Back to Archived Journals » Hypoxia » Volume 6
Born to sense: biophysical analyses of the oxygen sensing prolyl hydroxylase from the simplest animal Trichoplax adhaerens
Authors Lippl K, Boleininger A, McDonough MA , Abboud MI , Tarhonskaya H, Chowdhury R , Loenarz C , Schofield CJ
Received 19 May 2018
Accepted for publication 27 July 2018
Published 9 November 2018 Volume 2018:6 Pages 57—71
DOI https://doi.org/10.2147/HP.S174655
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Dörthe Katschinski
Kerstin Lippl, Anna Boleininger, Michael A McDonough, Martine I Abboud, Hanna Tarhonskaya, Rasheduzzaman Chowdhury, Christoph Loenarz, Christopher J Schofield
Chemistry Research Laboratory, University of Oxford, Oxford, UK
Background: In humans and other animals, the chronic hypoxic response is mediated by hypoxia inducible transcription factors (HIFs) which regulate the expression of genes that counteract the effects of limiting oxygen. Prolyl hydroxylases (PHDs) act as hypoxia sensors for the HIF system in organisms ranging from humans to the simplest animal Trichoplax adhaerens.
Methods: We report structural and biochemical studies on the T. adhaerens HIF prolyl hydroxylase (TaPHD) that inform about the evolution of hypoxia sensing in animals.
Results: High resolution crystal structures (≤1.3 Å) of TaPHD, with and without its HIFα substrate, reveal remarkable conservation of key active site elements between T. adhaerens and human PHDs, which also manifest in kinetic comparisons.
Conclusion: Conserved structural features of TaPHD and human PHDs include those apparently enabling the slow binding/reaction of oxygen with the active site Fe(II), the formation of a stable 2-oxoglutarate complex, and a stereoelectronically promoted change in conformation of the hydroxylated proline-residue. Comparison of substrate selectivity between the human PHDs and TaPHD provides insights into the selectivity determinants of HIF binding by the PHDs, and into the evolution of the multiple HIFs and PHDs present in higher animals.
Keywords: hypoxia, hypoxic response, oxygen sensing, hypoxia-inducible factor (HIF), evolution, dioxygenase, enzyme structure, PHD/EGLN prolyl hydroxylases, 2-oxoglutarate oxygenase, Trichoplax adhaerens
Introduction
Hypoxia inducible transcription factors (HIFs) play key roles in maintaining oxygen homeostasis in most, if not all, animals.1–4 Extensive work on the HIF mediated response to hypoxia in humans and other higher animals (Figure 1A and B) has revealed that the levels and transcriptional activity of the α,β-heterodimeric HIF complex are regulated by ferrous iron and 2-oxoglutarate (2OG) dependent oxygenases.4–6 Prolyl-4-hydroxylation of the oxygen dependent degradation domains (ODDs) in HIFα is catalyzed by the HIF prolyl hydroxylases (PHDs or EGLNs), and signals for HIFα degradation via the ubiquitin proteasome system. HIFα prolyl hydroxylation strongly promotes its binding to the von-Hippel Lindau protein (pVHL), a targeting component of an E3 ubiquitin ligase complex7–9 (Figure 1C). In some animals, a second 2OG oxygenase, factor inhibiting HIF (FIH), catalyzes asparaginyl-hydroxylation in the C-terminal transcriptional activation domain (CTAD) present in both HIF1α and HIF2α, but not in HIF3α;10,11 this modification inhibits the interaction between HIF and transcriptional coactivators, which are histone acetyl transferases (CBP/p300).11 FIH is active at lower oxygen concentrations than the PHDs12,13 and is likely less important from a fundamental hypoxia-sensing perspective.
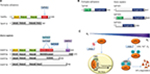  | Figure 1 Overview of the HIF system. Notes: (A) Comparison of domain architectures of HIFα and HIFβ in T. adhaerens and humans. In contrast to the multiple PHD/HIFα-isoforms and ODDs present in vertebrates, the prime components of the HIF system in T. adhaerens involve only one PHD and one HIFα, which has a single ODD.16 Arrows indicate assigned prolyl and asparaginyl hydroxylation sites. (B) Domain structures of HIF prolyl hydroxylases (PHDs) in T. adhaerens and humans. Domain acronyms: 2OG dioxygenase domain (oxygenase), MYeloid, Nervy, and DEAF-1 (MYND)-type zinc finger domain (MYND). (C) Outline of the conserved mechanism of the response to chronic hypoxia in animals. The HIF transcription factors are regulated by PHD catalyzed hydroxylation of prolyl residues in the ODD(s) of HIFα under normoxia. Recognition of the hydroxylated prolyl residues by pVHL is followed by ubiquitination by the E3 ubiquitin ligase, which tags HIFα for proteasomal degradation. In humans, FIH, which is only sporadically present in non-vertebrate animals,16 constitutes an additional oxygen dependent regulatory element of HIF activity. FIH catalyzes the hydroxylation of an asparagine residue in the CTAD of HIFα, thus disrupting its interaction with the CBP/p300 transcriptional co-activator, and hindering transcriptional activation.11 Abbreviations: PHD, Prolyl hydroxylases; HIF, hypoxia-inducible transcription factor; ODD, oxygen-dependent degradation domain; bHLH, basic helix-loop-helix motif; PAS, Per-ARNT-Sim domain; CTAD, C-terminal transactivation domain; PAC, PAS-associated C-terminal domain; FIH, factor inhibiting HIF. |
The PHDs and FIH belong to structurally distinct 2OG oxygenase subfamilies, with FIH being part of the Jumonji-C (JmjC) 2OG oxygenase subfamily IX,14 and the PHDs belonging to the PH (prolyl hydroxylase) subfamily VIII.15 However, unlike the HIFα-PHD-pVHL “hypoxia sensing triad”, the FIH-HIFα CTAD “diad” is not universally conserved, being only sporadically present in simple animals, implying it evolved subsequent to the PHD enabled sensing mechanism.16
Extensive genetic and chemical intervention studies support the physiological importance of the HIFα-PHD-pVHL triad in animals. Indeed, the human PHDs are current therapeutic targets, with PHD inhibitors for the treatment of anemia, via HIF mediated upregulation of erythropoietin, being in late stage clinical trials.17,18 In humans, there are three PHDs (PHD1-3) and three HIFα paralogues, but only one FIH. Animal studies imply that PHD2, which, unlike PHD1 and 3, contains a highly conserved Myeloid, Nervy, and DEAF-1 (MYND) finger domain, is the most important of the three mammalian PHDs from a physiological perspective.19,20 Unlike humans and other mammals, there is only one PHD present in (most) lower organisms, which is more similar to PHD2 than PHD1 or PHD3.16
The human HIF system regulates hundreds of different genes in a context and cell-type dependent manner, with emerging evidence suggesting different roles for HIF1α and HIF2α and the three PHDs.12,21–23 HIF1α and HIF2α each have two ODDs, the C-terminal and N-terminal (CODD and NODD), respectively, which manifest differential sensitivities to hypoxia.12,24 In contrast to the extensive studies conducted on HIF1α and HIF2α, the biological role of HIF3α, which contains a single ODD, is less understood, with a role for HIF3α splice variants in the regulation of the hypoxic response being proposed.25–27
The three human PHDs also have different ODD selectivities, with PHD1 and PHD2 accepting both NODD and CODD as substrates; however, PHD3 has a strong preference for CODD over NODD as a substrate.21,22 Biochemical studies on the PHDs are supportive of their roles as hypoxic sensors.21,28–30 An essential requirement for the proposed hypoxia sensing role of the PHDs is that their activity in cells is regulated by oxygen availability. Studies, both in cells and animals, support the proposed hypoxia sensing role of the PHDs,1–9 raising the question as to whether their hypoxia sensing role is manifested in specific biochemical properties. There is potential for various factors to help enable PHD activity to be limited by oxygen availability in cells, including regulation of localization, concentrations, or activities of the PHDs and HIFα isoforms. Several studies support the proposal that the kinetic properties of the PHDs are directly related to their sensing role.21,28–30 Strikingly, while PHD2, the most conserved of the human PHDs, binds Fe(II) and 2OG relatively tightly compared to other 2OG oxygenases, forming an unusually stable enzyme.Fe.2OG complex, the PHD2.Fe.2OG.ODD complex reacts unusually slowly with oxygen (Figure S1).30,31 Structural analyses on PHD2 have led to the proposal that its slow reaction with oxygen is related to the active site coordination chemistry, with substitution of an Fe(II) bound water by dioxygen being a limiting step in catalysis.32–36 To investigate the proposal that the apparently special kinetic properties of PHDs are a general feature of their hypoxia sensing roles, we have initiated studies on their evolution.
Bioinformatic studies suggest that the HIFα-PHD-pVHL triad only exists in animals, though homologues of the PHDs, but not apparently HIF, are present in earlier organisms.16,37,38 Roles for PHD homologues in early eukaryotes, ie, hydroxylation of S-phase kinase associated protein 1 (Skp1) in Dictyostelium,37 and in some bacteria, ie, hydroxylation of the Elongation Factor EF-Tu38 have been identified. However, while these enzymes also catalyze prolyl C-4 hydroxylation and their overall structures are clearly related to the PHDs, the available evidence is that these enzymes differ in their kinetic properties relative to the human PHDs,37–39 as is the case for the procollagen prolyl-4-hydroxylases.21,28,40–46
The extent to which there is structural conservation of the active sites between the PHDs from different animals has been unclear to date; we have reported experimental evidence for the presence of a functional HIFα-PHD-pVHL system in Trichoplax adhaerens, the simplest known animal (Figure S2).16 Interestingly, the genes encoding for TaHIFα and TaPHD are adjacent in the T. adhaerens genome, suggesting linked evolution, and TaPHD activity can be limited by oxygen availability in human cells.16 Here we report crystallographic and biochemical studies on the single PHD present in T. adhaerens (TaPHD, Uniprot ID: I6QVT6), the results of which reveal striking elements of structural and biochemical conservation between TaPHD and human PHD2 (HsPHD2) active sites, supporting oxygen binding/reaction as a key property in the oxygen sensing capability of the PHDs. The results also provide insights into the evolution of ODD binding by PHDs, and help describe how human PHDs achieve ODD selectivity.
Results
Steady-state kinetic studies of TaPHD in comparison to HsPHD2
The HIF system in T. adhaerens is apparently markedly simpler than in humans because it contains only one PHD (TaPHD), and one HIFα isoform with a single ODD (TaODD, Figure 1A).16 Although the sequences of human HIF1α CODD and NODD (HsHIF1α CODD/NODD), and TaODD differ substantially (Figure 2A), N-terminally truncated TaPHD (aa 64-300, TaPHD64-300) catalyzes prolyl hydroxylation of both the TaODD 25mer and the human HIF1α CODD and HIF1α NODD 19mer substrates, with the latter being hydroxylated less efficiently.16 To further investigate the substrate preferences of TaPHD64–300, its affinities for TaODD 25mer, human HIF sequences, and its co-substrate (2OG) were determined in steady-state kinetic studies and compared to the truncated human PHD isoform 2 (HsPHD2181-426). The kinetic parameters for the catalytic domain of HsPHD2181-426 have been shown to be similar to those of the full-length protein.31,59
  | Figure 2 Evidence for conservation of biochemical properties between TaPHD and HsPHD2. Notes: (A) Sequence alignment of TaHIFa, HsHIFa, and substrates of PHD-like enzymes in Dictyostelium discoideum (DdSkp1) and Pseudomonas putida (PpEF-Tu). (B) Kinetic parameters determined for TaPHD and HsPHD2 by MALDI-TOF-MS based assays, conditions: HsPHD2 or TaPHD (3.5 µM–7.0 µM), HsHIF1α CODD 19mer peptide (DLDLEMLAPYIPMDDDFQL-NH2, 100 µM) or TaHIFa ODD 25mer peptide (PINEKEDYDDLAPFVPPPSFDNRLY-NH2, 100 µM), (NH4)2Fe(II)(SO4)2 (50 μM), sodium L-ascorbate (4 mM) and 2OG disodium salt (300 µM) in Tris (50 mM), pH 7.5. Initial rates were determined by varying the concentrations of the respective peptide or 2OG. Peptide hydroxylation was analyzed by MALDI-MS; the apparent non-enzymatic Met oxidation was subtracted. Data were fitted with the Michaelis-Menten equation using GraphPad Prism® (errors are indicated as standard deviations, n=3). (C) Results of 1D CLIP HSQC [13C]-HsHIF1α CODD and [13C]-HsHIF1α NODD displacement experiments with a [13C]-prolyl-labelled reporter HsHIF1α CODD/NODD peptide reveal a higher binding affinity of TaPHD for HsHIF1α CODD over NODD (errors are indicated as standard deviations, n=3); conditions: [13C]-proline HsHIF1α CODD/NODD (DLDLEMLAPYIPMDDDFQL-NH2/DALTLLAPAAGDTIISLDF-NH2, 50 µM), TaPHD (50 µM), 2OG disodium salt (50 µM) buffered with Tris-D11 (50 mM), pH 7.5, in 10% D2O and 90% H2O. (D) Comparison of coupled and uncoupled (ie, in the absence of substrate) 2OG turnover by TaPHD and (E) HsPHD2. 2OG turnover was monitored by 1H CPMG NMR, conditions: TaPHD or HsPHD2 (20 μM), (NH4)2Fe(II)(SO4)2 (125 µM), sodium L-ascorbate (1 mM), HsHIF1α CODD (500 µM) or TaHIFa ODD substrate (500 µM) (where necessary), and 2OG disodium salt (400 µM), in 10% D2O and 90% H2O, Tris-D11 (50 mM), pH 7.5; [Succ]=Succinate. (F) Steady-state O2-dependence of TaPHD and (G) HsPHD2 (published in;29 conditions: 4 µM TaPHD/HsPHD2, 100 µM TaHIFa ODD/HsHIF1α CODD, (NH4)2Fe(II)(SO4)2 (50 µM), 2OG disodium salt (300 µM) and sodium L-ascorbate (4 mM) in Tris (50 mM), pH 7.5 were incubated at 37°C under different % O2. The extent of hydroxylation was analyzed by MALDI–ToF-MS (errors are indicated as standard deviations, n=3). |
Km and kcat values (Figure 2B) for HIF peptide substrates and 2OG were determined under optimized assay conditions described in “Experimental Methods” in Supplementary materials, with the extent of peptide hydroxylation being analyzed by Matrix-Assisted Laser Desorption/Ionization (MALDI) time-of-flight mass spectrometry (MS) (Figures 2B and S3–5). Consistent with previous reports,16 TaPHD64-300 efficiently catalyzes the hydroxylation of its natural substrate TaODD 25mer and the human HIF1α CODD 19mer peptide. The HsHIF1α NODD 19mer, by contrast, is not turned over above background level by TaPHD64-300, at least under the conditions/limits of detection of the assays employed here (Figure S5). TaPHD64-300 has a similar affinity to both these substrates, despite their differences in length, as judged by Km values (Km(TaODD 25mer, TaPHD64-300)=11.5±1.8 µM, Km(HsHIF1α CODD 19mer, TaPHD64-300)=15.1±2.2 µM). However, the catalytic efficiency of TaPHD64-300 as judged by kcat is lower with the human CODD sequence (kcat (HsHIF1α CODD 19mer, TaPHD64-300)=0.012±0.001 s−1, kcat (TaODD 25mer, TaPHD64-300)=0.026±0.001 s−1). HsPHD2181-426, by comparison, manifests a decreased affinity for the TaODD (Km(HsHIF1α CODD 19mer, HsPHD2181-426)=10.8±1.9 µM, Km(TaODD, HsPHD2181-426)=40.5±4.9 µM). HsPHD2181-426, however, catalyzes hydroxylation of HsHIF1α CODD 19mer and TaODD 25mer with comparable efficiency kcat (HsHIF1α CODD 19mer, HsPHD2181-426)=0.027±0.001 s−1, kcat (TaODD 25mer, HsPHD2181-426)=0.035±0.002 s−1).
Notably, the affinity of TaPHD64-300 for 2OG (Km(2OG, TaPHD64-300)=1.9±0.3 µM) is higher than for HsPHD2181-426 (Km(2OG, HsPHD2181-426)=15.8±2.6 µM), as judged by Km comparisons. The higher affinity of 2OG for TaPHD64-300 is in accord with the observation of formation of a stable TaPHD.Fe.2OG complex, with a half-life >24 h.16 The kinetic parameters reported here for the affinity of HsHIF1α CODD 19mer and 2OG for HsPHD2181-426 are consistent with previous studies,29 Km(HsHIF1α CODD 19mer, HsPHD2181-426)=9±3 µM, Km(2OG, HsPHD2181-426)=13±2 µM).
TaPHD binds HsHIF1α NODD less efficiently than HsHIF1α CODD
The observed preference of TaPHD64-300 for HsHIF1α CODD over HsHIF1α NODD is in agreement with the finding of one ODD in the genome of T. adhaerens, which is closer in sequence to the majority of CODD, compared to NODD sequences (Figure 2A).16 While a NODD-like sequence is present in all vertebrates, NODD is apparently not present in some non-vertebrates, eg, Placozoa and Cnidaria, suggesting that the CODD sequence might have evolved prior to that of NODD.16 Notably, like TaPHD64-300, one of the human PHD isoforms, HsPHD3, is highly selective for HsHIF1α/2α CODD over HsHIF1α/2α NODD.1,21,47
To more directly investigate the extent to which TaPHD64-300 can bind HsHIF1α NODD and CODD, we performed 1D CLIP HSQC NMR binding studies with HsHIF1α CODD and HsHIF1α NODD peptides labeled with [13C] at the target prolyl residues which undergo hydroxylation during catalysis (Figure 2C). The [13C]-labeled “reporter” HsHIF1α CODD/NODD peptides were incubated with Zn(II) and 2OG in the presence and absence of TaPHD64-300 (1:1 enzyme:ODD, for conditions see “Experimental methods” in Supplementary materials). The decrease of the reporter signal upon binding to the enzyme was measured to investigate the relative binding strength (Figure 2C). Consistent with the kinetic studies, the NMR results reveal that TaPHD64-300 strongly discriminates between binding of HsHIF1α CODD and HsHIF1α NODD, as ~40% of the HsHIF1α NODD peptide remained unbound, while HsHIF1α CODD was apparently completely bound to TaPHD64-300, showing that TaPHD64-300 binds HsHIF1α CODD better than HsHIF1α NODD.
Monitoring the coupled and uncoupled 2OG turnover of TaPHD
2OG dependent oxygenases catalyze the uncoupled turnover of 2OG to succinate and CO2 in the absence of their prime substrate to varying extents;48 in the case of HsPHD2, it has been shown that 2OG decarboxylation is strongly coupled to CODD hydroxylation.31 To directly compare the extent of uncoupled turnover in TaPHD64-300 and HsPHD2181-426, succinate formation and 2OG depletion were monitored by 1H-NMR49 in the presence and absence of their respective natural substrates (Figure 2D and E). The results demonstrate that the rate of uncoupled 2OG turnover is very low for both TaPHD64-300 and HsPHD2181-426; 2OG decarboxylation is strongly increased in presence of substrate, implying conservation of (at least some) kinetic properties between HsPHD2 and TaPHD. Overall, these results support the proposal that the strong binding of Fe(II) and 2OG, and the formation of a stable HsPHD2.Fe.2OG complex in the absence of HsHIFα, are conserved properties of the PHDs.
Oxygen dependence of TaPHD
In order for the PHDs to act as hypoxia sensors in cells, their activity must be limited by oxygen availability. This role is proposed to be reflected in the slow reaction of (at least) the HsPHD2.CODD/NODD.Fe.2OG complexes with oxygen.29,31 It is also proposed to manifest in the unusually high Km value (for 2OG oxygenases) of HsPHD2 for oxygen (Km(O2, HsPHD2) >400 µM).21,28,29,50 In order to study the O2 dependence of the TaPHD-catalyzed reaction, steady-state kinetics, using reported experimental conditions for HsPHD2;29 were conducted. TaPHD64-300 was reacted with its peptidic substrate in sealed glass-vials under varied oxygen concentrations,29 the initial rates of TaODD 25mer hydroxylation were determined by MALDI–TOF-MS (Figure 2F and G). No saturation of O2 was reached for the TaPHD64-300-catalyzed reaction with TaODD 25mer at 60% O2 level, resulting in a Km (O2, TaPHD64-300) >400 µM. This result suggests that TaPHD has the potential to react to small changes in atmospheric O2 partial pressure and could act as an effective oxygen sensor, indicating that the core biochemical features of HsPHD2 in the human oxygen sensing machinery may well be conserved in TaPHD. It has been reported that HsPHD2 reacts relatively slowly with oxygen, compared to other 2OG oxygenases as determined by stopped flow/UV-visible spectroscopy experiments.29,31 It was not possible to conduct a similar study on TaPHD for practical reasons (ie, neither TaPHD64-300, nor other tested constructs TaPHD1-300 and TaPHD79-300 could reach a sufficiently high protein concentration necessary for stopped-flow assays due to aggregation).
Crystallization of N-terminally truncated T. adhaerens PHD (TaPHD64-300)
In order to investigate the extent to which the structural features proposed to be necessary for oxygen sensing are conserved between HsPHD2 and TaPHD, we attempted to obtain a crystal structure of TaPHD in complex with Mn(II), a non-reactive substitute for Fe(II), using a TaPHD construct (aa 64-300, TaPHD64-300, MW=29.2 kDa) lacking the N-terminal MYND domain, which could be produced efficiently in Escherichia coli. The shorter construct (TaPHD64-300) yielded crystals that diffracted to 1.2 Å resolution (Table 1). The structure was solved by molecular replacement, using HsPHD2 (PDB: 2G19),32 as a search model. To gain insight into the substrate-binding mode of TaPHD, we then co-crystallized TaPHD64-300 with a 21mer peptide fragment of its substrate, the TaHIFα ODD (TaODD, residues E477-L497), Mn(II) and N-oxalylglycine (NOG), a close 2OG analog. The TaPHD.TaODD.Mn.NOG structure (Table 1) was solved to a resolution of 1.3 Å by molecular replacement, using the structure without TaODD as a search model. The substrate-bound and unbound forms of TaPHD crystallized in the P1 and P21 space groups, respectively, reflecting different packing, possibly relating to structural changes in TaPHD induced upon substrate binding.
Overall structure of TaPHD
The structure of TaPHD (Figure 3A) contains the conserved double-stranded β-helix core-fold (DSBH, or “jelly roll”-motif), which is present in all characterized 2OG dependent oxygenases, including the human PHDs.15 The DSBH is comprised of the major (βI, βVIII, βIII, βVI) and minor (βII, βVII, βIV, βV) β-sheets between which the metal and 2OG binding sites are sandwiched. As with HsPHD2, the major β-sheet core is stabilized by three α-helices (α1–3). The overall folds of the substrate-unbound TaPHD (TaPHD.Mn) and the substrate-bound TaPHD.TaODD complex are very similar (Figure 3A and B, RMSD for all Cα=0.15 Å). Eighteen residues of the 21mer TaODD peptide (A480TaODD-L497TaODD, Figure 3B) are visible in the TaPHD.TaODD structure.
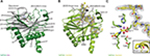  | Figure 3 Views from crystal structures of the Trichoplax adhaerens prolyl hydroxylases (PHD) without substrate bound (TaPHD) and in complex with a fragment of its substrate (TaPHD.TaODD). Notes: (A) Secondary structural elements in TaPHD comprise four α helices and ten β strands, eight of which form the double-stranded β-helix core fold (DSBH, gray, Roman numerals I–VIII). (B) Overall binding mode of TaODD to TaPHD in the TaPHD.TaODD complex structure showing the 2Fo-Fc electron density map for the peptidic substrate (gray mesh, contoured to 1.0 σ). (C) Active site close-up of TaPHD.TaODD reveals that the P486 TaODD C-4 methylene adopts an endo-conformation (2Fo-Fc density, gray mesh, contoured to 1.0 σ). The metal ion (manganese substituting for iron, purple sphere) is octahedrally coordinated by a triad of residues (H209TaPHD, D211TaPHD, and H270TaPHD), N-oxalylglycine (NOG), and a water molecule (W1, red sphere). The stable metal-water coordination observed here is conserved in HsPHD2,32,34 where it is proposed to enable the oxygen sensing ability in HsPHD2. |
Clear differences between the TaPHD.Mn and TaPHD.TaODD structures manifest in the mobile “β2/β3-finger-loop”,32–34 which is disordered in the substrate-unbound TaPHD structure (missing residues Q137TaPHD-R146TaPHD). In the substrate-bound structure, however, the β2/β3-finger-loop is ordered and makes extensive contacts with the substrate. By contrast, the β7(VI)/β8(V)-loop residue N245TaPHD is not observed in the TaPHD.TaODD structure, unlike in the substrate-unbound TaPHD.Mn structure, in which the electron density of the complete β7(VI)/β8(V)-loop is apparent (Figures 3–6, S4 and S6).
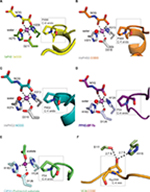  | Figure 6 Active site metal region details and comparison of the conformations of the target prolyl-residues in the enzyme-substrate complex structures of TaPHD, HsPHD2, PPHD, and CrP4H. Notes: (A) TaPHD.TaODD, (B) HsPHD2.CODD (PDB: 3HQR,34), (C) HsPHD2.NODD (PDB: 5L9V,34), (D) PPHD.EF-Tu, (PDB: 4IW3,38) and (E) CrP4H.proline-rich substrate (PDB: 3GZE,66) complex structures. Note that, in all the enzyme-substrate complexes, the proline C-4- methylene, that is hydroxylated, adopts the endo-conformation. (F) By contrast, Hyp564 in HsHIF1α CODD adopts the C-4 exo-conformation when bound to the VCB complex (PDB: 1LM8,55–57). In (A and D), N-oxalylglycine (NOG) acts as a 2OG analog. Note that the Zn (substituting for Fe) in the CrP4H active site is tetrahedrally coordinated, with an acetate binding instead of the 2OG co-substrate. The structures reveal conserved orientations of the “target” proline residues, which in each case, adopt a C-4 endo-conformation.55 Note that the metal bound water present in the TaPHD (and HsPHD2/PPHD) substrate complex structures is not observed in the CrP4H.(Ser-Pro)5 structure. |
Active site of TaPHD
The active site pocket of TaPHD, positioned at one end of the DSBH, deeply embeds the metal ion (Mn, substituting for Fe) and the co-substrate (NOG, substituting for 2OG, Figure 3B and C). The highly buried nature of the 2OG binding site likely contributes to the formation of a stable TaPHD.Fe.2OG complex, as reflected in the observed low rate of uncoupled 2OG turnover by TaPHD (Figures 2D and 3B).
In the TaPHD.Mn structure without TaODD, the manganese ion is octahedrally coordinated by a conserved triad of residues (H209TaPHD, D211TaPHD, and H270TaPHD), an acetate ion, and two water molecules.48,51 Deep within the 2OG binding pocket an additional acetate ion forms a salt bridge with R279TaPHD, apparently mimicking 2OG C5 carboxylate binding (Figure 3A).
The 6-coordinate state of the metal is retained in the TaPHD.TaODD complex. Metal-ion binding involves the highly conserved triad of metal-coordinating residues, and an ordered, metal-bound water molecule (W1) that is, in addition to the metal coordination, stabilized via hydrogen bonding with the metal-ligating side chain of D211TaPHD (Figures 3C and 6). Electron density corresponding to the 2OG analog inhibitor, NOG, is clearly visible in the TaPHD.TaODD complex structure. NOG coordinates to the metal ion in a bidentate manner and interacts with R279TaPHD by its C5 carboxylate. The 1-carboxylate of NOG is positioned at the coordination site closest to the hydroxylation target, P486TaODD, making the remaining metal coordination site, occupied by water W1, less accessible, and thus apparently hindering oxygen binding to the metal ion (Figure 6).33,34
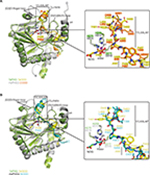
Crystal structures of the human PHD isoform 2 (HsPHD2) in complex with its substrates HsHIF1α CODD (referred to as HsPHD2.CODD, PDB: 3HQR34) and HsHIF1α NODD (referred to as HsPHD2.NODD, PDB: 5L9V33) have been reported (Figure 4). The overall active site arrangement observed in the TaPHD.TaODD complex is very similar to those observed in the HsPHD2.CODD/NODD complexes (Figures 4–6,33,34). Such conservation includes the octahedral coordination of the metal ion by H313HsPHD2, D315HsPHD2, and H374HsPHD2, NOG, and a metal-bound water molecule.33,34,52 The observation of a well-ordered metal bound water in both TaPHD and HsPHD2 structures is important from a hypoxia sensing perspective, because displacement of this water from the metal is required for oxygen to bind, and is proposed to be rate limiting in HsPHD2 catalysis.32,34,53 The buried nature of the metal, the 2OG analog NOG, and the associated ligating water molecule, as observed both in TaPHD and HsPHD2, are likely substantially responsible for the unusual stability of the HsPHD2.Fe.2OG complex,30 compared to other 2OG oxygenases such as FIH, which is active at lower oxygen concentrations.12,13,54
Taken together, the observation of conserved active site arrangements in the TaPHD.TaODD and HsPHD2.CODD crystal structures correlates well with the steady-state kinetic parameters determined for 2OG and oxygen, implying that core features necessary for the oxygen-sensing role of the enzymes are conserved between TaPHD and HsPHD2.
Comparison of ODD binding modes for TaPHD and HsPHD2
The main substrate binding elements in the TaPHD.TaODD complex structure, as in HsPHD2, comprise: the active site containing groove, the β2/β3-finger-loop, and the C-terminal α4-helix (Figure 3B). The TaODD peptide binds to TaPHD in an extended form; notably, the residues N-terminal to the target proline form a 310-helix. The target proline in TaODD and its N-terminally flanking residues form a YXXLAP motif, which differs by one residue (L->Y) compared to the consensus LXXLAP prolyl-hydroxylation site(s) present in all human, and many, but not all animal, HIFα proteins.16
Binding and positioning of the hydroxylation target P486TaODD directly adjacent to the metal in the active site of TaPHD is highly similar to that in the HsPHD2.CODD and HsPHD2.NODD complex structures (Figure 4A and B). Residues Q137TaPHD, L138TaPHD, and A139TaPHD in the β2/β3-finger-loop region (corresponding to Q239HsPHD2, L240HsPHD2, and V241HsPHD2) form H-bonds and hydrophobic contacts with TaODD residues P486TaODD, F487TaODD, A485TaODD (P564HsHIF1α CODD, A563HsHIF1α CODD, Y565HsHIF1α CODD and P402HsHIF1α NODD, A401HsHIF1α NODD, A403HsHIF1α NODD, respectively). The hydroxylation target proline is apparently further positioned via polar interactions with βII(β5)/βIII(β6) residues Y206TaPHD and R218TaPHD (Y310HsPHD2 and R322HsPHD2), and hydrophobic contacts are formed to residue W285TaPHD (W389HsPHD2) in the C-terminal strand βVIII(β11), (Table S1, Figure S4).
As observed in the HsPHD2.CODD/NODD structures, the substrate proline P486TaODD pyrrolidine ring adopts a C-4 endo-conformation, with the 4R C-H bond that is cleaved during hydroxylation, being positioned close to the metal center (distance: 4.5 Å, Figures 3C and 6,33,34); indeed, the conformations of the substrate target prolyl rings at the TaPHD and HsPHD2 active sites are nearly identical (Figure 4A and B). Upon hydroxylation by HsPHD2, the conformation of the pyrrolidine ring in CODD is proposed to switch from C-4 endo to C-4 exo55 (Figure 6). When bound to pVHL, the C-4 hydroxylated proline adopts the C-4 exo conformation as favored by stereoelectronic theory (the gauche effect);55–58 a role for this conformational change in promoting product release from HsPHD2 has been suggested.34
In the human PHDs, the β2/β3-finger-loop plays a significant role in substrate recognition and in determining NODD/CODD selectivity.33,34,59 The β2/β3-finger-loop is partially disordered without its substrate bound, likely reflecting flexibility relative to the enzyme-substrate complex.33 By contrast, in both the HsPHD2.CODD and HsPHD2.NODD structures, the β2/β3-loop residues fold to enclose the LXXLAP substrate motif at the active site (Figure 4A and B). Although, the β2/β3-finger–loop in the TaPHD.TaODD complex also constitutes one of the major substrate interaction sites, it is two residues shorter than in HsPHD2 between N141TaPHD-V142TaPHD (Figures S4 and 6). As a consequence, and by contrast to HsPHD2,33 the β2/β3- finger-loop in the TaPHD.TaODD structure is more condensed and stabilized by intramolecular interactions between loop residues, including backbone-backbone H-bonds and salt bridges between side chains (Figure S3). This structural difference might reflect the fact that there is only a single HIFα isoform with one ODD in T. adhaerens, which contrasts with the situation in humans where there are three HIFα isoform substrates; with both a NODD and a CODD in case of HIF1α and HIF2α (note other HsPHD substrates have also been reported).60–65 It is thus possible that binding of multiple HIF substrates to HsPHD233,34 requires a higher degree of flexibility in the β2/β3-finger-loop, compared to the apparently more rigid β2/β3-finger-loop in TaPHD.
In both the TaPHD.TaODD and HsPHD2.CODD34 structures, a salt-bridge between an arginine residue in the C-terminal α4-helix and an aspartate residue in the C-terminus of the respective peptide (R292TaPHD/D494TaODD, R396HsPHD2/D571HsHIF1α CODD) is formed (Figure S4). However, this salt-bridge is not present in the HsPHD2.NODD complex structure, where the HsHIF1α NODD peptide is positioned further from the C-terminal helix and interacts with the C-terminal region of HsPHD2 via hydrophobic contacts. Studies with the TaODD and HsPHD2 variants, including with HsPHD2 P317RHsPHD2 and R396THsPHD2, which are selective for CODD or NODD, respectively, support the proposal of a similar binding mode for TaODD and HsHIF1α CODD/NODD to HsPHD2 (Figure S4).
By contrast with HsHIF1α CODD/NODD peptides, the TaODD adopts a short 310-helix near its Pro-Pro-Pro motif (P489TaODD-491TaODD), which is C-terminal to the target proline P486TaODD residue (Figure 4A and B). Notably, the Pro-Pro-Pro motif and the adjacent 310-helix constitute the interaction surface with the C-terminal α4-helix of TaPHD (Figure 4A and B). In the TaPHD.TaODD complex structure, the TaODD is positioned particularly close to the α4-helix of TaPHD, compared to both HsHIF1α CODD and HsPHD2.NODD, whereas the latter is positioned even more distant from the C-terminus of HsPHD2 (Figure 4A and B).
Comparison of HIF- and non-HIF prolyl-4-hydroxylase structures
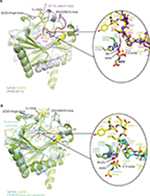
Crystal structures of the enzyme-substrate complex of a prolyl-4-hydroxylase from Chlamydomas reinhardtii (CrP4H), a member of the collagen-prolyl-4-hydroxylase-subfamily,66 and a Pseudomonas putida prolyl hydroxylase (PPHD), a clear prokaryotic PHD homologue, have been reported.38,66 Both CrP4H and PPHD act on non-HIFα substrates, ie (for consistency), CrP4H catalyzes the hydroxylation of a proline residue P6 in a proline-rich (Ser-Pro)5 substrate (PDB: 3GZE,66), and PPHD hydroxylates a proline residue (P54) in the translation elongation factor EF-Tu (PDB: 4IW3,38) (Figure 5).
Structure based sequence alignment of TaPHD with the human PHD isoforms, Chlamydomonas reinhardtii (CrP4H) and Pseudomonas putida (PPHD) was performed (Figure S2). Consistent with its essential role in catalysis, the 2OG-oxygenase domain (E90TaPHD-F287TaPHD) manifests the highest degree of sequence conservation among the HIF-and non-HIF-prolyl-4-hydroxylases. The N-terminal region is less well conserved, but like HsPHD2, TaPHD contains a conserved cysteine-rich MYND-type zinc finger domain. The function of the N-terminal domain of the PHDs is not fully understood; a role for the MYND domain in the interaction with heat shock protein 90 (HSP90), co-chaperones p23, and FKBP38 has been proposed.67
Analysis of the structural conservation between TaPHD and HsPHD2 (Figure S4, Table S2) reveals high overall similarity (RMSD for all Cα of TaPHDC73-Q297.TaODD - HsPHD2Q184-K408.CODD=0.54 Å, and RMSD for all Cα of TaPHDC73-Q297.TaODD - HsPHD2P189-Y403.NODD=0.39 Å). Comparisons with the other prolyl-4-hydroxylases imply that TaPHD shares a stronger structural conservation with PPHD (RMSD for all Cα of TaPHDC73-Q297.TaODD - PPHDH7-F207.EF-Tu=0.92 Å), than with CrP4H (RMSD for all Cα of TaPHDC73-Q297.TaODD - CrP4HW38-G250.(Ser-Pro)5=2.20 Å), suggesting a closer evolutionary relationship between HIF hydroxylases and PPHD than to the algal prolyl-4-hydroxylase CrP4H (Figure 4). The structural similarity between PPHD and TaPHD is higher than between PPHD and HsPHD2 (RMSD for all Cα of HsPHD2Q184-K408.CODD - PPHDH7-F207.EF-Tu=1.57 Å) (Figures 4 and 5).
In addition to their overall structural conservation, the prolyl-4-hydroxylases adopt strikingly similar substrate-binding features (Figures 4–6 and S5). The TaODD, HsHIF1α CODD/NODD, EF-Tu, and the proline-rich (Ser-Pro)5 substrate backbones are all oriented in the same N→C direction across the active site (Figure 5A and B). TaODD, HsHIF1α CODD, and HsHIF1α NODD all form a partial helical structure (310-helix), when bound to enzyme (Figure 4A and B), while isolated HsHIF1α CODD/NODD are predicted to be disordered in solution.33,34 By contrast, the EF-Tu switch loop retains loop secondary structure upon binding to PPHD, albeit after a large conformational change.38
Notably, the PHDs all employ a flexible β2/β3-finger-loop (G136TaPHD-D150TaPHD, S77CrP4H-S95CrP4H, A53PPHD-D70PPHD) that undergoes a major conformational change upon substrate binding and helps to enclose the target proline of the substrate in the active site cleft (Figures 5, 6 and S4).33,34 In TaPHD, HsPHD2 and PPHD, the C-terminus interacts with the substrate; it is unknown if this is the case for CrP4H, since the shorter peptide substrate used in the CrP4H.(Ser-Pro)5 structure does not appear to reach the C-terminus of the enzyme.66
Conservation of proline-ring conformation
Comparison of the binding modes of the prolyl-4-hydroxylase substrates in the immediate active sites reveals a high degree of conservation (Figures 5A, B and 6). The substrates all manifest a similar orientation toward the active site residues and the metal center, and the target prolyl residues all adopt the C-4 endo-conformation (Figure 5A and B).55 Similarly to the HIF prolyl-4-hydroxylases TaPHD and HsPHD2, PPHD and CrP4H both employ a HXD...H motif for metal-binding in the active site (H143CrP4H, D145CrP4H, H227CrP4H, and H124PPHD, D126PPHD, H183PPHD).38,66 In the PPHD crystal structure, the metal ion is octahedrally coordinated and one of the coordination sites is occupied by a water molecule (Figure 6), implying evolutionary conservation in the active site arrangement between the HIF prolyl-4-hydroxylases in animals and the bacterial non-HIF prolyl-4-hydroxylase PPHD. By contrast, in the CrP4H crystal structure,66 the metal ion is tetrahedrally coordinated; however, in this structure, binding of a water molecule to the metal is apparently blocked by an acetate ion (Figure 6E), and hence, at least in this detail, it may not be fully representative of the solution structure.
Discussion
It has been proposed that the ferrous iron dependent oxygenases may have evolved as a response to the advent of photosynthetically produced oxygen, possibly from precursors that used relatively bioavailable iron (comparable to eg, zinc) ions, predominantly in non-redox processes.68,69 The widespread occurrence of 2OG oxygenases in prokaryotes, coupled with their close relationship with two TCA cycle intermediates (2OG and succinate), suggests they may have early origins. The discovery that 2OG oxygenases play key roles in hypoxia sensing in higher animals was therefore of interest from a broad evolutionary perspective. We are interested in exploring the biological distribution of hypoxia sensing mechanisms, as well as 2OG oxygenases and related enzymes. We have found that there is a functional HIFα-PHD-pVHL triad in the simplest animal, T. adhaerens.16 Although bioinformatics and limited experimental analyses have not provided evidence for HIF transcription factors beyond animals,16 PHD-like enzymes are much more widely distributed (Figure S2).16,37,38
The PHDs are related to the pro-collagen prolyl hydroxylases and are part of a structurally distinct 2OG oxygenase subfamily; the PH VIII subfamily15 which includes other PHDs of biological interest. In the unicellular eukaryote Dictyostelium discoideum, a cytoplasmic PHD homologue, which is proposed to be involved in hypoxic responses, catalyzes hydroxylation of a proline-residue in Skp1, inducing further post-translational modification of Skp1 with a pentasaccharide group that is linked via the hydroxyproline residue alcohol.37,70 Recent studies have shown that PHD homologues also exist in bacteria, where a PHD present in Pseudomonas species catalyzes hydroxylation of the ribosome associated protein EF-Tu.38
Central to the hypoxia sensing ability of the PHDs is the capacity for their hydroxylase activity in many cell types to be limited by oxygen availability.12,29 It is proposed that this is in substantial part due to the slow reaction of oxygen with the active site Fe(II) of the PHDs.31 By contrast there is evidence that the human PHDs are (at least normally) relatively less sensitive to changes in Fe(II) or 2OG availability, consistent with the proposed specialized role for them as hypoxia sensors.30
The question then arises as to what extent are these apparently special properties of the human PHDs (especially HsPHD2) conserved in the PHDs/PHD-like enzymes present in animals and beyond. The results presented here provide evidence that the apparently unusual kinetic properties of HsPHD2 are conserved in TaPHD (Figure S7), likely as a consequence of the conservation of key structural features.
The overall folds of the catalytic domains of TaPHD and the HsPHD2 are very similar, including with respect to the elements involved in substrate binding, in particular the conformationally mobile β2/β3-finger-loop33 and the C-terminal helix (Figures 5 and 6). Detailed comparison of the PHD-substrate interactions reveals that the active site region interactions between TaPHD and TaODD more closely resemble those between HsPHD2 and HsHIF1α CODD, rather than those of HsPHD2 and HsHIF1α NODD (Figures 4 and 6). Thus, our results support the proposal that the PHD2/CODD-type ODD couple evolved first and is likely the most important of the PHD/ODD interactions in higher animals where there are multiple PHDs and ODDs.16 Given the likely relatively rigid nature of the Pro-Pro-Pro motif in TaODD, it also appears that TaODD binds to TaPHD in a less flexible manner than does HsHIF1α NODD to HsPHD2, and to a lesser extent, HsHIF1α CODD (Figure 4). Although solution studies are required to validate this proposal, these differences may further reflect the evolved roles of the HsPHDs (and likely those in other higher animals) in accepting multiple substrates (NODDs/CODDs of HIFα isoforms), and maybe, non-HIF substrates60–65 compared to TaPHD. The conservation in overall fold is also the case with respect to the prokaryotic PHD, PPHD, which acts on the highly abundant globular GTP-utilizing protein EF-Tu,38 rather than the disordered ODD regions of HIFα (Figure 5). (A role of PPHD in hypoxia sensing, if any, has yet to be identified).
Crucially, the structural elements which enable the unusually slow reaction of HsPHD2 with oxygen,31,34 (as well as tight 2OG/Fe(II) binding and low substrate uncoupled turnover), appear to be conserved in TaPHD as observed crystallographically and manifested in the available kinetic studies (Figures 2 and 6). Since humans and T. adhaerens are at near opposite ends of animal evolution, it seems possible that hypoxia sensing PHD homologues in organisms between them will have similar kinetic properties. Interestingly, TaPHD has a higher affinity for 2OG than HsPHD2 as observed by both Km comparisons and NMR binding studies. The difference in 2OG Km may reflect different 2OG metabolism in T. adhaerens and humans. Further, like HsPHD2, TaPHD forms an unusually stable TaPHD.Fe.2OG complex (half-life >24 h), consistent with both enzymes being specialized to preferentially respond to changes in oxygen, rather than 2OG/Fe(II), availability.11 Our overall results suggest that the evolution of the HIFα (with a TaODD/CODD type ODD)-PHD2-pVHL triad, may have been an important step in evolution of animals. A key element of this triad is a PHD2-type enzyme with appropriate kinetic properties, enabling it to “focus” on hypoxia sensing. The results also suggest that searching for appropriate kinetic and structural properties may be a general approach in helping to identify candidate (hypoxia) sensing enzymes.
Data availability
The TaPHD.Mn and TaPHD.TaODD structures are deposited in the RCSB PBD: entries 6EY1 and 6F0W, respectively.
Acknowledgments
This work was supported by the Wellcome Trust, Biotechnology and Biological Sciences Research Council (BBSRC), the Leverhulme Trust, the National Institutes of Health (NIH), Cancer Research UK (CRUK), the Biochemical Society Krebs Memorial Award and a Junior Research Fellowship from Kellogg College Oxford (to MIA), and the Abraham Newton Award. We thank the Diamond Synchrotron staff for access and assistance; (Data were collected under proposal mx1230).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. | ||
Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. | ||
Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. | ||
Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5(5):343–354. | ||
Chowdhury R, Hardy A, Schofield CJ. The human oxygen sensing machinery and its manipulation. Chem Soc Rev. 2008;37(7):1308–1319. | ||
Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405(1):1–9. | ||
Ivan M, Kondo K, Yang H, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. | ||
Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. | ||
Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. | ||
Hewitson KS, McNeill LA, Riordan MV, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277(29):26351–26355. | ||
Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295(5556):858–861. | ||
Tian YM, Yeoh KK, Lee MK, et al. Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J Biol Chem. 2011;286(15):13041–13051. | ||
Koivunen P, Hirsilä M, Günzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279(11):9899–9904. | ||
Elkins JM, Hewitson KS, McNeill LA, et al. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J Biol Chem. 2003;278(3):1802–1806. | ||
Aik WS, Chowdhury R, Clifton IJ, et al. Introduction to structural studies on 2-oxoglutarate-dependent oxygenases and related enzymes. In: Hausinger RP, Schofield CJ, editors. 2-Oxoglutarate-Dependent Oxygenases. 59–94. London: Royal Society of Chemistry; 2015:59–94. | ||
Loenarz C, Coleman ML, Boleininger A, et al. The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 2011;12(1):63–70. | ||
Chan MC, Holt-Martyn JP, Schofield CJ, Ratcliffe PJ. Pharmacological targeting of the HIF hydroxylases – a new field in medicine development. Mol Aspects Med. 2016;47-48:54–75. | ||
Yan L, Colandrea VJ, Hale JJ. Prolyl hydroxylase domain-containing protein inhibitors as stabilizers of hypoxia-inducible factor: small molecule-based therapeutics for anemia. Expert Opin Ther Pat. 2010;20(9):1219–1245. | ||
Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22(16):4082–4090. | ||
Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH. Placental but not heart defects are associated with elevated hypoxia-inducible factor α levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26(22):8336–8346. | ||
Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278(33):30772–30780. | ||
Huang J, Zhao Q, Mooney SM, Lee FS. Sequence determinants in hypoxia-inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J Biol Chem. 2002;277(42):39792–39800. | ||
Chan DA, Sutphin PD, Yen SE, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1α. Mol Cell Biol. 2005;25(15):6415–6426. | ||
Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 2001;20(18):5197–5206. | ||
Heikkilä M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell Mol Life Sci. 2011;68(23):3885–3901. | ||
Makino Y, Cao R, Svensson K, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414(6863):550–554. | ||
Yamashita T, Ohneda O, Nagano M, et al. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol. 2008;28(4):1285–1297. | ||
Ehrismann D, Flashman E, Genn DN, et al. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J. 2007;401(1):227–234. | ||
Tarhonskaya H, Chowdhury R, Leung IK, et al. Investigating the contribution of the active site environment to the slow reaction of hypoxia-inducible factor prolyl hydroxylase domain 2 with oxygen. Biochem J. 2014;463(3):363–372. | ||
McNeill LA, Flashman E, Buck MR, et al. Hypoxia-inducible factor prolyl hydroxylase 2 has a high affinity for ferrous iron and 2-oxoglutarate. Mol Biosyst. 2005;1(4):321–324. | ||
Flashman E, Hoffart LM, Hamed RB, Bollinger JM, Krebs C, Schofield CJ. Evidence for the slow reaction of hypoxia-inducible factor prolyl hydroxylase 2 with oxygen. FEBS J. 2010;277(19):4089–4099. | ||
McDonough MA, Li V, Flashman E, et al. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc Natl Acad Sci U S A. 2006;103(26):9814–9819. | ||
Chowdhury R, Leung IK, Tian YM, et al. Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases. Nat Commun. 2016;7:12673. | ||
Chowdhury R, McDonough MA, Mecinović J, et al. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17(7):981–989. | ||
Flashman E, Hoffart LM, Hamed RB, Bollinger JM, Krebs C, Schofield CJ. Evidence for the slow reaction of hypoxia-inducible factor prolyl hydroxylase 2 with oxygen. FEBS J. 2010;277(19):4089–4099. | ||
Tarhonskaya H, Chowdhury R, Leung IK, et al. Investigating the contribution of the active site environment to the slow reaction of hypoxia-inducible factor prolyl hydroxylase domain 2 with oxygen. Biochem J. 2014;463(3):363–372. | ||
van der Wel H, Ercan A, West CM. The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-α class of animal prolyl 4-hydroxylases. J Biol Chem. 2005;280(15):14645–14655. | ||
Scotti JS, Leung IK, Ge W, et al. Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation. Proc Natl Acad Sci U S A. 2014;111(37):13331–13336. | ||
Xu Y, Brown KM, Wang ZA, et al. The Skp1 protein from Toxoplasma is modified by a cytoplasmic prolyl 4-hydroxylase associated with oxygen sensing in the social amoeba Dictyostelium. J Biol Chem. 2012;287(30):25098–25110. | ||
Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med. 2008;40(6):402–417. | ||
Myllyharju J. Intracellular post-translational modifications of collagens. In: Brinckmann J, Notbohm H, Müller PK, editors. Collagen: Primer in Structure, Processing and Assembly. 115-147 Heidelberg: Springer-Verlag Berlin Heidelberg; 2005:115–147. | ||
Kukkola L, Hieta R, Kivirikko KI, Myllyharju J. Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J Biol Chem. 2003;278(48):47685–47693. | ||
Myllyharju J, Kivirikko KI. Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 1997;16(6):1173–1180. | ||
Nokelainen M, Nissi R, Kukkola L, Helaakoski T, Myllyharju J. Characterization of the human and mouse genes for the α subunit of type II prolyl 4-hydroxylase Eur J Biochem. 2001;268:5300–5309. | ||
Hirsilä M, Koivunen P, Xu L, Seeley T, Kivirikko KI, Myllyharju J. Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 2005;19(10):1308–1310. | ||
Koivunen P, Hirsilä M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J Biol Chem. 2006;281(39):28712–28720. | ||
Appelhoff RJ, Tian YM, Raval RR, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279(37):38458–38465. | ||
Hausinger RP. FeII/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39(1):21–68. | ||
Abboud MI, McAllister TE, Leung IKH, et al. 2-Oxoglutarate regulates binding of hydroxylated hypoxia-inducible factor to prolyl hydroxylase domain 2. Chemical Commun. 2018; 54(25):3130–3133. | ||
Dao JH, Kurzeja RJ, Morachis JM, et al. Kinetic characterization and identification of a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase 2 using a time-resolved fluorescence resonance energy transfer-based assay technology. Anal Biochem. 2009;384(2):213–223. | ||
Hangasky JA, Taabazuing CY, Martin CB, Eron SJ, Knapp MJ. The facial triad in the α-ketoglutarate dependent oxygenase FIH: A role for sterics in linking substrate binding to O2 activation. J Inorg Biochem. 2017;166:26–33. | ||
Wilkins SE, Abboud MI, Hancock RL, Schofield CJ. Targeting protein-protein interactions in the HIF system. ChemMedChem. 2016;11(8):773–786. | ||
Neidig ML, Brown CD, Light KM, et al. CD and MCD of CytC3 and taurine dioxygenase: role of the facial triad in α-KG-dependent oxygenases. J Am Chem Soc. 2007;129(46):14224–14231. | ||
Tarhonskaya H, Hardy AP, Howe EA, et al. Kinetic Investigations of the Role of Factor Inhibiting Hypoxia-inducible Factor (FIH) as an Oxygen Sensor. J Biol Chem. 2015;290(32):19726–19742. | ||
Loenarz C, Mecinović J, Chowdhury R, McNeill LA, Flashman E, Schofield CJ. Evidence for a stereoelectronic effect in human oxygen sensing. Angew Chem Int Ed Engl. 2009;48(10):1784–1787. | ||
Min JH, Yang H, Ivan M, Gertler F, Kaelin WG, Pavletich NP. Structure of an HIF-1α -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296(5574):1886–1889. | ||
Hon WC, Wilson MI, Harlos K, et al. Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature. 2002;417(6892):975–978. | ||
Domene C, Jorgensen C, Vanommeslaeghe K, Schofield CJ, Mackerell A. Quantifying the Binding Interaction between the Hypoxia-Inducible Transcription Factor and the von Hippel-Lindau Suppressor. J Chem Theory Comput. 2015;11(8):3946–3954. | ||
Flashman E, Bagg EA, Chowdhury R, et al. Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases. J Biol Chem. 2008;283(7):3808–3815. | ||
Place TL, Domann FE. Prolyl-hydroxylase 3: evolving roles for an ancient signaling protein. Hypoxia. 2013;2013(1):13–27. | ||
Kuznetsova AV, Meller J, Schnell PO, et al. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci U S A. 2003;100(5):2706–2711. | ||
Luo W, Hu H, Chang R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. | ||
Xie L, Xiao K, Whalen EJ, et al. Oxygen-Regulated β(2)-Adrenergic Receptor Hydroxylation by EGLN3 and Ubiquitylation by pVHL. Science Signal. 2009;2(78):ra33. | ||
Anderson K, Nordquist KA, Gao X, et al. Regulation of cellular levels of Sprouty2 protein by prolyl hydroxylase domain and von Hippel-Lindau proteins. J Biol Chem. 2011;286(49):42027–42036. | ||
Guo J, Chakraborty AA, Liu P, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science. 2016;353(6302):929–932. | ||
Koski MK, Hieta R, Hirsilä M, Rönkä A, Myllyharju J, Wierenga RK. The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J Biol Chem. 2009;284(37):25290–25301. | ||
Song D, Li LS, Heaton-Johnson KJ, Arsenault PR, Master SR, Lee FS. Prolyl hydroxylase domain protein 2 (PHD2) binds a Pro-Xaa-Leu-Glu motif, linking it to the heat shock protein 90 pathway. J Biol Chem. 2013;288(14):9662–9674. | ||
Taylor CT, Mcelwain JC. Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology. 2010;25(5):272–279. | ||
Barlow JN, Baldwin JE, Clifton IJ, et al. Studies on non-haem ferrous-dependent oxygenases and oxidases. Biochem Soc Trans. 1997;25(1):86–90. | ||
West CM, Wang ZA, van der Wel H. A cytoplasmic prolyl hydroxylation and glycosylation pathway modifies Skp1 and regulates O2-dependent development in Dictyostelium. Biochim Biophys Acta. 2010;1800(2):160–171. |
 © 2018 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
© 2018 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.