Back to Journals » Drug Design, Development and Therapy » Volume 10
Bulgecin A as a β-lactam enhancer for carbapenem-resistant Pseudomonas aeruginosa and carbapenem-resistant Acinetobacter baumannii clinical isolates containing various resistance mechanisms
Authors Skalweit M, Li M
Received 9 April 2016
Accepted for publication 30 June 2016
Published 20 September 2016 Volume 2016:10 Pages 3013—3020
DOI https://doi.org/10.2147/DDDT.S110193
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Wei Duan
Marion J Skalweit,1–5 Mei Li2
1Department of Medicine, 2Research Section, 3Infectious Diseases Section, Louis Stokes Cleveland Department of Veterans, 4Department of Medicine, 5Department of Biochemistry, Case Western Reserve University School of Medicine, Cleveland, OH, USA
Abstract: Genetic screening of Pseudomonas aeruginosa (PSDA) and Acinetobacter baumannii (ACB) reveals genes that confer increased susceptibility to β-lactams when disrupted, suggesting novel drug targets. One such target is lytic transglycosylase. Bulgecin A (BlgA) is a natural product of Pseudomonas mesoacidophila and a lytic transglycosolase inhibitor that works synergistically with β-lactams targeting PBP3 for Enterobacteriaceae. BlgA also weakly inhibits di-Zn2+ metallo-β-lactamases like L1 of Stenotrophomonas maltophilia. We hypothesized that because of its unique mechanism of action, BlgA could restore susceptibility to carbapenems in carbapenem-resistant PSDA (CR-PSDA) and carbapenem-resistant ACB, as well as ACB resistant to sulbactam. A BlgA-containing extract was prepared using a previously published protocol. CR-PSDA clinical isolates demonstrating a variety of carbapenem resistance mechanisms (VIM-2 carbapenemases, efflux mechanisms, and AmpC producer expression) were characterized with agar dilution minimum inhibitory concentration (MIC) testing and polymerase chain reaction. Growth curves using these strains were prepared using meropenem, BlgA extract, and meropenem plus BlgA extract. A concentrated Blg A extract combined with low concentrations of meropenem, was able to inhibit the growth of clinical strains of CR-PSDA for strains that had meropenem MICs ≥8 mg/L by agar dilution, and a clinical strain of an OXA-24 producing ACB that had a meropenem MIC >32 mg/L and intermediate ampicillin/sulbactam susceptibility. Similar experiments were conducted on a TEM-1 producing ACB strain resistant to sulbactam. BlgA with ampicillin/sulbactam inhibited the growth of this organism. As in Enterobacteriaceae, BlgA appears to restore the efficacy of meropenem in suppressing the growth of CR-PSDA and carbapenem-resistant ACB strains with a variety of common carbapenem resistance mechanisms. BlgA extract also inhibits VIM-2 β-lactamase in vitro. BlgA may prove to be an exciting adjunctive compound to extend the life of carbapenems against these vexing pathogens.
Keywords: carbapenem resistance, metallo-β-lactamase, efflux pumps
Introduction
The Infectious Diseases Society of America has instituted a campaign for antimicrobial development called “10x’20” with the hope that ten new antibiotics will be developed by 2020.1 The number of multidrug-resistant pathogens has increased both in hospital and outpatient settings including long-term care facilities, as well as in the community. Many experts in the area of infectious diseases are pessimistic about the ability to meet emerging resistance challenges with new agents.1–5 Patients who are elderly, immunocompromised, who have diabetes, pulmonary, and vascular disease are especially vulnerable to infections with drug-resistant gram-negative bacteria.6–9 Resistant pathogens are even being found in the community.10–12 New approaches, such as developing antibiotic enhancers that can restore the activity of antibiotics to which resistance exists, may allow for continued use of antimicrobials already available in the clinical arena. Bulgecin A (BlgA), a noncovalent inhibitor of bacterial lytic transglycosolase (Ltg) cell wall enzymes, may be a candidate antibiotic enhancer.
BlgA is a “β-lactam enhancer”
The gram-negative cell wall is a complex structure which makes development of new antimicrobials challenging. Penicillin binding proteins (PBPs) and Ltgs are located in the inner membrane, the periplasmic space, and on the inner leaflet of the outer membrane,13 respectively. In addition to these cell wall proteins, β-lactamases such as AmpC cephalosporinases, metallo- β-lactamases (MBLs), and other carbapenemases are found in the periplasmic space. Chromosomal AmpC β-lactamases are typically produced at low levels in gram-negative bacteria unless there is production of 1,6-anhydro-muropeptides from altered peptidoglycan production by PBPs in the presence of β-lactam antibiotics and constitutive expression of AmpC β-lactamase or “de-repression”.13 BlgA is a novel compound that can enhance the function of β-lactams potentially by several unique mechanisms: 1) inhibition of Ltgs; 2) preventing de-repression of AmpC enzymes; and 3) inhibition of MBLs. Genetic screening of Acinetobacter baumannii (ACB) and Pseudomonas aeruginosa (PSDA) reveals a variety of genes that confer increased susceptibility to β-lactams when these genes are disrupted.14,15 One such gene product identified is the Ltg family of enzymes found in many gram-negative organisms.14–18 Ltgs catalyze the lytic cell wall reaction MurNAc–GlucNAc 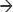 MurNAc + GlucNAc. Synthesis and lysis of peptidoglycan are strictly coordinated during cell division, and any imbalance can lead to cell lysis. In a clinical ACB complex strain (later identified as Acinetobacter nosocomialis), the Ltg was also associated with cellular motility and biofilm formation.19,20
MurNAc + GlucNAc. Synthesis and lysis of peptidoglycan are strictly coordinated during cell division, and any imbalance can lead to cell lysis. In a clinical ACB complex strain (later identified as Acinetobacter nosocomialis), the Ltg was also associated with cellular motility and biofilm formation.19,20
BlgA (Figure 1) is an inhibitor of the soluble Escherichia coli Ltg, Slt70,21 and has been found to dramatically lower the minimum inhibitory concentrations (MICs) of ampicillin and cefmenoxime when combined with these agents and tested against E. coli and Helicobacter pylori strains.16,22 BlgA is a natural product derived from Pseudomonas mesoacidophila, a nonpathogenic strain that also produces monobactam antibiotics.22–24 The name derives from the bulge-like morphologic changes observed in Enterobacteriaceae when such organisms are grown in the presence of β-lactams and BlgA.22,25 Structurally, it contains a GlcNAC moiety like the substrate of Ltgs, and thus acts as a transition state inhibitor of Ltgs.26 BlgA appears to exert its effect when combined with β-lactams that target PBP3 in Enterobacteriaceae as a noncompetitive inhibitor of Ltgs.17,21 X-ray structures of Slt70 of E. coli demonstrate that the hydroxymethyl side chain of the pyrrolidine ring hydrogen bonds to the catalytic Glu478 in the manner of a transition state analog of the GlcNAC–MurNAC substrate.13,21
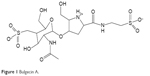  | Figure 1 Bulgecin A. |
In addition to its inhibitory effects on Ltgs, BlgA has also been found to act as an inhibitor of di-Zn2+ MBLs.27 Zn2+ MBLs are Class B β-lactamases that have carbapenemase activity, as well as activity against penicillins and cephalosporins, found in Pseudomonads. In the US, rare instances of VIM-2–producing PSDA have been noted; worldwide, imipenemase (IMP), Verona imipenemase (VIM), and even New Delhi metallo-beta-lactamase (NDM) types are common in PSDA isolates.28,29 Among the Class B MBLs, most require two Zn2+ ions for activity. BlgA had micromolar inhibitory activity against the di-Zn2+ forms of MBLs, Bce II of Bacillus cereus and L1 of Stenotrophomonas maltophilia. A model of BlgA docked into the active site of the L1 MBL shows an interaction between the second (ZNII) Zn2+ site and the sulfonate group of the sugar moiety of BlgA. This occurs when BlgA is 50%–100% deprotonated, as it should be at physiologic pH. This docking mechanism is similar to what is seen with MBL and angiotensin converting enzyme inhibitors such as D- and L-captopril. In addition, there is an interaction between the Asp14 and the proline moiety of BlgA, regardless of the protonation state of the inhibitor.27 Further studies of MBL inhibition by BlgA were halted due to a lack of availability of this drug.
Our pilot studies described in this paper suggest that BlgA can be a broad-spectrum adjunctive agent that is able to augment the activity of β-lactams to inhibit the growth of resistant gram-negative bacteria including ACB and PSDA. By enhancing the activity for a given β-lactam or β-lactam–β-lactamase inhibitor combination, BlgA may be able to overcome resistance mediated by several mechanisms including efflux, lower-affinity PBPs, and production of β-lactamases. It may also impact biofilm formation in ACB infections. Through this early report, we hope to inspire and stimulate further development of BlgA and analogs that can be used as “antibiotic assistants” to treat infections with ACB and PSDA isolates with different resistance genotypes/phenotypes. By doing so, we can define the antimicrobial spectrum of BlgA and determine the most effective partner β-lactam. In addition, we can determine whether BlgA prevents the de-repression of AmpC β-lactamases, or if BlgA provides inhibitory activity against MBLs in organisms that express these resistance determinants.
Materials and methods
Strains used in this study
Clinical strains of PSDA (a VIM-2 producing strain R9630 and Cleveland Clinic CL231 PDC-5 hyperproducing strain31) were a kind gift of R Bonomo; an efflux strain 860 was collected at the Cleveland VA hospital. ACB strains UH83 (OXA-24/40+) and UH1026,32 were obtained as a kind gift from R Bonomo. E. coli MC1061 (Thermo Fisher Scientific, Waltham, MA, USA) was used as a positive control to assess BlgA activity with aztreonam.33
MIC determination
Agar dilution MIC determination for the clinical strains used in these studies was performed in triplicate according to the Clinical and Laboratory Standards Institute guidelines34 using a 0.5 McFarland inoculum plated onto Mueller–Hinton agar supplemented with antibiotics (meropenem [AstraZeneca plc, London, UK], imipenem [Merck & Co., Inc., Whitehouse Station, NJ, USA], aztreonam [Bristol-Myers Squibb, New York, NY, USA]). To assess for efflux, agar dilution MIC determination of carbapenem-resistant PSDA (CR-PSDA) clinical isolates (identified by the VA clinical microbiology laboratory Vitek II™ system [bioMérieux, Inc., Durham, NC, USA] as meropenem R and imipenem S) was also performed in triplicate using a 0.5 McFarland inoculum onto Mueller–Hinton agar supplemented with meropenem (1–32 mg/L) ± Phe-Argβ-naphthylamide (PAβN; Sigma-Aldrich Co., St Louis, MO, USA) 50 mg/L efflux inhibitor.35 Susceptibility results using Microscan™ (Beckman-Coulter Inc, Brea, CA, USA) reported by the University Hospitals of Cleveland clinical microbiology laboratory as sensitive, intermediate, or resistant (S, I, R) were utilized for ampicillin–sulbactam.
BlgA extract
BlgA extract was prepared by adapting the methods of Shinagawa et al.24 Briefly, P. mesoacidophila (ATCC® 31433) is grown in ATCC® Medium 3 (Manassas, VA, USA) in an overnight culture at 28°C. After pelleting, the supernatant broth pH is adjusted to ten using 1 M NaOH to hydrolyze sulfazecin and IVY proteins,36 and kept at 25°C for 2 hours. For our initial studies, we prepared a bulgecin extract by neutralizing the hydrolyzed supernatant to pH 7.0 with addition of 1 M HCl. The treated supernatant was concentrated 10× at room temperature and used in growth inhibition experiments.
Growth curves
Growth curves were constructed for control strain E. coli MC1016 according to the method of Heidrich et al33 as follows: 1 μL of a 1:10 dilution of an overnight culture was added to 95 μL of super optimal broth (SOB) medium (~105 colony forming units [cfu]/mL). An initial OD600 nm was obtained using an enzyme-linked immunosorbent assay plate reader, and then the sample was allowed to grow for 100 minutes at 37°C. After 100 minutes incubation, another OD600 nm reading was obtained and then either saline (null), BlgA extract alone (final v:v [BlgA] =10%), aztreonam (final concentration 0.01 mg/L), or 10% (v/v) bulgecin extract with 0.01 mg/L aztreonam was added to the well (in 100 μL total volume) and growth was further monitored at OD600 nm at various time points. Growth curves for the clinical bacterial strains were obtained in a similar manner using appropriate partner antibiotics for the particular resistance phenotype and adjusting the inoculum or antibiotic concentration to allow growth of the organism. For the multidrug resistant (MDR) ACB clinical strain, UH83,26 1 μL of a 1:10 dilution of an overnight culture was used with meropenem (0.02 mg/L) ± 10% (v/v) bulgecin extract in 100 μL total volume. For the sulbactam-resistant ACB clinical strain UH10,32 1 μL of a 1:10 dilution of an overnight culture was used with ampicillin–sulbactam (0.03/0.015 mg/L) ± 10% (v/v) bulgecin extract in 100 μL total volume. UH83 produces OXA-24 carbapenemase and is resistant to carbapenems (MIC ≥32 mg/L), cephalosporins, and intermediate to sulbactam.26 UH10 has been completely sequenced32 and produces TEM-1 β-lactamase as the basis of its sulbactam resistance. The PSDA strains included CL231, a PDC-5 AmpC hyperproducing strain isolated from the sputum of a patient at the Cleveland Clinic in the mid-2000s;31 a VIM-2 producing strain;30 and a PSDA urinary tract isolate, resistant to meropenem/susceptible to imipenem that effluxes meropenem but has no other identifiable carbapenem resistance mechanism (this study). For PSDA strains, growth assays were performed (mentioned earlier in this section), by adding 1 μL of an overnight culture (cx) of a 1:1, 1:2, or 1:10 dilution thereof to 95 μL volumes of SOB medium. After 100 minutes incubation, either saline (null), BlgA extract alone (final v:v [BlgA] =10%), meropenem (final 0.03 mg/L), or BlgA 10% + meropenem 0.03 mg/L was added to the well and growth was monitored at OD600 nm using an enzyme-linked immunosorbent assay plate reader at various time points.
Characterization of resistance determinants
For additional verification of the PSDA isolates, we performed polymerase chain reaction analysis to look for the presence of β-lactamases, for example, SHV, TEM, CTX-M, PER, VEB, GES, IMP, VIM, and OXA β-lactamases, as well as MEX efflux pumps using the methods and primers of Hujer et al.37 To screen for efflux phenotypes, we did agar dilution MICs of CR-PSDA clinical isolates using a 0.5 McFarland inoculum onto Mueller–Hinton agar supplemented with meropenem (1–32 mg/L) ± PAβN 50 mg/L efflux inhibitor.35
VIM-2 IC50 measurements
Purified VIM-2 β-lactamase was a kind gift from Dr J-D Docquier.38 Substrate hydrolysis rates with nitrocefin were measured using an Agilent Technologies (Santa Clara, CA, USA) 8453 ultraviolet–visible spectrophotometer at 482 nm and recorded using ultraviolet–visible ChemStation Online (Agilent Technologies). Reactions are performed in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–50 μM ZnCl2 at pH 7.4 with 8 nM concentrations of β-lactamase protein per total reaction volume of 500 μL. Using 200 μM nitrocefin as the reporter substrate, initial hydrolysis rates are measured in the presence of 0%–1.5% BlgA extract. Enzyme is preincubated with BlgA for 5 minutes before 50% inhibitory concentration (IC50) determination. To determine a crude IC50, expressed as a percent concentration of BlgA extract, fractional activity (v/vo) was calculated from the initial rate measurements for substrate hydrolysis and plotted as a function of inhibitor percent concentration: v/vo = 1/[1+([I]/IC50)], where v is the reaction velocity, vo the reaction velocity without inhibitor, [I] the inhibitor concentration, and IC50 is the concentration of inhibitor where v = vo/2.
Results and discussion
Minimum inhibitory concentrations
Agar dilution MICs for the strains included in the BlgA assays are shown in Table 1 for each organism used in this study, for comparison of the resistance phenotypes. Porin downregulation and AmpC hyperproduction were not assessed in this study, and may contribute additional resistance in these strains. The PSDA CL231 strain is known to hyperproduce the AmpC enzyme PDC-5.31
Polymerase chain reaction
Polymerase chain reaction assays confirmed the presence of resistance determinants as shown in Table 1. All the PSDA isolates tested positive for MEX A and MEX C, except for the VIM-2–producing strain R96, which also was not inhibited by PAβN.
Preparation of BlgA extracts and growth assays
We prepared crude extracts of P. mesoacidophila isolate ATCC® 31433 and the growth curves for the ACB and PSDA clinical strains and the E. coli control strain as shown in Figures 2 and 3.
BlgA extract showed good activity against the control strain E. coli MC1061 when combined with aztreonam (Figure 2A).33 In clinical ACB strain UH83,26 growth was suppressed by a bulgecin–meropenem combination as shown in Figure 2B. In clinical ACB strain UH10,32 growth was inhibited by the bulgecin/ampicillin/sulbactam combination (Figure 2C), however, not as well. We hypothesize that the TEM-1 production is still sufficient to hydrolyze the sulbactam, and that the Ltgs inhibited by BlgA may not interact as well with the PBPs that bind sulbactam in ACB, compared to those inhibited by meropenem.
BlgA extract inhibited the growth of clinical PSDA strains – AmpC hyperproducer PDC-5 CL231 (Figure 3A), meropenem effluxing strain 860 (Figure 3B and C), and VIM-2-producing strain R96 (Figure 3D)-when combined with meropenem. Of interest, when examining the growth curves of PSDA strains R96 (Figure 3D), which is meropenem resistant on the basis of VIM-2 production, and the VA clinical strain 860 (Figure 3B and C), which has an efflux mechanism of resistance for meropenem, the growth curves differ. For strain 860, we studied two different inocula of bacteria, with ~5×105 cfu/mL (Figure 3B) and with ~105 cfu/mL (Figure 3C). Differences in the curves show that there is an inoculum effect to the growth suppression, but there are also differences in the growth suppression depending on the mechanism of resistance. This suggests one of two possibilities within the limitations of the assay: either BlgA is acting by a second mechanism, that is, inhibition of the VIM-2 β-lactamase (in strain R96), or BlgA is effluxed to some extent (strain 860), or both mechanisms may be operating. Strain 860 may also have other mechanisms of resistance such as OprD loss which have not been characterized. This prompted us to examine whether BlgA extract could inhibit VIM-2 in vitro.
Inhibition of VIM-2 MBL
In order to demonstrate whether BlgA extract could inhibit VIM-2 MBL, we measured the IC50 of the extract expressed as a percent concentration for VIM-2 MBL of PSDA39 (Figure 4) using a kinetic assay (see the “Materials and methods” section). BlgA is hypothesized to bind in a reversible manner to di-Zn2+ MBLs27 by coordinating its GlcNAC sulfate group to the Zn2+ ions. The measured IC50 for our extract is 0.5%±0.1%. BlgA has an IC50 of 150 μM for the structurally similar L1 di-Zn2+ enzyme of S. maltophilia.27 A concentration of 10% BlgA extract was used in the growth inhibition experiments, or a 50-fold higher concentration in the medium, compared to the estimated IC50 % in the in vitro enzyme assay. So, it is reasonable to assume that some inhibition of the PSDA R96 strains is due to the activity of BlgA extract against VIM-2. Further studies with pure BlgA are needed to demonstrate this effect, as well as further characterization of strain 860, to determine whether efflux of BlgA may occur or if OprD loss plays a role in resistance to BlgA.
  | Figure 4 IC50 expressed as percent concentration of bulgecin A extract to inhibit VIM-2 β-lactamase hydrolysis activity. |
One of the limitations of the study is that thus far, we only have crude extract available for testing. We attempted to perform disk diffusion and broth MIC determinations using the extract, but the results showed only one dilution change in most instances in the MIC (data not shown). Because a single MIC determination can vary by one dilution either way, this was not deemed a significant change. Our current studies are limited by our ability to produce sufficiently pure BlgA to confirm the activity of this agent against highly resistant clinical pathogens. However, these data are highly suggestive that a bulgecin compound has β-lactam–enhancing capabilities in these bacteria, either through inhibition of Ltgs or MBLs or both.
Conclusion
BlgA extract is active against carbapenem-resistant ACB and CR-PSDA isolates including those that produce OXA-24/40 and VIM-2 β-lactamases, efflux meropenem, and have AmpC hyperproduction. Furthermore, our BlgA extract has an IC50 of 0.5% concentration versus VIM-2 β-lactamase. BlgA extract is also effective at suppressing the growth of ACB strains resistant to sulbactam. It is unknown whether it is a bactericidal or bacteriostatic agent due to the limitations of our current BlgA purity and the assay conditions. Further studies are needed with pure material before this can be determined. Because BlgA-containing extract appears to have a wide range of activity versus numerous MDR and extensively drug resistant (XDR) isolates, it may be an adjunctive therapy to extend the life of β-lactam antibiotics versus MDR ACB and PSDA.
Further work is needed to completely purify the compound and determine the best partner β-lactam with which to treat these resistant pathogens.
Acknowledgments
This research was supported by a Department of Veterans Affairs VISN 10 RIP Program grant, as well as funding from the Steris Foundation. Dr Skalweit is an employee of the Department of Veterans Affairs and Dr Li was an employee of the VA Research and Education Foundation. The authors thank Drs Robert A Bonomo and J-D Docquier for their generous contributions of material (strains, VIM-2 β-lactamase).
Disclosure
The authors report no conflicts of interest in this work.
References
Spellberg B, Guidos R, Gilbert D, et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46(2):155–164. | ||
Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! an update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. | ||
Laxminarayan R, Rex JH. Antibacterial R&D incentives. Nat Rev Drug Discov. 2011;10(10):727–728. | ||
David M Shlaes. Antibiotics: The Perfect Storm. Available from: https://bfosinfo.files.wordpress.com/2010/09/9048190568antibiotics.pdf. Accessed August 24, 2016. | ||
Spellberg B, Sharma P, Rex JH. The critical impact of time discounting on economic incentives to overcome the antibiotic market failure. Nat Rev Drug Discov. 2012;11(2):168. | ||
Decramer M, Janssens W, Miravitlles M. Chronic obstructive pulmonary disease. Lancet. 2012;379(9823):1341–1351. | ||
Everaert K, Lumen N, Kerckhaert W, Willaert P, van Driel M. Urinary tract infections in spinal cord injury: prevention and treatment guidelines. Acta Clin Belg. 2009;64(4):335–340. | ||
Hannaman MJ, Ertl MJ. Patients with immunodeficiency. Med Clin North Am. 2013;97(6):1139–1159. | ||
Peters EJ, Lipsky BA. Diagnosis and management of infection in the diabetic foot. Med Clin North Am. 2013;97(5):911–946. | ||
Carmeli Y, Akova M, Cornaglia G, et al. Controlling the spread of carbapenemase-producing Gram-negatives: therapeutic approach and infection control. Clin Microbiol Infect. 2010;16(2):102–111. | ||
El Salabi A, Walsh TR, Chouchani C. Extended spectrum beta-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. Crit Rev Microbiol. 2013;39(2): 113–122. | ||
Savard P, Perl TM. A call for action: managing the emergence of multidrug-resistant Enterobacteriaceae in the acute care settings. Curr Opin Infect Dis. 2012;(25):371–377. | ||
Johnson JW, Fisher JF, Mobashery S. Bacterial cell-wall recycling. Ann N Y Acad Sci. 2013;1277:54–75. | ||
Lee H, Vazquez-Laslop N, Klyachko KA, Neyfakh AA. Isolation of antibiotic hypersusceptibility mutants of Acinetobacter spp. by selection for DNA release. Antimicrob Agents Chemother. 2003;47(4):1267–1274. | ||
Potvin E, Lehoux DE, Kukavica-Ibrulj I, et al. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol. 2003;5(12):1294–1308. | ||
Bonis M, Williams A, Guadagnini S, Werts C, Boneca IG. The effect of bulgecin A on peptidoglycan metabolism and physiology of Helicobacter pylori. Microb Drug Resist. 2012;18(3):230–239. | ||
Templin MF, Edwards DH, Holtje JV. A murein hydrolase is the specific target of bulgecin in Escherichia coli. J Biol Chem. 1992;267(28):20039–20043. | ||
van Asselt EJ, Thunnissen AM, Dijkstra BW. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999;291(4):877–898. | ||
Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology. 2011;157(Pt 9):2534–2544. | ||
Carruthers MD, Harding CM, Baker BD, et al. Draft genome sequence of the clinical isolate Acinetobacter nosocomialis strain M2. Genome Announc. 2013;1(6):e00906–e00913. | ||
Thunnissen AM, Rozeboom HJ, Kalk KH, Dijkstra BW. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. implications for the enzymatic mechanism. Biochemistry. 1995;34(39):12729–12737. | ||
Imada A, Kintaka K, Nakao M, Shinagawa S. Bulgecin, a bacterial metabolite which in concert with beta-lactam antibiotics causes bulge formation. J Antibiot (Tokyo). 1982;35(10):1400–1403. | ||
Gwynn MN, Box SJ, Brown AG, Gilpin ML. MM 42842, a new member of the monobactam family produced by Pseudomonas cocovenenans. I. Identification of the producing organism. J Antibiot (Tokyo). 1988;41(1):1–6. | ||
Shinagawa S, Maki M, Kintaka K, Imada A, Asai M. Isolation and characterization of bulgecins, new bacterial metabolites with bulge-inducing activity. J Antibiot (Tokyo). 1985;38(1):17–23. | ||
Nakao M, Yukishige K, Kondo M, Imada A. Novel morphological changes in gram-negative bacteria caused by combination of bulgecin and cefmenoxime. Antimicrob Agents Chemother. 1986;30(3):414–417. | ||
Perez F, Endimiani A, Ray AJ, et al. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J Antimicrob Chemother. 2010;65(8):1807–1818. | ||
Simm AM, Loveridge EJ, Crosby J, Avison MB, Walsh TR, Bennett PM. Bulgecin A: a novel inhibitor of binuclear metallo-beta-lactamases. Biochem J. 2005;387(Pt 3):585–590. | ||
Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19(12):588–595. | ||
Perez F, Ray AJ, Dumford D, Jacobs M, Bonomo RA. C2-720 Metallo-b-lactamases in the Heartland of the United States: International Strain of VIM-2 producing Pseudomonas aeruginosa in a Community Hospital in Northeast Ohio. In: 52nd Interscience Conference on antimicrobial agents and chemotherapeutics; 2012; San Francisco; CA. | ||
Perez F, Hujer AM, Marshall SH, et al. Extensively drug-resistant pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in Northeast Ohio. Antimicrob Agents Chemother. 2014;58(10):5929–5935. | ||
Winkler ML, Papp-Wallace KM, Hujer AM, et al. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015;59(2):1020–1029. | ||
Wright MS, Haft DH, Harkins DM, et al. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio. 2014;5(1):e00963–e00913. | ||
Heidrich C, Templin MF, Ursinus A, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41(1):167–178. | ||
CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. 9th ed. CLSI document M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. | ||
Mesaros N, Glupczynski Y, Avrain L, Caceres NE, Tulkens PM, Van Bambeke F. A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa. J Antimicrob Chemother. 2007;59(3):378–386. | ||
Clarke CA, Scheurwater EM, Clarke AJ. The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J Biol Chem. 2010;285(20):14843–14847. | ||
Hujer KM, Hujer AM, Hulten EA, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006;50(12):4114–4123. | ||
Garcia-Saez I, Docquier JD, Rossolini GM, Dideberg O. The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J Mol Biol. 2008;375(3):604–611. | ||
Docquier JD, Lamotte-Brasseur J, Galleni M, Amicosante G, Frère JM, Rossolini GM. On functional and structural heterogeneity of VIM-type metallo-b-lactamases. J Antimicrob Chemother. 2003;51(2):257–266. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



