Back to Journals » OncoTargets and Therapy » Volume 12
CD24 isoform a promotes cell proliferation, migration and invasion and is downregulated by EGR1 in hepatocellular carcinoma
Authors Li L, Chen J, Ge C, Zhao F, Chen T, Tian H, Li J , Li H
Received 30 November 2018
Accepted for publication 18 January 2019
Published 28 February 2019 Volume 2019:12 Pages 1705—1716
DOI https://doi.org/10.2147/OTT.S196506
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Liangyu Li,1,* Jing Chen,2,* Chao Ge,2 Fangyu Zhao,2 Taoyang Chen,3 Hua Tian,2 Jinjun Li,2 Hong Li2
1Key Laboratory of Medical Molecular Virology, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China; 2State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 3Qi Dong Liver Cancer Institute, Qi Dong, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Introduction: CD24 is known as a heavily glycosylated cell surface molecule that is highly expressed in a wide variety of human malignancies. Previous studies have shown that CD24 plays an important role in self-renewal, proliferation, migration, invasion and drug resistance of hepatocellular carcinoma (HCC). However, little is known about the expression and function of CD24 isoform a (CD24A) and CD24 isoform b (CD24B) in HCC.
Materials and methods: Quantitative real-time polymerase chain reaction (qPCR) and Western blotting were performed to detect CD24 and EGR1 expression in HCC cells and tissue. The function of CD24 in cell proliferation was verified with MTT assays, colony formation assays and tumor xenograft models. Wound healing assays and invasion assays were performed to clarify the function of CD24 in the regulation of cell migration and invasion in HCC. A dual luciferase reporter assay and chromatin immunoprecipitation assay were used to analyze the regulation mechanism of CD24A.
Results: CD24A but not CD24B, which was barely detected by qPCR and Western blotting, is significantly upregulated in HCC tissue. Both CD24A and CD24B contribute to HCC cell proliferation, migration and invasion, but CD24A is more effective than CD24B. EGR1 downregulates CD24A and exerts transcription-promoting activity on the CD24A promoter. Furthermore, EGR1 represses HCC cell proliferation via downregulation of CD24A.
Conclusion: CD24A is the predominant CD24 isoform in HCC and plays a major role in cell proliferation, migration, and invasion. EGR1 can exert its antitumor effect through transcriptional downregulation of CD24A in HCC.
Keywords: CD24A, CD24B, EGR1, proliferation, hepatocellular carcinoma
Introduction
Liver cancer is predicted to be the sixth most commonly diagnosed cancer in 2018 and ranks fifth in terms of global cases and second in terms of deaths for males.1,2 Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is a highly therapy-resistant and thus difficult to treat cancer;3 although systemic therapies have clinical benefits, only a few patients with HCC (<10%) are cured.3 Thus, it is of great significance to reveal molecular alterations in HCC and find novel therapy targets for HCC.
CD24 is a highly glycosylated cell surface glycoprotein expressed on the surface of most B lymphocytes4 and is highly expressed in a wide variety of human malignancies.5–14 The CD24 gene is located on chromosome 6q21 and contains five variants; variant 1, variant 2, variant 3 and variant 7 encode CD24A preproprotein, while variant 4 encodes CD24B. CD24B (129aa) has a distinct N-terminus and is longer than CD24A (83aa). In HCC, CD24 was found to be a functional liver tumor-initiating cell marker15,16 that drives tumor-initiating cell genesis through STAT3-mediated NANOG regulation,16 and Twist2 augments liver cancer stem-like cell self-renewal in a CD24-STAT3-Nanog-dependent manner.17 CD24 induces sorafenib resistance by activating autophagy in HCC,18 and CD24-targeted therapy may be a promising therapeutic strategy for treatment of HCC.18–20 Two CD24 isoforms are encoded by different variants and only share approximately 47% amino acid identity; however, previous studies have focused on total CD24, and thus far, very little has been done to investigate the expression and role of the two CD24 isoforms in HCC. Hence, identification of the predominant CD24 isoform may have therapeutic implications for CD24-targeted treatment of HCC.
Our study identified CD24A as a direct target gene of EGR1 (Early growth response protein 1). EGR1, a nuclear transcription factors, binds to the GC enrichment region of DNA sequences to play its role as a transcriptional regulator.21 Abnormal expression of EGR1 is often correlated with ischemic injury, atherosclerosis, inflammation and tumors.22–25 EGR1 plays complicated roles in HCC. Several studies have stated that EGR1 is overexpressed in HCC tissues, enhances drug resistance by promoting hypoxia-induced autophagy26 and accelerates the progression of HCC;27–29 however, data from several independent laboratories have demonstrated that EGR1 inhibits HCC cell motility and invasion.30–32
In our present study, we found that CD24A was the predominant CD24 isoform in HCC and plays a major role in cell proliferation, migration, and invasion. EGR1 regulated CD24A expression directly and exerted its antitumor effect through downregulation of CD24A in HCC.
Materials and methods
Human liver specimens and TCGA cohort
Ninety paired human primary HCC and matched adjacent non-cancerous liver tissue specimens were obtained from Qidong Liver Cancer Institute (Qidong, China). All tissues were frozen at −80°C until mRNA and protein were extracted. This study was approved by the Research Ethics Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine. Informed consent was signed by all patients, and all procedures were conducted in accordance with the Declaration of Helsinki. TCGA data (mRNA expression data for 50 paired cancer/non-cancerous tissues were available) were accessed from the website (https://tcga-data.nci.nih.gov/tcga/).
Cell lines and cell culture
The human HCC cell lines SMMC-7721 and BEL-7402 and immortalized normal liver L-02 cells were purchased from the Cell Bank of the Institute of Biochemistry and Cell Biology, China Academy of Sciences (Shanghai, People’s Republic of China). Li7 was purchased from SXBIO Corporation (Shanghai, People’s Republic of China). Huh6 and Huh7 cell lines were obtained from Riken Cell Bank (Tsukuba, Japan). The HCC-LY5 and HCC-LY10 cell lines were established in our laboratory. MHCC-LM3, MHCC-97, MHCC-97H, and MHCC-97L cell lines were obtained from Liver Cancer Institute, Zhongshan Hospital of Fudan University (Shanghai, People’s Republic of China). H2P and H2M cell lines were kindly provided by the University of Hong Kong (Hong Kong, People’s Republic of China). Other cell lines not specifically mentioned here were all purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich Co., St Louis, MO, USA) containing 10% fetal bovine serum (HyClone, Logan, UT, USA) at 37°C in 5% CO2. The use of all lines was approved by the Research Ethics Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine.
Quantitative real time polymerase chain reaction (qPCR)
Total RNA was extracted from HCC tissues and cells using Trizol (Thermo Fisher Scientific, Waltham, MA, USA), and reverse transcription was performed with a PrimeScript™ RT Reagent Kit (Perfect Real Time) (Takara, Dalian, People’s Republic of China). qPCR was performed with SYBR Premix Ex Taq II (Takara, Dalian, People’s Republic of China) according to the manufacturer’s protocol. The expression levels were normalized using human GAPDH (glyceraldehyde-3-phos-phate dehydrogenase). The primer sequences are listed in Table S1.
Western blotting
Proteins extracted from HCC tissues and cells were separated on 12% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Merck Millipore, Billerica, MA, USA). Mouse anti-CD24 (SAB-1402713, Sigma-Aldrich Co.), rabbit anti-EGR1 (SC-189, Santa Cruz Biotechnology Inc., Dallas, TX, USA) and mouse anti-β-Actin (A3854, Sigma-Aldrich Co.) antibodies were incubated separately with the membranes at 4°C overnight after blocking with 5% nonfat milk, and the membranes were then probed with corresponding HRP-conjugated secondary antibody for 1.5 hours at room temperature.
Plasmid construction, lentivirus production, and cell transfection
The full-length human CD24A and CD24B open reading frame cDNA sequences were separately amplified and cloned into a pWPXL plasmid (Addgene, Cambridge, MA, USA) using BamHI and EcoRI. The full-length human EGR1 open reading frame cDNA sequence was amplified and cloned into pWPXL using MluI and EcoRI. The CD24A promoter, which spans a 2,100 bp-region (−1,900 bp to +200 bp based on the first ATG), and mutant were separately cloned into PGL3-Enhancer (Promega Corporation, Fitchburg, WI, USA). The primer sequences are listed in Table S1.
For lentivirus production and cell transfection, after co-transfection of HEK 293 T cells with the pWPXL-CD24A vector, pWPXL-CD24B vector or pWPXL-EGR1 vector with psPAX2 and pMD2.G (Addgene) using Lipofectamine 2000 (Thermo Fisher Scientific) for 48 hours, viruses were harvested and used to infected target cells in the presence of 6 μg/mL polybrene (Sigma-Aldrich Co.).
MTT assays
A total of 800–1,000 cells per well were seeded into 96-well plates and cultured for 7 days. MTT reagent (5 mg/mL, Sigma-Aldrich Co.) was added and detected according to the manufacturer’s instructions. All of the experiments were performed in triplicate.
Colony formation assays
A total of 500–1,000 cells per well were seeded into 6-well plates and cultured for approximately 2 weeks. Then, the cells were fixed with 10% formaldehyde for 30 minutes and stained with Giemsa solution (Sigma-Aldrich Co.) for 40 minutes at room temperature. All of the experiments were performed in triplicate.
Wound healing assays
Cells were seeded into 6-well plates and cultured to approximately 90% confluence in 24 hours. Then, wound healing assays were performed as described previously.33
Transwell invasion assays
Approximately 1×105 cells in serum-free DMEM were seeded into the top chamber of transwell chambers (8 mm pore, Merck Millipore) precoated with Matrigel (BD Biosciences, San Jose, CA, USA), while 600 μL DMEM containing 10% FBS was added to the bottom chamber. After 24–48 hour of incubation, cells adhering to the lower membrane of the inserts were fixed with 10% formaldehyde for 30 minutes and stained with Giemsa solution. Cells in five randomly selected fields were counted.
Dual luciferase reporter assay
Cells were seeded into 96-well plates overnight and co-transfected with the relevant reporter plasmids using Lipofectamine 2000 (Thermo Fisher Scientific). A PRL-TK reporter construct was used as the internal reference. After 48 hours of incubation, firefly luciferase activity and Renilla luciferase activity were detected according to the manufacturer’s instructions (Promega).
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed with SMMC-7721 and Hep3B cells as previously described.33 Rabbit anti-EGR1 antibody (SC-189, Santa Cruz Biotechnology) or rabbit IgG were used to immunoprecipitate DNA-containing complexes. The isolated DNA samples were subjected to PCR analyses. The primer sequences are listed in Table S1.
Tumor xenograft models
All animal experiments were approved by the Shanghai Cancer Institute Experimental Animal Care Commission prior to commencement of the study and performed following the guidelines and regulations of Shanghai Cancer Institute Experimental Animal Care Commission. The 6–8-week-old male Balb/c (nu/nu) mice were divided randomly into groups. Approximately 2×106 Li7 cells stably expressing CD24A, EGR1 or control (pWPXL) were injected subcutaneously into each mouse. After approximately 7 weeks, all mice were sacrificed. Xenograft tumors were weighed and frozen at −80°C.
Statistical analyses
The data are presented as the mean ± standard deviation (SD) and were analyzed using Student’s t-test. Statistical analyses were performed using GraphPad Prism 5 software. P<0.05 was considered statistically significant.
Results
CD24A is overexpressed in human HCC clinical specimens and cell lines
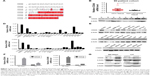
Amino acid sequence alignment34 of CD24A with CD24B showed that CD24B has a distinct N-terminus and shares approximately 47% amino acid identity with CD24A (Figure 1A). We designed different primers to detect the expression of CD24A and CD24B, and the results showed that the CD24A mRNA level in HCC tissues was much higher than that in matched non-cancerous liver tissues (Figure 1B). However, CD24B was barely detected in HCC tissues and their matched non-cancerous liver tissues by qPCR using SYBR Premix Ex Taq II. The same results were obtained in HCC and matched non-cancerous liver tissues with Western blotting (Figure 1C). We also detected the expression of CD24 in HCC cell lines via qPCR and Western blotting. As shown in Figure 1D, the mRNA expression of CD24A was much higher than that of CD24B. The Western blotting results showing CD24 protein levels in HCC cell lines were in agreement with the above results (Figure 1E), confirming that only CD24A could be detected. Taken together, these findings indicate that CD24A is the predominant CD24 isoform and is significantly upregulated in HCC tissues.
CD24A promotes HCC cell proliferation, migration, and invasion in vitro
To better investigate the function of CD24A and CD24B in HCC progression, we selected two CD24 low-expressing cell lines, SK-Hep1 and Li7, to construct stable overexpressing cell lines via lentiviral infection. The efficiency of CD24A and CD24B overexpression was verified by qPCR and Western blotting (Figure 1F and G).
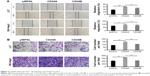
MTT assays and colony formation assays showed that both CD24 isoforms contributed to HCC cell growth, but overexpression of CD24A had a stronger effect on cell proliferation in vitro than overexpression of CD24B (Figure 2A and B). To further clarify the role of CD24A in HCC cell growth in vivo, Li7 cells stably expressing CD24A or the control (pWPXL) were subcutaneously injected into male Balb/c (nu/nu) mice. After 7 weeks, the weight of tumors revealed that overexpression of CD24A significantly promoted the tumorigenicity of Li7 cells in vivo (Figure 2C). Western blotting analysis of CD24A protein levels showed that the tissues of xenografts overexpressing CD24A maintained a high expression level of CD24A (Figure 2D).
Then, we performed wound healing assays and transwell assays to detect the effect of the CD24 isoforms on cell migration and invasion. The results showed that CD24A enhanced cell migration and invasion more effectively than CD24B in vitro (Figure 3A and B). Therefore, these findings indicate that CD24A but not CD24B effectively facilitates HCC proliferation, migration, and invasion.
EGR1 represses CD24A expression in human HCC cells
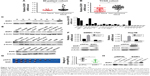
To understand the regulatory mechanism of CD24A expression, bioinformatics analysis was performed and indicated that there were several functional binding sites for EGR1 on the CD24A promoter region. The mRNA level of EGR1 in 90 pairs of human primary HCC tissues and matched adjacent non-cancerous liver tissues from our lab was analyzed by qPCR. The results showed that EGR1 mRNA expression was significantly downregulated in HCC tissues compared with matched adjacent non-cancerous liver tissues (Figure 4A), which is consistent with the analysis of TCGA data (Figure 4B). Western blotting analyses also showed that EGR1 protein expression was significantly downregulated in HCC tissues (Figure 4C).
To explore whether EGR1 can regulate CD24A expression, we detected the mRNA and protein expression profiles of EGR1 in HCC cell lines (Figure 4D and E) and chose SMMC-7721 and Hep3B cell lines to overexpress EGR1 (Figure 4F). qPCR and Western blotting showed that overexpression of EGR1 repressed CD24A expression (Figure 4F). Considering the complicated role of EGR1 in HCC, we also clarified the effect of EGR1 on cell growth in vivo and found that tumor weights were remarkably decreased in the EGR1-overexpressing group compared with the control group (Figure 4G). EGR1 protein levels in the xenograft tumors were analyzed by Western blotting (Figure 4H). Overall, EGR1 is significantly downregulated in HCC tissues and may inhibit HCC cell proliferation by repressing CD24A expression.
CD24A is a direct target of EGR1
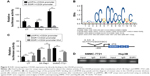
To clarify whether EGR1 can transcriptionally regulate CD24A expression directly, we analyzed the binding site for EGR1 on the CD24A promoter using the JASPAR database (http://jaspar.genereg.net/) and cloned the main full-length CD24A promoter region (from −1900 bp to +200 bp). Dual luciferase reporter assays showed that overexpression of EGR1 significantly reduced the activity of the CD24A promoter in Li7, SK-Hep1 and SMMC-7721 cell lines (Figure 5A). According to the sequence of the potential EGR1 binding site in JASPAR, we constructed a mutant of the CD24A promoter (Figure 5B). The inhibitory effect of EGR1 on the activity of the CD24A promoter was obviously weakened by the mutation (Figure 5C). A ChIP assay further verified that EGR1 can bind the site on the CD24A promoter (Figure 5D). Therefore, CD24A is a direct transcriptional target of EGR1.
EGR1 suppresses HCC cell proliferation by downregulating CD24A
To further investigate whether downregulation of CD24A is responsible for the tumor suppressive function of EGR1 in HCC, CD24A was overexpressed in Li7 and SK-Hep1 cells with stable overexpression of EGR1 (Figure 6A). MTT assays and colony formation assays in vitro showed that overexpression of EGR1 significantly inhibited cell proliferation, and restoration of CD24A expression reversed the EGR1-induced inhibition of cell proliferation (Figure 6B and C). In conclusion, these data indicate that EGR1 suppresses HCC cell proliferation by repressing CD24A.
Discussion
CD24 is a glycosylphosphatidylinositol-anchored membrane protein reported to be overexpressed in many tumor types, including colorectal cancer,13 ovarian cancer,7 bladder cancer,14 prostate cancer,5,11 lung cancer,12 and breast cancer.9 CD24 plays important roles in tumorigenesis,35–37 progression38–40 and drug resistance.18,41 CD24 is considered to be a negative cancer stem cell marker, specifically in breast cancer.42–44 Intriguingly, mounting evidence has shown that CD24-positive tumor cells are tumor-initiating cells for some cancers, such as colon cancer,45 pancreatic cancer,46,47 cholangiocarcinoma,48 gastric cancer,49 cervical cancer,50,51 and ovarian cancer.52,53 Similarly, CD24 promotes HCC progression and endows HCC cells with stem cell and drug-resistant properties.10,16,18 Therefore, increasing evidence suggests that identification of CD24 signaling pathways may provide an attractive therapeutic strategy against HCC. However, these previous studies did not investigate whether CD24 isoforms have distinct expression patterns and roles, especially in HCC.
Two CD24 isoforms are encoded by different variants and share approximately 47% amino acid identity. This study is the first to demonstrate that CD24A is the predominant isoform and significantly upregulated in HCC tissues, while CD24B could barely be detected by qPCR using SYBR Premix Ex Taq II or by Western blotting in HCC tissues and cell lines. Moreover, CD24A had a stronger effect on proliferation, invasion and migration of HCC cells in vitro than CD24B. Therefore, the expression and role of CD24A are consistent with previous reports of CD24 in HCC.10 Intermediate and advanced HCC are highly heterogeneous,10,54 and thus, whether CD24B is expressed in a very small population of cells in HCC tissues should be further investigated.
Considering the predominant expression and role of CD24A in HCC cells, we further explored the regulation of CD24A in HCC cells. The promoter region of CD24A was analyzed using the JASPAR database, and an EGR1 binding site located in the CD24A promoter region was found. EGR1 plays complex and often contradictory roles in human malignancies.55,56 Based on previous reports, EGR1 also displays both facilitating and repressing effects in HCC.26–32 For instance, EGR1 functions as an oncogene, promoting HCC growth and migration/invasion27,29,31 and enhancing the drug resistance of HCC cells, likely through autophagy.26 Conversely, EGR1 can inhibit HCC progression by repressing the EGFR-MAPK/AKT pathway32 and promoting PTEN transcription.30 In our current study, EGR1 expression in a 90-patient cohort from our laboratory and a 50-patient cohort from TCGA was analyzed, and we found that EGR1 expression was significantly downregulated in HCC tissues compared with matched adjacent non-cancerous liver tissues. The effect of EGR1 on the proliferation of HCC cells in vivo was detected, and the results showed that overexpression of EGR1 remarkably inhibited HCC cell growth. Further investigation into the mechanism showed that EGR1 repressed the activity of CD24A by directly binding to the CD24A promoter, and restoration of CD24A expression reversed the EGR1-induced inhibition of cell proliferation. Therefore, our data support the hypothesis that EGR1 acts as a tumor suppressor in HCC.
Conclusion
Our findings demonstrate that CD24A, the predominant CD24 isoform, plays the main role in cell proliferation, invasion and migration. EGR1 is a key player in transcriptional control of CD24A and inhibits cell proliferation by downregulating CD24A. These data provide evidence supporting CD24-targeted treatment of HCC.
Acknowledgments
This work was supported in part by grants from the National Natural Science Foundation of China (81672832), the Project of Shanghai Municipal Commission of Health and Family Planning (2018BR20), and the State Key Laboratory of Oncogenes and Related Genes (91-17-18).
Disclosure
The authors report no conflicts of interest in this work.
References
Bray F, Ferlay J, Soerjomataram I, et al. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: Cancer J Clinicians. 2018;68(1):7–30. | ||
Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15(10):599–616. | ||
Gilliam DT, Menon V, Bretz NP, Pruszak J. The CD24 surface antigen in neural development and disease. Neurobiol Dis. 2017;99:133–144. | ||
Zhang W, Yi B, Wang C, et al. Silencing of CD24 enhances the PRIMA-1-Induced restoration of mutant p53 in prostate cancer cells. Clin Cancer Res. 2016;22(10):2545–2554. | ||
Sung CO, Park W, Choi YL, et al. Prognostic significance of CD24 protein expression in patients treated with adjuvant radiotherapy after radical hysterectomy for cervical squamous cell carcinoma. Radiother Oncol. 2010;95(3):359–364. | ||
Tarhriz V, Bandehpour M, Dastmalchi S, Ouladsahebmadarek E, Zarredar H, Eyvazi S. Overview of CD24 as a new molecular marker in ovarian cancer. J Cell Physiol. 2019;234(3):2134–2142. | ||
Majores M, Schindler A, Fuchs A, et al. Membranous CD24 expression as detected by the monoclonal antibody SWA11 is a prognostic marker in non-small cell lung cancer patients. BMC Clin Pathol. 2015;15(1):19. | ||
Kristiansen G, Winzer KJ, Mayordomo E, et al. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res. 2003;9(13):4906–4913. | ||
Yang XR, Xu Y, Yu B, et al. CD24 is a novel predictor for poor prognosis of hepatocellular carcinoma after surgery. Clin Cancer Res. 2009;15(17):5518–5527. | ||
Zhang Y, Li B, Zhang X, et al. CD24 is a genetic modifier for risk and progression of prostate cancer. Mol Carcinog. 2017;56(2):641–650. | ||
Kristiansen G, Schlüns K, Yongwei Y, Denkert C, Dietel M, Petersen I. CD24 is an independent prognostic marker of survival in nonsmall cell lung cancer patients. Br J Cancer. 2003;88(2):231–236. | ||
Wang JL, Guo CR, Su WY, Chen YX, Xu J, Fang JY. CD24 overexpression related to lymph node invasion and poor prognosis of colorectal cancer. Clin Lab. 2018;64(4):497–505. | ||
Ooki A, Vandenbussche CJ, Kates M, et al. CD24 regulates cancer stem cell (CSC)-like traits and a panel of CSC-related molecules serves as a non-invasive urinary biomarker for the detection of bladder cancer. Br J Cancer. 2018;119(8):961–970. | ||
Wang R, Li Y, Tsung A, et al. iNOS promotes CD24+ CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci U S A. 2018;115(43):E10127–E10136. | ||
Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24 (+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated Nanog regulation. Cell Stem Cell. 2011;9(1):50–63. | ||
Liu AY, Cai Y, Mao Y, et al. Twist2 promotes self-renewal of liver cancer stem-like cells by regulating CD24. Carcinogenesis. 2014;35(3):537–545. | ||
Lu S, Yao Y, Xu G, et al. CD24 regulates sorafenib resistance via activating autophagy in hepatocellular carcinoma. Cell Death Dis. 2018;9(6):646. | ||
Wan X, Cheng C, Shao Q, Lin Z, Lu S, Chen Y. CD24 promotes HCC progression via triggering Notch-related EMT and modulation of tumor microenvironment. Tumor Biol. 2016;37(5):6073–6084. | ||
Li B, Shao Q, Ji D, Li F, Guo X, Chen G. Combined aberrant expression of N-myc downstream-regulated gene 2 and CD24 is associated with disease-free survival and overall survival in patients with hepatocellular carcinoma. Diagn Pathol. 2014;9(1):209. | ||
Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci. 1989;86(22):8737–8741. | ||
Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 2018;78(20):5877–5890. | ||
Gomez-Martin D, Diaz-Zamudio M, Galindo-Campos M, Alcocer-Varela J. Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmun Rev. 2010;9(6):454–458. | ||
Sun M, Nie FQ, Zang C, et al. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing Egr1 and RhoB. Mol Ther. 2017;25(3):739–751. | ||
Kim J, Kang HS, Lee YJ, et al. Egr1-dependent PTEN upregulation by 2-benzoyloxycinnamaldehyde attenuates cell invasion and EMT in colon cancer. Cancer Lett. 2014;349(1):35–44. | ||
Peng WX, Xiong EM, Ge L, et al. Egr-1 promotes hypoxia-induced autophagy to enhance chemo-resistance of hepatocellular carcinoma cells. Exp Cell Res. 2016;340(1):62–70. | ||
Zhang Q, Song G, Yao L, et al. miR-3928v is induced by HBx via NF-κB/EGR1 and contributes to hepatocellular carcinoma malignancy by down-regulating VDAC3. J Exp Clin Cancer Res. 2018;37(1):14. | ||
Lu D, Han C, Wu T. Microsomal prostaglandin E synthase-1 promotes hepatocarcinogenesis through activation of a novel EGR1/β-catenin signaling axis. Oncogene. 2012;31(7):842–857. | ||
Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of snail. Embo J. 2006;25(15):3534–3545. | ||
Tian H, Ge C, Li H, et al. Ribonucleotide reductase M2B inhibits cell migration and spreading by early growth response protein 1-mediated phosphatase and tensin homolog/Akt1 pathway in hepatocellular carcinoma. Hepatology. 2014;59(4):1459–1470. | ||
Ozen E, Gozukizil A, Erdal E, Uren A, Bottaro DP, Atabey N. Heparin inhibits hepatocyte growth factor induced motility and invasion of hepatocellular carcinoma cells through early growth response protein 1. PLoS One. 2012;7(8):e42717. | ||
Wang L, Sun H, Wang X, et al. Egr1 mediates miR-203a suppress the hepatocellular carcinoma cells progression by targeting HOXD3 through EGFR signaling pathway. Oncotarget. 2016;7(29):45302–45316. | ||
Jiang J, Liu Z, Ge C, et al. NK3 homeobox 1 (NKX3.1) up-regulates forkhead box O1 expression in hepatocellular carcinoma and thereby suppresses tumor proliferation and invasion. J Biol Chem. 2017;292(47):19146–19159. | ||
Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. | ||
Agarwal N, Dancik GM, Goodspeed A, et al. Gon4l drives cancer growth through a YY1-androgen receptor-CD24 axis. Canc Res. 2016;76(17):5175–5185. | ||
Wang YC, Wang JL, Kong X, et al. CD24 mediates gastric carcinogenesis and promotes gastric cancer progression via STAT3 activation. Apoptosis. 2014;19(4):643–656. | ||
Naumov I, Zilberberg A, Shapira S, et al. CD24 knockout prevents colorectal cancer in chemically induced colon carcinogenesis and in APC Min/CD24 double knockout transgenic mice. Int J Canc. 2014;135(5):1048–1059. | ||
Okabe H, Aoki K, Yogosawa S, Saito M, Marumo K, Yoshida K. Downregulation of CD24 suppresses bone metastasis of lung cancer. Cancer Sci. 2018;109(1):112–120. | ||
Zeng C, Chen T, Zhang Y, Chen Q. Hedgehog signaling pathway regulates ovarian cancer invasion and migration via adhesion molecule CD24. J Cancer. 2017;8(5):786–792. | ||
Thomas S, Harding MA, Smith SC, et al. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Canc Res. 2012;72(21):5600–5612. | ||
Onishi H, Suyama K, Yamasaki A. CD24 modulates chemosensitivity of MCF-7 breast cancer cells. Anticancer Res. 2017;37(2):561–565. | ||
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci. 2003;100(7):3983–3988. | ||
Bauerschmitz GJ, Ranki T, Kangasniemi L, et al. Tissue-specific promoters active in CD44+CD24−/low breast cancer cells. Canc Res. 2008;68(14):5533–5539. | ||
Hardt O, Wild S, Oerlecke I, et al. Highly sensitive profiling of CD44+/CD24− breast cancer stem cells by combining global mRNA amplification and next generation sequencing: evidence for a hyperactive PI3K pathway. Cancer Lett. 2012;325(2):165–174. | ||
Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci U S A. 2010;107(8):3722–3727. | ||
Zhu J, He J, Liu Y, Simeone DM, Lubman DM. Identification of glycoprotein markers for pancreatic cancer CD24+ CD44+ stem-like cells using nano-LC–MS/MS and tissue microarray. J Proteome Res. 2012;11(4):2272–2281. | ||
Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Canc Res. 2007;67(3):1030–1037. | ||
Wang M, Xiao J, Shen M, et al. Isolation and characterization of tumorigenic extrahepatic cholangiocarcinoma cells with stem cell-like properties. Int J Canc. 2011;128(1):72–81. | ||
Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+ CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137(11):1679–1686. | ||
Zhang J, Chen X, Bian L, Wang Y, Liu H. CD44+/CD24+-expressing cervical cancer cells and radioresistant cervical cancer cells exhibit cancer stem cell characteristics. Gynecol Obstet Invest. 2018:1–9. | ||
Liu H, Wang YJ, Bian L, Fang ZH, Qy Z, Cheng JX. CD44+/CD24+ cervical cancer cells resist radiotherapy and exhibit properties of cancer stem cells. Eur Rev Med Pharmacol Sci. 2016;20(9):1745–1754. | ||
Burgos-Ojeda D, Wu R, McLean K, et al. CD24+ ovarian cancer cells are enriched for cancer-initiating cells and dependent on JAK2 signaling for growth and metastasis. Mol Cancer Ther. 2015;14(7):1717–1727. | ||
Gao MQ, Choi YP, Kang S, Youn JH, Cho N-H. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29(18):2672–2680. | ||
Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676. | ||
Liu M, Wang X, Peng Y, Shen S, Li G. Egr-1 regulates the transcription of NGX6 gene through a Sp1/Egr-1 overlapping site in the promoter. BMC Mol Biol. 2014;15(1):14. | ||
Cheng JC, Chang HM, Leung PCK. Egr-1 mediates epidermal growth factor-induced downregulation of E-cadherin expression via SLUG in human ovarian cancer cells. Oncogene. 2013;32(8):1041–1049. |
Supplementary material
  | Table S1 Primers for qPCR, ChIP and clone |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.