Back to Journals » Infection and Drug Resistance » Volume 16
Brachybacterium muris Detected in a Hepatocellular Carcinoma Patient with Pleural Effusion: A Case Report
Authors Wu Y, Wang S, Wu G, Zhang J, Liu S
Received 9 February 2023
Accepted for publication 9 May 2023
Published 15 May 2023 Volume 2023:16 Pages 3003—3006
DOI https://doi.org/10.2147/IDR.S406259
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
YuJing Wu,1,* Sumei Wang,1,* Guangye Wu,2 Jiandong Zhang,1 Shuye Liu1
1The Third Central Hospital of Tianjin, Tianjin Key Laboratory of Extracorporeal Life Support for Critical Diseases, Artificial Cell Engineering Technology Research Center, Tianjin Institute of Hepatobiliary Disease, Tianjin, People’s Republic of China; 2Second Teaching Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin Traditional Chinese Medicine Rehabilitation Center, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shuye Liu, Tel +86-022-84112332, Fax +86 022-24384350, Email [email protected]
Background: Brachybacterium muris is a species of Gram positive and strictly aerobic bacterium. It was first reported in 2003 after being isolated from the liver of a laboratory mouse strain. It was also found on human skin and nasal cavity. Herein, we present the first case pleural effusion infection in humans caused by Brachybacterium muris.
Case Presentation: A 65-year-old man was admitted to our hospital for a 4-week history of fever, accompanied by chills, occasional abdominal pain, occasional chest tightness and shortness of breath. On the day of hospitalization, thoracentesis was performed and 1000mL of yellow cloudy fluid was released. Result of pleural fluid culture was positive and B. muris was identified using 16S rDNA amplification and sequence comparisons.
Conclusion: To our knowledge, this is the first report of pleural effusion infection caused by B. muris. B. muris can be pathogenic in humans.
Keywords: Brachybacterium muris, pleural effusion, infection
Background
Genus Brachybacterium was first discovered and isolated in 1988.1 Up to now, there were 25 child taxa with a validly published and correct name (https://lpsn.dsmz.de/genus/brachybacterium). Genus Brachybacterium can be isolated from various environmental samples, such as seawater,2 plant roots and branches,3,4 river,5 surface of cheeses,6 coastal sand7 and soil.8 A few studies have also reported the isolation of this genus from human samples.9–11
Brachybacterium muris was first reported in 2003 after being isolated from the liver of a laboratory mouse strain.12 In 2017, Brachybacterium muris was also isolated from Chaka Salt Lake (Qinghai, China).13 Brachybacterium muris was also found in human samples, such as human nasal cavity14 and skin.15 However, to our knowledge, there are no reports of the species Brachybacterium muris infecting humans and causing disease. We herein report the first case of Brachybacterium muris isolated from pleural effusion.
Case Presentation
A man with a medical history of hepatitis B for more than ten years was diagnosed with hepatocellular carcinoma in June 2021. The patient accepted treatment with radiofrequency on July 1, 2021. But in March 2022, the hepatocellular carcinoma recurred and he underwent radiofrequency treatment again. In November 2022, this 65-year-old man presented to Tianjin Third Central Hospital (Tianjin), with a 4-week history of fever, accompanied by chills, occasional abdominal pain, chest tightness and shortness of breath. On admission, this patient presented chronic liver disease face, liver palm and loss of breath sounds in the right lung. The patient had decompensated cirrhosis of the liver with a Child-Pugh classification of B and a small amount of ascites.
On the day of hospitalization, thoracentesis was performed and 1000mL of yellow cloudy fluid was released. Laboratory tests of whole blood showed the following results: white blood cell count 2.17 × 109/L (neutrophil 77.1%), red blood cell count 3.06 × 1012/L, hemoglobin 90 g/L and platelet count 46 × 109/L. His serum hepatitis B surface antigen and hepatitis B core antibody were 13.41 IU/mL and 8.78 IU/mL, respectively. His procalcitonin level in serum was 0.33 ng/mL. The result of G test was 114.30pg/mL. The G test detects (1,3)-b-D-glucan, a component of the cell wall of fungi. Pleural effusion tests revealed a white blood cell count of 2.3 × 109/L with 20% neutrophils and 80% lymphocytes. The specific gravity of the pleural fluid was 1.015 and the Rivalta test result was positive. The above results suggest that the pleural fluid was exudative and the patient had a pleural fluid infection. Empirical anti-infective treatment with piperacillin/tazobactam was initiated after collection of pleural fluid for culture. A 4.5g of piperacillin/tazobactam was dissolved in 100mL of saline and administered intravenously every 8 hours, three times a day. The next day the patient underwent thoracentesis drainage and released 700 mL of pleural fluid. Two days after the patient received anti-infection and intermittent pleural fluid release treatment, the patient’s temperature returned to normal.
The infecting agent was isolated from pleural fluid by performing in vitro culture and then the infecting agent was purified in blood agar culture. Next, the organism was subjected to matrix-assisted laser desorption/ionization time-of-fight mass spectrometry (MALDI-TOF MS) analysis. We used a Vitek MS system. Unfortunately, our database did not include this microorganism. We outsourced microbial sequencing to Beijing RuiBio BioTech Co. The microorganism was identified using 16S rDNA amplification and sequence comparisons. Two primers were used. Their sequences were “AGAGTTTGATCATGGCTC” and “TAGGGTTACCTTGTTACGACTT”. Comparison with sequences in the BLAST database (http://www.ncbi.nlm.nih.gov/BLAST) revealed that the sequence has 99.131% similarity to the Brachybacterium muris sequence (GenBank accession number NR 024571.1). The sequence was attached in Supplementary 1. As shown in Figure 1, culture on blood agar plate grew white, moist, and smooth colonies after 24 h of incubation at 35 °C in a 5% CO2 atmosphere.
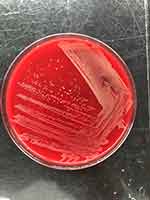 |
Figure 1 Culture on blood agar plate. |
Discussion and Conclusion
Brachybacterium muris is a species of Gram positive, strictly aerobic, yellow-pigmented bacterium. The cells are coccoid during the stationary phase, and irregular rods during the exponential phase. It was first isolated from the liver of a laboratory mouse. The species was first described in 2003, and the name is derived from the Latin muris (mouse). The optimum growth temperature for B. muris is 25–37 °C. It can grow in the 15–42 °C range and in pH 6.0–9.0.12 B. muris have been reported in the literature to be found in human skin/faeces.15,16 However, there are few reports of human diseases caused by this bacterium.
In this case, the patient had a history of hepatitis B-related liver cancer and was treated with radiofrequency. The infecting organism may have come from the environment or from the patient’s skin. It was not entirely clear that how he got the infection. Empirical treatment with piperacillin/tazobactam seemed to be effective, as he experienced significant relief of the infection two days after the administration of the drug. We identified the isolated strains by MALDI-TOF and 16S rRNA sequencing.
In summary, we present the first case of pleural effusion infection caused by B. muris. The organism was identified by 16S ribosomal RNA gene-sequencing analysis. Although human infection is rare, B. muris can be pathogenic in humans.
Consent for Publication
Written informed consent was provided by the patient for the publication of the case details and images. Institutional approval was obtained to publish the case details.
Funding
This work is supported by Tianjin Key Medical Discipline (Specialty) Construction Project (Grants No. TJYXZDXK-047A).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Collins MD, Brown J, Jones D. Brachybacterium faecium gen. nov., sp. nov., a coryneform bacterium from poultry deep litter. Int J Syst Bacteriol. 1988;38:45–48. doi:10.1099/00207713-38-1-45
2. Kaur G, Kumar N, Mual P, et al. Brachybacterium aquaticum sp. nov., a novel actinobacterium isolated from seawater. Int J Syst Evol Microbiol. 2016;66(11):4705–4710. doi:10.1099/ijsem.0.001414
3. Gontia I, Kavita K, Schmid M, et al. Brachybacterium saurashtrense sp. nov., a halotolerant root-associated bacterium with plant growth-promoting potential. Int J Syst Evol Microbiol. 2011;61(Pt 12):2799–2804. doi:10.1099/ijs.0.023176-0
4. Liu Y, Zhai L, Yao S, et al. Brachybacterium hainanense sp. nov., isolated from noni (Morinda citrifolia L.) branch. Int J Syst Evol Microbiol. 2015;65(11):4196–4201. doi:10.1099/ijsem.0.000559
5. Alam SA, Saha P. Biodegradation of p-nitrophenol by a member of the genus Brachybacterium, isolated from the river Ganges. Biotech. 2022;12(9):213. doi:10.1007/s13205-022-03263-7
6. Schubert K, Ludwig W, Springer N, et al. Two coryneform bacteria isolated from the surface of French Gruyère and Beaufort cheeses are new species of the genus Brachybacterium: brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov. Int J Syst Bacteriol. 1996;46(1):81–87. doi:10.1099/00207713-46-1-81
7. Chou JH, Lin KY, Lin MC, et al. Brachybacterium phenoliresistens sp. nov., isolated from oil-contaminated coastal sand. Int J Syst Evol Microbiol. 2007;57(Pt 11):2674–2679. doi:10.1099/ijs.0.65019-0
8. Singh H, Du J, Yang JE, et al. Brachybacterium horti sp. nov., isolated from garden soil. Int J Syst Evol Microbiol. 2016;66(1):189–195. doi:10.1099/ijsem.0.000696
9. Renvoise A, Aldrovandi N, Raoult D, et al. Helcobacillus massiliensis gen. nov., sp. nov., a novel representative of the family Dermabacteraceae isolated from a patient with a cutaneous discharge. Int J Syst Evol Microbiol. 2009;59(Pt 9):2346–2351. doi:10.1099/ijs.0.003319-0
10. Kuete E, Mbogning Fonkou MD, Mekhalif F, et al. Brachybacterium timonense sp. nov., a new bacterium isolated from human sputum. New Microbes New Infect. 2019;29(31):100568. doi:10.1016/j.nmni.2019.100568
11. Tamai K, Akashi Y, Yoshimoto Y, et al. First case of a bloodstream infection caused by the genus Brachybacterium. J Infect Chemother. 2018;24(12):998–1003. doi:10.1016/j.jiac.2018.06.005
12. Buczolits S, Schumann P, Weidler G, et al. Brachybacterium muris sp. nov., isolated from the liver of a laboratory mouse strain. Int J Syst Evol Microbiol. 2003;53(Pt 6):1955–1960. doi:10.1099/ijs.0.02728-0
13. Gu D, Jiao Y, Wu J, et al. Optimization of EPS Production and Characterization by a Halophilic Bacterium, Kocuria rosea ZJUQH from Chaka Salt Lake with Response Surface Methodology. Molecules. 2017;22(5):814. doi:10.3390/molecules22050814
14. Ruokolainen L, Paalanen L, Karkman A, et al. Significant disparities in allergy prevalence and microbiota between the young people in Finnish and Russian Karelia. Clin Exp Allergy. 2017;47(5):665–674. doi:10.1111/cea.12895
15. Leung MHY, Tong X, Bastien P, et al. Changes of the human skin microbiota upon chronic exposure to polycyclic aromatic hydrocarbon pollutants. Microbiome. 2020;8(1):100. doi:10.1186/s40168-020-00874-1
16. Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389(10075):1218–1228. doi:10.1016/S0140-6736(17)30182-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
