Back to Journals » OncoTargets and Therapy » Volume 13
Caspase Recruitment Domain-Containing Protein 9 Expression is a Novel Prognostic Factor for Lung Adenocarcinoma
Authors Miwa N, Nagano T , Jimbo N, Dokuni R , Kiriu T , Mimura C, Yasuda Y, Katsurada M , Yamamoto M , Tachihara M , Tanaka Y, Kobayashi K, Itoh T, Maniwa Y, Nishimura Y
Received 1 June 2020
Accepted for publication 28 August 2020
Published 10 September 2020 Volume 2020:13 Pages 9005—9013
DOI https://doi.org/10.2147/OTT.S265539
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Nanako Miwa,1 Tatsuya Nagano,1 Naoe Jimbo,2 Ryota Dokuni,1 Tatsunori Kiriu,1 Chihiro Mimura,1 Yuichiro Yasuda,1 Masahiro Katsurada,1 Masatsugu Yamamoto,1 Motoko Tachihara,1 Yugo Tanaka,3 Kazuyuki Kobayashi,1 Tomoo Itoh,2 Yoshimasa Maniwa,3 Yoshihiro Nishimura1
1Division of Respiratory Medicine, Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan; 2Department of Diagnostic Pathology, Kobe University Graduate School of Medicine, Kobe, Japan; 3Division of Thoracic Surgery, Kobe University Graduate School of Medicine, Kobe, Japan
Correspondence: Tatsuya Nagano Tel +81 78 382 5660
Fax +81 78 382 5661
Email [email protected]
Purpose: Caspase recruitment domain-containing protein 9 (CARD9) is expressed at high levels in bone marrow cells and has a crucial role in innate immunity. Current studies indicate that CARD9 also plays a key role in tumor progression, but there are few reports on the role of CARD9 in lung cancer. The aim of this study was to clarify the role of CARD9 in lung adenocarcinoma.
Patients and Methods: Lung adenocarcinoma tumor samples from 74 patients who underwent complete resection at Kobe University Hospital from January 2014 to December 2014 were analyzed by immunohistochemistry. The role of CARD9 in cancer cells was analyzed using lung cancer cell lines treated with CARD9 siRNA.
Results: High expression of CARD9 was observed in 32.4% of tumors, and compared to low expression of CARD9, high expression was associated with poorer overall survival (P = 0.0365). Univariate and multivariate analyses showed that high expression of CARD9 was an independent prognostic factor. Knockdown of CARD9 in lung adenocarcinoma cells inhibited proliferation but did not increase apoptosis. In addition, CARD9 activated the NF-κB pathway in a lung adenocarcinoma cell line.
Conclusion: CARD9 was shown to be an independent prognostic factor of poor outcome for lung cancer and may represent a molecular target for treatment.
Keywords: CARD9, lung adenocarcinoma, spiral array, immunohistochemistry
Introduction
To date, certain clinical factors and tumor stages have been used as prognostic tools for non-small-cell lung cancer (NSCLC). However, prognosis can vary widely even among patients with tumors of the same stage. Most reports show that 20% of patients with stage I NSCLC experience recurrence after surgery,1 and the efficacy of adjuvant treatment for stage I patients is controversial.2 Some patients with stage I NSCLC receive adjuvant chemotherapy, which is similar to what patients at stage II or higher receive.3 Therefore, it is important to identify the prognostic markers that predict patient prognosis in NSCLC.
Caspase recruitment domain-containing protein 9 (CARD9) is a central regulator of innate immunity.4 Myeloid cells, including macrophages and dendritic cells, express high levels of CARD9.4 Pathogens can activate CARD9 signaling, which regulates innate immune responses and affects adaptive immunity.5,6 Current research reports that CARD9 plays a key role in tumor progression,7 although there have been few reports on the role of CARD9 in lung cancer and even fewer on its potential as a prognostic biomarker for lung cancer.
Therefore, the aim of this study was to clarify the prevalence and clinical significance of CARD9 expression in NSCLC. In this study, we will introduce a spiral array containing 74 NSCLC postoperative specimens, which was used in our analysis.
Patients and Methods
Patients and Spiral Array Construction
A lung cancer spiral array was constructed with surgical specimens from 74 consecutive patients who underwent surgery for lung cancer and were diagnosed with adenocarcinoma at Kobe University Hospital from January 2014 to December 2014. The protocol for constructing the spiral array was described previously.8 The methods of data collection and analysis were approved by the institutional review board (permission number: 160,117), and written informed consent was obtained from all the patients.
Immunohistochemistry
Tissue sections from spiral array blocks were deparaffinized and rehydrated using standard procedures.9 Briefly, slides were deparaffinized and treated to a heat-induced antigen retrieval procedure by incubating them for 20 min in pH 7.8 Tris-EDTA-citrate buffer in an autoclave at 121°C. A primary antibody against CARD9 (1:200) (polyclonal; Sigma-Aldrich, MO) was applied at 37°C for 60 min. We classified the stained specimens into two groups, low immunostaining and high immunostaining, for statistical analysis (Figure 1). CARD9 high was defined as a case in which there were 50% more positive cells than there were in normal tissue.
 |
Figure 1 Immunohistochemical staining of CARD9 protein in lung adenocarcinoma. |
Cell Culture and siRNA Interference
Lung adenocarcinoma cell lines A549 and PC9, human large cell lung cancer cell line H460, human lung squamous cell carcinoma cell line H520, and human small cell lung cancer cell lines DMS53 and DMS114 were purchased from ATCC (Manassas, VA, USA). The lung adenocarcinoma cell line II18 was purchased from RIKEN Cell Bank (Riken, Tsukuba, Japan). These cells were cultured at 37°C with 5% CO2 in RPMI-1640 medium (Wako, Japan) containing 10% fetal calf serum. The tumor cells were transfected with a CARD9 siRNA (Thermo Fisher Scientific, MA) or with Silencer® Select Negative Control No. 1 siRNA (Thermo Fisher Scientific, MA), which had no effect on any gene expression.
Real-Time PCR Analysis
RNA was isolated from various lung cancer cell lines (A549, PC9, II18, H460, H520, DMS53 and DMS114) according to the protocol of the manufacturer of Sepasol®-RNAI Super G (NACALAI TESQUE, INC. Japan). Extracted and purified RNA was reverse transcribed to generate cDNA using a PrimeScript RT Reagent Kit (Takara, Japan). Real-time PCR was performed using Thermal CyclerDice® Real Time System II (Takara, Japan). Relative mRNA levels were calculated with the ∆∆Ct method using GAPDH mRNA as an internal control. The primers used in this study were as follows: 5ʹ-CCCTCACGCATCACACCTTAC-3ʹ and 5ʹ-GCACACCCACTTTCCGTTTG-3ʹ for CARD9 and 5ʹ-CCACTCCTCCACCTTTGAC-3ʹ and 5ʹ-ACCCTGTTGCTGTAGCCA-3ʹ for GAPDH.
Cell Proliferation Assay
A549, PC9 and II18 cells in the logarithmic growth phase were seeded at a density of 5 × 103 cells/well in a 96-well plate and were precultured for 24 h in a 5% CO2 incubator. The cells were transfected with a CARD9 siRNA to knockdown CARD9 and then were cultured for 0, 24 and 48 h at 37°C with 5% CO2. A total of 10 µL of Cell Counting Kit-8 solution (CCK-8; Dojindo, Kumamoto, Japan) was added to each well, and the cells were incubated for another 30 min. Absorbance was measured at 450 nm using a spectrophotometer (Bio-Rad, iMark microplate reader, CA, USA).
Flow Cytometry
PC9 cells were transfected with a CARD9 siRNA or Silencer® Select Negative Control siRNA and cultured at 37°C with 5% CO2 for 48 h. Detection of apoptosis was performed by using Annexin V-FITC and propidium iodide (PI) and a BD FACSVerse System (BD Bioscience, Franklin Lakes, NJ).
Western Blot Analysis
A detailed protocol for Western blotting has been described previously.10 Antibodies against the following proteins were used in this study: β-actin (#4967), NF-κB p65 (#8242) and phospho-NF-κB p65 (#3033). These antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA).
Statistical Analysis
All statistical analyses were performed by using EZR version 1.37 (Saitama Medical Center Jichi Medical University; http://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmed.html; Kanda, 2018) and a graphical user interface for R version 3.4.1 (The R Foundation for Statistical Computing, Vienna, Austria).9,11 Differences between the two groups were analyzed by Pearson χ2 tests or Fisher exact tests. For the univariate analysis, all cumulative survival was estimated by the Kaplan–Meier method, and differences were assessed by Log rank test. A multivariate regression analysis was conducted according to the Cox proportional hazard model. All P values reported are 2-sided, and a P < 0.05 was considered significant.
Results
Patient Characteristics
The median age of patients was 71.5 years old (range 49–85 years old). Forty-one of the included patients were men, and 33 of the patients were women. Regarding the pathological data, 26 tumors were T1, 36 were T2 and 1 was T3. Although 59 patients were negative for lymph node metastasis, 8 patients had tumors that were categorized as N1 and 7 were N2 (Table 1).
 |
Table 1 Association Between CARD9 Expression and Clinicopathological Variables in Lung Adenocarcinoma |
Relationship Between CARD9 Expression and Clinicopathologic Factors
CARD9 was expressed in the cytoplasm of adenocarcinoma cells, as detected by immunohistochemistry. Figure 1 shows a typical CARD9 immunostaining result. In adenocarcinoma, high expression of CARD9 was seen in 32.4% of tumors, and low expression was seen in 67.6% of analyzable tumor samples. Compared to samples with low expression of CARD9, those with high levels correlated with the median survival (P = 0.0365; Figure 2). The results of univariate and multivariate analyses are shown in Tables 2 and 3, respectively. High expression of CARD9 was revealed to be an independent prognostic factor of poor outcome. Due to the relatively small number of cases in this study, lymph node metastasis was not found to be significantly different between the groups following univariate analysis.
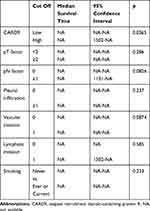 |
Table 2 Univariate Analysis of CARD9 Expression and Clinical Features of Lung Adenocarcinoma Cases |
 |
Table 3 Multivariate Analysis of CARD9 Expression and Clinical Features of Lung Adenocarcinoma Cases |
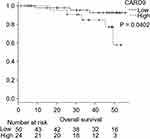 |
Figure 2 Kaplan–Meier curves of the overall survival of lung adenocarcinoma patients. |
Effect of CARD9 on Lung Cancer Cells
To confirm the effect of CARD9 on lung cancer cell lines, CARD9 mRNA levels in various lung cancer cell lines (A549, PC9, II18, H460, H520, DMS53 and DMS114) were analyzed by real-time PCR (Figure 3). The results suggested that CARD9 has a possible role in various types of lung cancer cell lines.
 |
Figure 3 mRNA expression levels of CARD9 in various lung cancer cells. |
Proliferation of Lung Adenocarcinoma Cells
A549, PC9 and II18 cells were transfected with CARD9 siRNA, and mRNA expression was analyzed 24 hours later. In all these cell lines, the mRNA expression of CARD9 was significantly reduced compared to that in the control cells (Figure 4A). Then, the proliferation of these cell lines was analyzed by the CCK8 method. Growth inhibition was observed in CARD9 knockdown PC9 cells. No clear growth inhibition was observed in A549 and II18 cells. (Figure 4B). This result suggested that CARD9 may promote cell proliferation in PC9 cells.
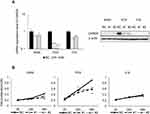 |
Figure 4 Knockdown efficiency of CARD9 siRNA and tumor growth inhibition test. |
Effect of CARD9 on Apoptosis in Lung Adenocarcinoma Cells
PC9 cells were transfected with CARD9 siRNA, and apoptosis was analyzed by flow cytometry. The proportion of apoptotic cells was almost the same between the CARD9 knockdown PC9 cells and control cells (Figure 5A). This suggests that CARD9 does not have a role in suppressing apoptosis in lung adenocarcinoma.
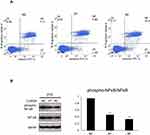 |
Figure 5 Apoptosis assay. |
Effect of CARD9 on the NF-κB Pathway in Lung Adenocarcinoma Cells
Western blot assays were performed to determine the activity of the NF-κB pathway. In PC9 cells transfected with CARD9 siRNA, p-NF-κB levels were lower relative to total NF-κB than they were in cells treated with the negative control siRNA (Figure 5B). This demonstrated that CARD9 promotes activation off the NF-κB pathway in a lung adenocarcinoma cell line.
Discussion
In this paper, we show for the first time that CARD9 is expressed in lung cancer cells and is a prognostic indicator of poor outcome.
To date, there have been many reports of prognostic biomarkers in NSCLC, but few have been used in clinical practice. It is a well-known fact that DNA microarray methods have the advantage of enabling assessment of a large amount of gene expression,12,13 but the disadvantage is that information about proteins cannot be obtained. We applied spiral array immunostaining as a simpler and faster method of predicting prognosis.
The human CARD9 locus has long been known to be an inflammatory bowel disease susceptibility factor, and the basic mechanism of CARD9 signaling activity has been elucidated.7,14 In addition, in recent years, CARD9 has been implicated in tumorigenesis, as CARD9 promotes antitumor T cell responses through various mechanisms, thereby activating proinflammatory carcinogenic STAT3 or antitumor suppressor functions in malignant cells.15 It has also been found to play a facilitating role in tumorigenesis.16,17
According to immunohistochemical staining results in a previous study, compared to that in normal tissues, CARD9 expression is increased mainly in tumor-infiltrating leukocytes in colon cancer tissues but not in cancer cells.18 Furthermore, CARD9 immunoreactivity was histologically observed in macrophages, and CARD9 was reported in patients with colon cancer. CARD9 expression is associated with tumor progression as well as liver metastasis of colon carcinoma cells.18
Regarding the distribution of CARD9 in cells, a previous study revealed that CARD9 is expressed in the cytoplasm of oral squamous cell carcinoma cells (OSCC) and OSCC cell lines.19 This previous study also showed that, with respect to the role of CARD9, downregulation of CARD9 is associated with suppression of OSCC growth and migration and that this is regulated through the NF-κB pathway. The arguments in this previous study seem to support our conclusions.
However, another study found that CARD9 suppressed MDSC expansion and IDO production in mice and prevented lung cancer by suppressing the noncanonical NF-κB pathway in CARD9 knockdown mice and inflammatory cells, and the mechanism was reported. There are two known signaling pathways that lead to inactivation of NF-κB: canonical and noncanonical.20–23 A common regulatory mechanism in these two pathways is the activation of the IκB kinase (IKK) complex, which phosphorylates IκB. The signals that stimulate the classical pathway are the binding of ligands (such as LPS) to the Toll-like receptor (TLR) superfamily and the binding of a ligand (TNF-α) to the TNF receptor.24 These proteins recruit adapter proteins, such as TRAFs, to the intracellular domain of the receptor. This adapter also recruits the IKK complex. In the classical pathway the IKK complex is composed of a homodimer or heterodimer consisting of IKKα and IKKβ and a scaffold protein NEMO (NF-κB essential modulator).25
IκB that is bound to the NF-κB dimer is degraded by the proteasome system upon phosphorylation by the IKK complex. After the degradation of IκB, the activated NF-κB dimer exposes its nuclear localization signal sequence (NLS), which allows its translocation into the nucleus.26 After it translocates into the nucleus NF-κB, induces the expression of various target genes.
Nonclassical pathways function in the development of lymphoid organs to produce B and T cells. Known factors that stimulate these pathways are lymphotoxin β and BAFF (B cell activating factor).27 When a ligand binds to a cell surface receptor, NIK (NF-κB inducing kinase) is activated and phosphorylates the IKK complex. The IKK complex in this pathway is a homodimer of IKKα, which does not include NEMO. The phosphorylated and activated IKK complex phosphorylates p100 (IκB domain) of the p100/RelB complex, which triggers the p100/RelB complex to undergo limited degradation to generate the active p52/RelB complex. This complex translocates to the nucleus and induces the expression of various target genes.25
In the current study, we demonstrated that CARD9 enhances the canonical NF-κB pathway to promote cell proliferation, as observed in the lung cancer cell line PC9. Therefore, inconsistencies with previous reports may be because the mechanism of NF-κB activation in the noncanonical pathway seen in mice is defined by the balance with the mechanism of NF-κB activation in the canonical pathway that is seen in human cell lines.
The limitation of the present study is the relatively small sample size. In addition, relatively few deaths occurred. However, we were convinced that the clinical data were supported by in vitro data.
Conclusion
CARD9 was shown to be an independent prognostic factor for poor outcome in lung adenocarcinoma. CARD9 may be a promising molecular target for lung cancer, and adjuvant therapy may be considered for CARD9-positive lung cancer.
Acknowledgments
The authors thank the members of the Division of Respiratory Medicine, Department of Internal Medicine, Kobe University Graduate School of Medicine for their helpful discussions.
Disclosure
This work was supported by grant 17K09614 from JSPS KAKENHI to Tatsuya Nagano. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors report no other potential conflicts of interest for this work.
References
1. Lou F, Huang J, Sima CS, Dycoco J, Rusch V, Bach PB. Patterns of recurrence and second primary lung cancer in early-stage lung cancer survivors followed with routine computed tomography surveillance. J Thorac Cardiovasc Surg. 2013;145(1):75–81. doi:10.1016/j.jtcvs.2012.09.030
2. Group NM-aC. Preoperative chemotherapy for non-small-cell lung cancer: a systematic review and meta-analysis of individual participant data. Lancet. 2014;383(9928):1561–1571. doi:10.1016/S0140-6736(13)62159-5
3. Arriagada R, Auperin A, Burdett S, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet. 2010;375:1267–1277.
4. Ruland J. CARD9 signaling in the innate immune response. Ann N Y Acad Sci. 2008;1143(1):35–44. doi:10.1196/annals.1443.024
5. Hsu YM, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8(2):198–205. doi:10.1038/ni1426
6. Hara H, Ishihara C, Takeuchi A, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8(6):619–629. doi:10.1038/ni1466
7. Zhong X, Chen B, Yang L, Yang Z. Molecular and physiological roles of the adaptor protein CARD9 in immunity. Cell Death Dis. 2018;9(2):52. doi:10.1038/s41419-017-0084-6
8. Nakamura S, Hayashi K, Imaoka Y, et al. Intratumoral heterogeneity of programmed cell death ligand-1 expression is common in lung cancer. PLoS One. 2017;12(10):e0186192. doi:10.1371/journal.pone.0186192
9. Ito T, Ishii G, Nagai K, et al. Low podoplanin expression of tumor cells predicts poor prognosis in pathological stage IB squamous cell carcinoma of the lung, tissue microarray analysis of 136 patients using 24 antibodies. Lung Cancer. 2009;63(3):418–424. doi:10.1016/j.lungcan.2008.06.008
10. Hatakeyama Y, Kobayashi K, Nagano T, et al. Synergistic effects of pemetrexed and amrubicin in non-small cell lung cancer cell lines: potential for combination therapy. Cancer Lett. 2014;343(1):74–79. doi:10.1016/j.canlet.2013.09.019
11. Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. doi:10.1038/bmt.2012.244
12. Ling B, Liao X, Huang Y, et al. Identification of prognostic markers of lung cancer through bioinformatics analysis and in vitro experiments. Int J Oncol. 2020;56:193–205.
13. Huang R, Gao L. Identification of potential diagnostic and prognostic biomarkers in non-small cell lung cancer based on microarray data. Oncol Lett. 2018;15:6436–6442.
14. Luo P, Yang Z, Chen B, Zhong X. The multifaceted role of CARD9 in inflammatory bowel disease. J Cell Mol Med. 2020;24(1):34–39. doi:10.1111/jcmm.14770
15. Zhong X, Chen B, Liu M, Yang Z. The Role of Adaptor Protein CARD9 in Colitis-Associated Cancer. Mol Ther Oncolytics. 2019;15:1–6. doi:10.1016/j.omto.2019.08.007
16. Zhong X, Chen B, Yang L, Yang Z. Card9 as a critical regulator of tumor development. Cancer Lett. 2019;451:150–155. doi:10.1016/j.canlet.2019.03.001
17. Hartjes L, Ruland J. CARD9 Signaling in Intestinal Immune Homeostasis and Oncogenesis. Front Immunol. 2019;10:419. doi:10.3389/fimmu.2019.00419
18. Yang M, Shao JH, Miao YJ, et al. Tumor cell-activated CARD9 signaling contributes to metastasis-associated macrophage polarization. Cell Death Differ. 2014;21(8):1290–1302. doi:10.1038/cdd.2014.45
19. Ye LJ, Zhou XC, Yin XJ, et al. CARD9 downregulation suppresses the growth of oral squamous cell carcinoma by regulating NF-κB. Oral Dis. 2019;25(8):1886–1896. doi:10.1111/odi.13157
20. Shih VF, Tsui R, Caldwell A, Hoffmann A. A single NFκB system for both canonical and non-canonical signaling. Cell Res. 2011;21:86–102. doi:10.1038/cr.2010.161
21. Tergaonkar V. NFkappaB pathway: a good signaling paradigm and therapeutic target. Int J Biochem Cell Biol. 2006;38(10):1647–1653. doi:10.1016/j.biocel.2006.03.023
22. Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25(51):6685–6705. doi:10.1038/sj.onc.1209934
23. Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18(49):6867–6874. doi:10.1038/sj.onc.1203219
24. Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi:10.1101/gad.183434.111
25. Fusella F, Seclì L, Cannata C, Brancaccio M. The One Thousand and One Chaperones of the NF-κB Pathway. Cell Mol Life Sci. 2002;77(12):2275–2288. doi:10.1007/s00018-019-03402-z
26. Manavalan B, Basith S, Choi YM, Lee G, Choi S. Structure-function Relationship of Cytoplasmic and Nuclear IκB Proteins: an in Silico Analysis. PLoS One. 2010;5(12):e15782. doi:10.1371/journal.pone.0015782
27. Sun SC. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21(1):71–85. doi:10.1038/cr.2010.177
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
