Back to Journals » OncoTargets and Therapy » Volume 12
Biological Functions of TNKS1 and Its Relationship with Wnt/β-Catenin Pathway in Astrocytoma
Authors Chen M, Tang B, Xie S, Yan J, Yang L, Zhou X, Zeng E
Received 20 February 2019
Accepted for publication 5 November 2019
Published 11 December 2019 Volume 2019:12 Pages 10841—10850
DOI https://doi.org/10.2147/OTT.S206142
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Min Chen,1 Bin Tang,2 Shenhao Xie,2 Jian Yan,2 Le Yang,2 Xinhui Zhou,2 Erming Zeng2
1Department of Neurosurgery, The Second Affiliated Hospital of Nanchang University, Nanchang 330006, People’s Republic of China; 2Department of Neurosurgery, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi 330006, Republic of China
Correspondence: Bin Tang
Department of Neurosurgery, The First Affiliated Hospital of Nanchang University, 17 Yongwai Street, Nanchang 330006, People’s Republic of China
Tel +8679188692748
Email [email protected]
Background: Tankyrase1 (TNKS1), which often shows abnormal expression in many malignant tumor cells, plays an important role in tumor progression. In our previous study, we found that TNKS1 is also closely related to pathologic grade in human astrocytoma and its expression level is positively correlated with the Wnt/β-catenin pathway. This study is aimed to further elucidate the biological functions of TNKS1 as well as its relationship with the Wnt/β-catenin pathway.
Methods: TNSK1 overexpression and knockdown vectors were constructed and transfected into glioblastoma cell lines U251 MG and U87, respectively. Viability, apoptosis, cell cycle and cell invasiveness in the treated cells were investigated.
Results: In comparison with untreated cells, U251 and U87 cells overexpressing TNSK1 showed significantly increased cell viability and decreased apoptosis, while the TNKS1 knockdown U251 and U87 cells had reduced cell invasive ability and increased apoptosis, respectively. In addition, immunoprecipitation study showed that TNKS1 could be detected by β-catenin antibody after pull-down, indicating that TNKS1 directly interacts with β-catenin, further indicating that TNKS1 could be regarded as a positive regulator of the Wnt/β-catenin pathway in astrocytoma. Moreover, knockdown of TNKS1 in U251 and U87 cells also leads to suppressed Wnt/β-catenin signaling, and subsequent decrease of cell growth and proliferation, reduced invasion ability and increased apoptosis.
Conclusion: Our findings suggest that TNKS1 might be a potential new therapeutic target for human astrocytoma in gene therapy.
Keywords: TNKS1, Wnt/β-catenin, astrocytoma, U251, U87, siRNA, ADP, diphosphate
Background
Astrocytoma is a type of tumor composed of astrocytes. It is a very common neurotic tumor.1 The characteristics of malignant astrocytoma include quick proliferation, induction of neurodegeneration and diffuse invasion. Although a number of comprehensive treatment measures are available such as surgical resection, postoperative radiotherapy, chemotherapy and immunotherapy, the prognosis is still poor. Therefore, it remains a hard nut to effectively improve the diagnosis and treatment of astrocytoma.2 Studies have shown that the occurrence and development of astrocytoma are closely related to the overexpression of oncogenes, inactivation of tumor suppressor genes, and abnormal activation or mutation of the signal pathways.3 Wnt/β-catenin pathway plays an important role in a variety of physiological behaviors, such as embryonic development and tissue balance, and its abnormal activation or mutation that is associated with developmental defects, tumors and many other diseases.4 Studies have shown that abnormal activation or mutation of Wnt/β-catenin pathway is closely related to the occurrence and development of astrocytoma.5–9
Tankyrase (TNKS) is a member of the growing family of poly-adenosine diphosphate (ADP)-ribose (PAR) polymerases (PARP), which is responsible for PARsylation of target proteins using nicotinamide adenine dinucleotide (NAD)+ as a substrate.10 TNKS has been found to be involved in the Wnt/β-catenin pathway. It stabilizes β-catenin through binding to and destabilizing Axin via Axin PARsylation and ubiquitination.11 There are two TNKS genes, TNKS1 and TNKS2, both have indistinguishable properties in the Wnt/β-catenin pathway. As a major member of the TNKS family, TNKS1 is abnormally and highly expressed in malignant tumor cells and has a role in tumor progression.12–18 In our previous study, we have found that the expression of TNKS1 is correlated with pathologic grade and Wnt/β-catenin pathway in human astrocytoma,19 providing additional evidence for the involvement of the TNKS1 gene and the Wnt/β-catenin pathway in the genesis and progression of astrocytoma. However, the biological functions of TNKS1 in astrocytoma and its relationship with the Wnt/β-catenin pathway are still unclear.
In this study, we up- and down-regulated the expression of the TNKS1 gene in U251 and U87 glioblastoma cell lines to investigate its effects on the proliferation, cell cycle, invasion, and to reveal mechanisms and regulatory role of the TNKS1 gene in Wnt/β-catenin pathway. The findings would provide insight into the pathogenesis, diagnosis and better therapeutic targets of astrocytoma.
Materials and Methods
Cell Lines
U251 and U87 cell lines (cat. nos. BNC337874 and BNCC337874) were purchased in the Cell Bank, Shanghai Academy of Sciences, Shanghai, China and maintained in RPMI-1640 medium at 37°C in 5% CO2 incubator.
Reagents and Instruments
TNKS1-siRNAs and negative control (Table 1) were synthesized at Generalbio, Beijing, China; Lipofectamine 3000 (cat. no. 18882752) was purchased from Invitrogen, USA; OPTI-MEM I (cat. no. 331985-062) was purchased from GIBCO, USA; F12 complete medium (cat. no. KGM21700S-500) and RPMI-1640 medium (cat. no. KGM31800S-500) were purchased from Keygen, Beijing, China; FBS (cat. no. 04-001-1ACS) was purchased from BI, USA; mini plasmid extraction kit (cat. no. DP103-02) was a product of Trangen, Beijing, China; Trizon Reagent (cat. no. CW0580S), Ultrapure RNA extraction kit (cat. no. CW0581M), HiFiScript first-strand cDNA synthesis kit (cat. no. CW2569M) and UltraSYBR Mixture (cat. no. CW0957M) were purchased from CWBIO, Beijing, China; Cell Cycle Staining kit (cat. no. CCS012) was purchased from MULTI SCIENCES, Beijing, China; rabbit antibodies against β-catenin (cat. no. ab32572, 1:1000), axin 2 (cat. no. ab109307, 1:800), tankyrase (cat. no. ab86279, 1:1000), cyclin D1 (cat. no. ab13417, 1:1000), MMP7 (cat. no. ab207299, 1:1000), phospho Y142 (cat. no. ab27798,1:500) and c-Myc (cat. no. ab32072, 1:10,000) were purchased from Abcam, US. Fluorescent cell imager (ZOE) was obtained from Bio-rad, USA. Nucleic acid quantizer (Nanodrop 2000) and flow cytometry (MoFlo Astrios EQ) were obtained from Beckman-Culter, USA. Fluorescent quantitative PCR Instrument (CFX Connect™) and ultrasensitive chemiluminescence imaging system (Chemi DocTM XRS+) were purchased from Bio-Rad, USA.
 |
Table 1 TNKS1 siRNA Sequences |
Cell Culture
Cells were cultured in RPMI-1640 medium to a confluency of 70–80%, harvested, washed with 3–5 mL PBS and digested with trypsin containing EDTA for 3 mins at room temperature. Digested cells were suspended in 5A complete medium containing 10% FBS and pelleted. The pelleted cells were re-suspended in the 5A medium and seeded on plates and cultured at 37°C in a 5% CO2 incubator. The medium was refreshed every 2 days, and the cells were subcultured when reaching a confluency of 70–80%, as described above.
Construction of TNKS1 RNAi Vectors
Three potential siRNA sequences (Table 1) targeting TNKS1 were designed using online tools (http://rnaidesigner.thermofisher.com/rnaiexpress/) based on the TNKS1 gene sequence from the NCBI databases and synthesized.
Construction of Overexpression Vectors
TNKS1 and β-catenin sequences were obtained from the NCBI database and chemically synthesized after adding BamHI and XbaI sites to facilitate cloning. The gene fragments were ligated to pcDNA3.1 to construct overexpression vectors.
Transfection
Transfection of cells was performed using a Lipofectamine 3000 Transfection Reagent according to the manufacturer’s instructions. Briefly, U251 and U87 cells were seeded at a concentration of 1× 105/mL in six-well culture plates. After 24 hrs (70% confluence), the cells were transfected with 2.5 µg plasmid DNA of three TNKS1-siRNA (TNKS1-siRNA), TNKS1-NC siRNA (TNKS1-siRNA NC), TNKS1 overexpression vector (TNKS1) and TNKS1 empty vector (TNKS1 NC). Non-transfected cells were used as control. The transfected cells were cultured in RPMI-1640 medium with 20% FBA for 48 hrs and harvested for subsequent analysis.
Fluorescent Quantitative PCR
Total RNA isolated from cells using Trizon Reagent and Ultrapure RNA extraction kits and reversely transcripted into cDNA using HiFiScript first-strand cDNA synthesis kit according to manufacturer’s recommendations. PCR was carried out in a total volume of 10 μL containing 1.5 μL of cDNA, 10 μL of UltraSYBR Mixture and 1 μL of primers (TNKS1-F, GTAAAGAGGCTGGTGGACG, TNKS1-R GGCATTATGAAGCGGGAT, GAPDH-F, GAAGGTCGGAGTCAACGGAT, GAPDH-R, CCTGGAAGATGGTGATGGG) on fluorescent quantitative PCR Instrument (CFX Connect™). The cycling conditions were 50°C for 2 mins, 95°C for 10 mins followed by 40 cycles, each one consisting of 30 s at 95°C and 1 min at 57°C, with final extension at 72°C for 30 s. Samples were run in triplicate and the mean value was calculated for each case. Human glyceraldehyde-3-phosphate dehydrogenase, GADPH (Hs03929097_g1), was used as an internal control. The data were managed using the Applied Biosystems software RQ Manager v1.2.1. Relative expression was calculated by using comparative Ct method and obtaining the fold change value (2−ΔΔCt) according to previously described protocol.20
Western Blot Analysis
Cells were harvested, washed twice with cold PBS and lysed with RIPA buffer that containing protease and phosphatase inhibitors cocktail (Roche, UK). The supernatants were collected and assayed for protein content using the BCA method. Fifty microgram of protein was applied to polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a PVDF membrane, and then detected by the proper primary and secondary antibodies before visualization with a chemiluminescence kit. The gels were imaged with ultrasensitive chemiluminescence imaging system. The intensity of blot signals was quantitated using ImageQuant TL analysis software (General Electric, UK).
Cell Viability Assay
The cell viability was measured by MTT assay. In brief, cells were plated in 96-well culture plates at the density of 1–1.5 × 104 per well in complete medium. After 24 hrs incubation, cells were treated with 20 µL/well MTT solution (5 mg/mL) and incubated for 4 hrs. The optical density at 490 nm was measured using a plate reader.
Cell Cycle Analysis
Cells were harvested, washed twice with PBS and digested with 0.25% EDTA-trypsin at 37°C for 4 hrs. Floating and adherent cells were collected, suspended in PBS and stained with Cell Cycle Staining following the manufacturer’s instruction. MoFlo Astrios EQ flow cytometer was used to assess the cell cycles.
Detection of Apoptosis by Flow Cytometry
Cells were harvested, washed twice with PBS and digested with 0.25% EDTA-trypsin, washed with PBS and fixed with ice-cold 70% ethanol at 4°C for 2 hrs. DNA was labeled with Annexin V-FITC and PI and the fluorescence was measured with a MoFlo Astrios EQ flow cytometer. Data collection and analysis of the cell cycle distribution were performed using CellQuest and the Modfit software (Becton Dickinson).
Caspase 3/7 Activity Apoptosis Assay
Cells were harvested, washed twice with PBS and assayed for Caspase 3/7 activity using Caspase 3/7 activity apoptosis assay kit (cat. no. E607103-0200, Sangon Biotech, Shanghai, China) according to the supplier's instructions. Fluorescence was measured at ex/em wavelength of 490/525 nm.
Cell Invasion Assays
For the assessment of invasion, 5×105 transfected and viable cells in serum-free medium were placed into the upper chamber of an insert coated with Matrigel (BD Bioscience, USA). Media containing 10% FBS were added to the lower chamber. After 24 hrs of incubation, the cells remaining on the upper membrane were removed with cotton wool, whereas the cells that had migrated or invaded through the membrane were stained with 0.2% crystal violet in 25% methanol/PBS at room temperature for 20 mins, imaged and counted using an inverted microscope.
Co-Immunoprecipitation
Non-transfected cells or cells transfected with TNKS1 overexpression vector and empty vector were transfected with pCDNA3.1-β-catenin. The co-transfected cells were lysed within ODG buffer and the protein was quantified with the BCA method. Equal amount of protein was pre-treated with protein A/G-plus agarose beads and then incubated with β-catenin antibody. Fifty microgram of incubated protein was applied to SDS-PAGE, transferred to a PVDF membrane, and then detected by the proper primary and secondary antibodies before visualization with a chemiluminescence kit.
Statistical Analysis
All data were expressed as mean ± standard error of the mean (SEM) obtained from at least three independent experiments. Statistical comparisons between experimental and control groups were assessed using the Student’s t-test. P < 0.05 was considered statistically significant.
Results
TNKS1 Over Expression and Silencing
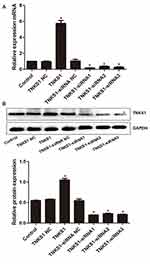
Transfected cells were examined for the expression of the TNKS1 gene at mRNA and protein levels. RT PCR and Western blot analyses showed that compared with the control, the expression of TNKS1 was significantly higher or lower after the cells were transfected with the overexpression or with three TNKS1-siRNAs vector (P < 0.05), particularly with TNKS1-siRNA1 (Figure 1A and B). Therefore, TNKS1-siRNA1 was selected for subsequent experiments.
Cell Viability
MTT assays showed that the viability of U251 and U87 cells was significantly higher and lower after transfection with TNKS1 overexpression vector and TNKS1-siRNA (P < 0.05, Figure 2A and B), respectively.
 |
Figure 2 Viability of U251 and U87 cells following transfection with overexpression and siRNA vectors: (A) U251 cells and (B) U87 cells. * denotes P < 0.05 vs control. |
Apoptosis
Flow cytometry studies showed that U251 and U87 cells had significantly lower and higher apoptosis after transfection with TNKS1 overexpression vector and TNKS1-siRNA (P < 0.05, Figure 3A–D), respectively. Caspase 3/7 activity assay showed that the activity was significantly lower and higher after transfection with TNKS1 overexpression vector and TNKS1-siRNA (P < 0.05, Figure 4A and B)
Cell Cycle
Flow cytometry studies also showed that there were less U251 cells in G1 stage and more in S stage following transfection with TNKS1 overexpression vector. On the other hand, TNKS1-siRNA transfection resulted in more U251 and U87 cells in G1 stage and less in S stage (P < 0.05, Figure 5A–D).
Cell Invasion Ability
Tranwell assays showed that U251 and U87 cells had increased invasion ability following transfection with TNKS1 overexpression vector and the ability was reduced after transfection with TNKS1-siRNA (P < 0.05, Figure 6A–D).
Gene Expression
We then examined the protein expression of TNKS1, cyclinD1, MMP-7, β-catenin, p-β-catenin and c-Myc in U251 and U87 cells using Western blot analysis. The results showed that compared with the control, cells transfected with TNKS1 overexpression vector increased the levels of TNKS1, cyclinD1, MMP-7, β-catenin, p-β-catenin and c-Myc, while these proteins were all down-regulated following TNKS1-siRNA1 treatment (P < 0.05, Figure 7A–D).
Interaction Between TNKS1and β-Catenin
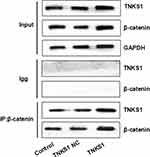
Co-immunoprecipitation assays showed that β-catenin antibody was able to detect TNKS1 in U251 cell lysates before transfection or transfected with empty vector or TNKS1 overexpression vector (Figure 8).
Discussion
Glioma is a type of tumor that starts in the glial cells of the brain or the spine.21 Gliomas comprise about 30% of all brain tumors and central nervous system tumors, and 80% of all malignant brain tumors.22 Astrocytoma is the most common type of glioma. The tumor does not have obvious envelope and invasively grows in the white matter. It is believed that the malignant progression of astrocytoma is likely to involve multiple gene mutations, abnormal activation of cell signaling pathways, inhibition of apoptosis pathway, and cell immortalization. Therefore, gene therapy has become an important therapeutic option for astrocytoma.23
TNKS1 is a member of the PARP family, consisting of four structurally unique domains: the HPS domain, the anchor protein repeat domain, the SAM domain and the PARP domain from the N-terminal to the C-terminal. In the PARP domain, polyADP ribose in NAD+ can be added to the receptor protein to ribosylate poly adenosine diphosphate, which is a transient post-translational modification. Ribosylated poly adenosine diphosphate participates in various physiological activities, such as DNA replication, repair, gene expression, chromosome separation, cell malignant transformation, cell differentiation and apoptosis. Therefore, the active domain is the most important domain in TNKS1. TNKS1 relies on the domain to play a variety of important biological functions, such as the positive regulation of telomere length by binding with TRF1 to ribosylate polyadenosine diphosphate.24 TNKS1 is widely expressed in human tissues and is highly conserved in mammalian.25 Recent studies have shown that TNKS1 is abnormally and highly expressed in malignant tumor cells such as malignant hematopoietic cell tumors.12–18 Inhibiting the PARP activity of TNKS1 and blocking the expression of TNKS1 gene are shown to have anti-tumor activity,26,27 which suggest that TNKS1 is involved in the occurrence and development of tumor and TNKS1 can be used as a molecular target for gene therapy of cancer. In addition, TNKS1 is shown to regulate Wnt/β-catenin pathways through its PARP activity. TNKS1 and TNSK2 bind to the tankyrase-binding domain (TBD) of Axin to glycosylate polyadenosine diphosphate ribose, resulting in degradation of Axin by the ubiquitin-proteasome system. Once Axin is degraded, the β-catenin destruction complex is degraded and cytoplasmic unphosphated β-catenin is accumulated and translocated to nuclei to form a complex with transcriptional factor TCF/LEF that triggers the expression of downstream target genes and activate the Wnt/-catenin pathway.11
Our results show that overexpression of TNKS1 increased the level of the TNKS1 protein, while siRNA decreased the level of the protein in U251 and U87 cells, indicating that the expression of the gene was successfully manipulated in U251 and U87 cells. Matsutani et al found that the expression of TNKS1 mRNA was up-regulated in gastric cancer tissue,28 which was confirmed by Gao et al29 at both mRNA and protein levels. And they further demonstrated that the level expression of TNKS1 is related to tissue differentiation and tumor staging. In this study, cells transfected with TNKS1 overexpression vector were found to significantly increase cell viability, while TNKS1-siRNA significantly decreased the viability. These results suggest that TNKS1-siRNA may be used to slow down the growth of U251 and U87 cells and reduce their proliferation ability. Since we also found that Caspases 3/7 activity is increased by TNKS1-siRNA and decreased by TNKS1 overexpression, the increased Caspases 3/7 activity is likely the reason that leads to increased apoptosis of U251 and U87 cells after TNKS1 knockdown.
A major characteristic of malignant astrocytoma is that it grows invasively in the white matter, and is therefore difficult to resect completely, resulting in frequent relapse easily and poor prognosis.30,31 Therefore, inhibiting the invasion ability of the tumor cells is of great significance in improving the therapeutic effect of astrocytoma. Our study shows that the number of invaded cells decreased significantly after silencing the TNKS1 gene, suggesting that siRNA-mediated silencing of the TNKS1 gene may effectively inhibit the invasiveness of U25l cells. Since the invasion assays might be influenced by the cell number, we used the same number of viable cells in the assays to minimize the impact. Therefore, the changes in the invaded cells in the invasion assay are a reliable reflection of invasion ability. Furthermore, TNKS1-siRNA treatment generated more cells arrested at G1 stage and less in S1 stage. Reduced cells at S1 stage imply less DNA synthesis in the cells and slower cell proliferation. On the other hand, TNKS1 overexpression vector gave rise to the opposite results. To further validate the results, we determined the apoptosis of U251 and U87 cells following up- and down-regulation of TNKS1 and found that apoptosis was reduced or increased when TNKS1 was up-regulated or silenced with siRNA, respectively, suggesting that silencing of TNKS1 gene may induce apoptosis of U251 and U87 cells.
Studies have shown that TNKS1 participates in the Wnt/β-catenin signaling pathway and plays an important regulatory role in the occurrence and development of tumors.32 Wnt/β-catenin signaling pathway plays an important role in a variety of physiological behaviors,33,34 and abnormal activation or mutation of the Wnt/β-catenin signaling pathway is closely related to the occurrence and development of astrocytoma.5,9 Our co-immunoprecipitation assays confirmed the interaction between TNKS1 and β-catenin. Furthermore, we showed that the overexpression of TNKS1 resulted in increased β-catenin and p-β-catenin levels, while these levels were reduced once TNKS1 was silenced, suggesting that TNKS1 may be a positive regulator in Wnt/β-catenin signaling pathway. High-level TNKS1 might prevent the degradation of β-catenin by its degradation complex, leading to the abnormal accumulation of β-catenin in the cytoplasm and translocation into the nucleus, where it activates the downstream target genes to initiate Wnt/β-catenin signaling pathway, leading to the occurrence of the tumor. Once Wnt/β-catenin signaling pathway is activated, the expressions of the downstream target genes cyclinD1, MMP-7 and c-Myc are induced, leading to abnormal proliferation, apoptosis and differentiation of the cells, and the formation of tumors.35,36 Up-regulation of cyclinD1 and c-Myc expression accelerates cell proliferation through the interaction with cyclin-dependent kinases (CDKs) in G1 stage and up-regulation of MMP-7 promotes cell invasion by degrading extracellular matrix.37 We found that silencing TNKS1 inhibited the Wnt/β-catenin signaling pathway, thereby inhibiting the expression of cyclinD1, MMP-7 and c-Myc genes. Huang et al found that small molecule inhibitor XAV939 effectively inhibits the clonal proliferation of Wnt/β-catenin pathway dependent-colon cancer cell line DLD-1.11 Kaner et al showed that TNKS1 plays an important role in the development of rat kidneys as a regulatory factor of the Wnt/β-catenin pathway.38 Bao et al demonstrated that after silencing of TNKS1 with siRNA, Wnt/β-catenin pathway is inactivated in breast cancer cell line.39 These results are consistent with our findings, confirming that TNKS1 is a positive regulator of the Wnt/β-catenin signaling pathway.
Conclusion
We have demonstrated that TNKS1 is a positive regulator of the Wnt/β-catenin signaling pathway, silencing of TNKS1 inhibits the growth, reduces the invasion ability of U251 and U87 cells with increased apoptosis via suppression of Wnt/β-catenin signaling pathway. Therefore, TNKS1 may be a new target of gene therapy for astrocytoma.
Availability of Data and Material
Data are available from the authors upon reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Fuller GN, Scheithauer BW. The 2007 revised World Health Organization (WHO) classification of tumours of the central nervous system: newly codified entities. Brain Pathol. 2007;17(3):304–307. doi:10.1111/j.1750-3639.2007.00084.x
2. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi:10.1056/NEJMra0708126
3. Collins VP. Brain tumours: classification and genes. J Neurol Neurosurg Psychiatry. 2004;75 Suppl 2(Suppl 2):ii2–ii11. doi:10.1136/jnnp.2004.040337
4. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi:10.1016/j.cell.2006.10.018
5. Yu JM, Jun ES, Jung JS, et al. Role of Wnt5a in the proliferation of human glioblastoma cells. Cancer Lett. 2007;257(2):172–181. doi:10.1016/j.canlet.2007.07.011
6. Pu P, Zhang Z, Kang C, et al. Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther. 2009;16(4):351–361. doi:10.1038/cgt.2008.78
7. Sareddy GR, Panigrahi M, Challa S, Mahadevan A, Babu PP. Activation of Wnt/beta-catenin/Tcf signaling pathway in human astrocytomas. Neurochem Int. 2009;55(5):307–317. doi:10.1016/j.neuint.2009.03.016
8. Yang Z, Wang Y, Fang J, et al. Expression and aberrant promoter methylation of Wnt inhibitory factor-1 in human astrocytomas. J Exp Clin Cancer Res. 2010;29:26. doi:10.1186/1756-9966-29-26
9. Zhang LY, Jiang LN, Li FF, et al. Reduced beta-catenin expression is associated with good prognosis in astrocytoma. Pathol Oncol Res. 2010;16(2):253–257. doi:10.1007/s12253-009-9219-0
10. Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90(1):83–92. doi:10.1016/j.biochi.2007.07.012
11. Huang SM, Mishina YM, Liu S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi:10.1038/nature08356
12. Xu D, Zheng C, Bergenbrant S, et al. Telomerase activity in plasma cell dyscrasias. Br J Cancer. 2001;84(5):621–625. doi:10.1054/bjoc.2000.1655
13. MacNamara B, Wang W, Chen Z, et al. Telomerase activity in relation to pro- and anti-apoptotic protein expression in high grade non-Hodgkin’s lymphomas. Haematologica. 2001;86(4):386–393.
14. Klapper W, Krams M, Qian W, Janssen D, Parwaresch R. Telomerase activity in B-cell non-hodgkin lymphomas is regulated by hTERT transcription and correlated with telomere-binding protein expression but uncoupled from proliferation. Br J Cancer. 2003;89(4):713–719. doi:10.1038/sj.bjc.6601112
15. Gelmini S, Poggesi M, Distante V, et al. Tankyrase, a positive regulator of telomere elongation, is over expressed in human breast cancer. Cancer Lett. 2004;216(1):81–87. doi:10.1016/j.canlet.2004.05.010
16. Gelmini S, Poggesi M, Pinzani P, et al. Distribution of Tankyrase-1 mRNA expression in colon cancer and its prospective correlation with progression stage. Oncol Rep. 2006;16(6):1261–1266.
17. Gelmini S, Quattrone S, Malentacchi F, et al. Tankyrase-1 mRNA expression in bladder cancer and paired urine sediment: preliminary experience. Clin Chem Lab Med. 2007;45(7):862–866. doi:10.1515/CCLM.2007.133
18. Shervington A, Patel R, Lu C, et al. Telomerase subunits expression variation between biopsy samples and cell lines derived from malignant glioma. Brain Res. 2007;1134(1):45–52. doi:10.1016/j.brainres.2006.11.093
19. Tang B, Wang J, Fang J, et al. Expression of TNKS1 is correlated with pathologic grade and Wnt/beta-catenin pathway in human astrocytomas. J Clin Neurosci. 2012;19(1):139–143. doi:10.1016/j.jocn.2011.08.013
20. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
21. Mamelak AN, Jacoby DB. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601). Expert Opin Drug Deliv. 2007;4(2):175–186. doi:10.1517/17425247.4.2.175
22. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi:10.1016/j.cancergen.2012.10.009
23. Norden AD, Wen PY. Glioma therapy in adults. Neurologist. 2006;12(6):279–292. doi:10.1097/01.nrl.0000250928.26044.47
24. Zhu L, Smith S, de Lange T, Seldin MF. Chromosomal mapping of the tankyrase gene in human and mouse. Genomics. 1999;57(2):320–321. doi:10.1006/geno.1999.5771
25. Scherthan H, Jerratsch M, Li B, et al. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell. 2000;11(12):4189–4203. doi:10.1091/mbc.11.12.4189
26. Zou LH, Shang ZF, Tan W, et al. TNKS1BP1 functions in DNA double-strand break repair though facilitating DNA-PKcs autophosphorylation dependent on PARP-1. Oncotarget. 2015;6(9):7011–7022. doi:10.18632/oncotarget.3137
27. Johannes JW, Almeida L, Daly K, et al. Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering. Bioorg Med Chem Lett. 2015;25(24):5743–5747. doi:10.1016/j.bmcl.2015.10.079
28. Matsutani N, Yokozaki H, Tahara E, et al. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int J Oncol. 2001;19(3):507–512.
29. Gao J, Zhang J, Long Y, Tian Y, Lu X. Expression of tankyrase 1 in gastric cancer and its correlation with telomerase activity. Pathol Oncol Res. 2011;17(3):685–690. doi:10.1007/s12253-011-9369-8
30. Merzak A, Koocheckpour S, Pilkington GJ. CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res. 1994;54(15):3988–3992.
31. Wick W, Platten M, Weller M. Glioma cell invasion: regulation of metalloproteinase activity by TGF-beta. J Neurooncol. 2001;53(2):177–185. doi:10.1023/A:1012209518843
32. Tian X, Bai S, Fan J. Down-regulation of tankyrase 1 mediated by RNAi leads to the apoptosis of SH-SY5Y cells. Acta Anatom Sin. 2014;45(2):196–203.
33. Liang S, Zhang S, Wang P, et al. LncRNA, TUG1 regulates the oral squamous cell carcinoma progression possibly via interacting with Wnt/beta-catenin signaling. Gene. 2017;608:49–57. doi:10.1016/j.gene.2017.01.024
34. McCracken KW, Aihara E, Martin B, et al. Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541(7636):182–187. doi:10.1038/nature21021
35. Zhou Y, Tao H, Li Y, et al. Berberine promotes proliferation of sodium nitroprusside-stimulated rat chondrocytes and osteoarthritic rat cartilage via Wnt/beta-catenin pathway. Eur J Pharmacol. 2016;789:109–118. doi:10.1016/j.ejphar.2016.07.027
36. Shin S, Im HJ, Kwon YJ, et al. Human steroid sulfatase induces Wnt/beta-catenin signaling and epithelial-mesenchymal transition by upregulating Twist1 and HIF-1alpha in human prostate and cervical cancer cells. Oncotarget. 2017;8(37):61604–61617. doi:10.18632/oncotarget.18645
37. Mao XW, Xiao JQ, Xu G, et al. CUL4B promotes bladder cancer metastasis and induces epithelial-to-mesenchymal transition by activating the Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8(44):77241–77253. doi:10.18632/oncotarget.20455
38. Karner CM, Merkel CE, Dodge M, et al. Tankyrase is necessary for canonical Wnt signaling during kidney development. Dev Dyn. 2010;239(7):2014–2023. doi:10.1002/dvdy.22340
39. Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Inhibition of tankyrases induces axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS One. 2012;7(11):e48670. doi:10.1371/journal.pone.0048670
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.