on eosinophils: mechanism of eosinophilia in allergic inflammation. Allergy. 2006;61:878–885. doi:10.1111/j.1398-9995.2006.01102.x
21. Chung SD, Chen PY, Lin HC, Hung SH. Comorbidity profile of chronic rhinosinusitis: a population-based study. Laryngoscope. 2014;124:1536–1541. doi:10.1002/lary.24581
22. Rosenfeld RM. Clinical practice guideline on adult sinusitis. Otolaryngol Head Neck Surg. 2007;137:365–377. doi:10.1016/j.otohns.2007.07.021
23. Brumbaugh CD, Kim HJ, Giovacchini M, Pourmand N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC Bioinform. 2011;12:479. doi:10.1186/1471-2105-12-479
24. Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;94:25B–10.
25. Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer: New York; 2009.
26. Johansson MW, Annis DS, Mosher DF. αMβ2 integrin-mediated adhesion and motility of IL-5-stimulated eosinophils on periostin. Am J Respir Cell Mol Biol. 2013;48:503–510. doi:10.1165/rcmb.2012-0150OC
27. Vorup-Jensen T, Jensen RK. Structural immunology of complement receptors 3 and 4. Front Immunol. 2018;9:2716. doi:10.3389/fimmu.2018.02716
28. Fischer E, Capron M, Prin L, Kusnierz JP, Kazatchkine MD. Human eosinophils express CR1 and CR3 complement receptors for cleavage fragments of C3. Cell Immunol. 1986;97:297–306. doi:10.1016/0008-8749(86)90400-4
29. Skubitz KM, Campbell KD, Skubitz AP. CD50 monoclonal antibodies inhibit neutrophil activation. J Immunol. 1997;159:820–828.
30. Hua S. Targeting sites of inflammation: intercellular adhesion molecule-1 as a target for novel inflammatory therapies. Front Pharmacol. 2013;4:127. doi:10.3389/fphar.2013.00127
31. Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi:10.1016/S0092-8674(02)00971-6
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.