Back to Journals » International Journal of Nanomedicine » Volume 11
Biofabrication of a novel biomolecule-assisted reduced graphene oxide: an excellent biocompatible nanomaterial
Authors Zhang X, Gurunathan S
Received 8 September 2016
Accepted for publication 15 October 2016
Published 8 December 2016 Volume 2016:11 Pages 6635—6649
DOI https://doi.org/10.2147/IJN.S121847
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Xi-Feng Zhang,1 Sangiliyandi Gurunathan2
1College of Biological and Pharmaceutical Engineering, Wuhan Polytechnic University, Wuhan, People’s Republic of China; 2Department of Stem Cell and Regenerative Biotechnology, Konkuk University, Seoul, Republic of Korea
Abstract: Graphene has been shown much interest, both in academics and industry due to its extraordinary physical, chemical, and biological proprieties. It shows great promises in biotechnological and biomedical applications as an antibacterial and anticancer agent, nanocarrier, sensor, etc. However, many studies demonstrated the toxicity of graphene in several cell lines, which is an obstacle to its use in biomedical applications. In this study, to improve the biocompatibility of graphene, we used nicotinamide (NAM) as a reducing and stabilizing agent to catalyze the reduction of graphene oxide (GO) to reduced graphene oxide (rGO). The resulted smaller-sized GO (NAM-rGO) showed excellent biocompatibility with mouse embryonic fibroblast cells, evidenced by various cellular assays. Furthermore, NAM-rGO had no effect on mitochondrial membrane permeability and caspase-3 activity compared to GO. Reverse transcription polymerase chain reaction analysis allowed us to identify the molecular mechanisms responsible for NAM-rGO-induced biocompatibility. NAM-rGO significantly induced the expression of genes encoding tight junction proteins (TJPs) such as zona occludens-1 (Tjp1) and claudins (Cldn3) without any effect on the expression of cytoskeleton proteins. Furthermore, NAM-rGO enhances the expression of alkaline phosphatase (ALP) gene, and it does this in a time-dependent manner. Overall, our study depicted the molecular mechanisms underlying NAM-rGO biocompatibility depending on upregulation of TJPs and ALP. This potential quality of graphene could be used in diverse applications including tissue regeneration and tissue engineering.
Keywords: biocompatibility, graphene oxide, nicotinamide, reduced graphene oxide, tight junction proteins, alkaline phosphatase
Introduction
Graphene is a monolayer of carbon atoms composed of sp2-hybridized carbon atoms tightly packed into a two-dimensional honeycomb structure. Interest in graphene among several other nanomaterials is rapidly growing in the field of nanotechnology, including in the fields of nanomedicine, regenerative medicine, and biomedical applications. Graphene possesses unique and exceptional physical and chemical properties such as high electrical conductance, high fracture strength, thermal conductivity, mechanical strength and stiffness, unique optical properties, extreme chemical stability, fast mobility of charge carriers, large surface area, elasticity, biocompatibility, cytocompatibility, and degradability.1–8 Besides these characteristics, graphene has other fascinating properties, including high tunable amphiphilicity, photoconversion efficiency, excellent drug loading capacity, flexibility in size, and low cytotoxicity used for nanomaterial-based biomedical applications such as cancer and stem cell therapies.1–12
Recently, both graphene oxide (GO) and reduced graphene oxide (rGO) have been used in several biomedical applications.13 GO can be prepared in bulk quantities through extensive oxidation/exfoliation of graphite.14 On the other hand, rGO presents enhanced optical, electrical, and biological properties compared to GO; therefore, the use of rGO has immensely increased. Various methods have been developed, including physical, chemical, and biological approaches, to reduce GO to rGO. Although chemical methods are simple, they require the use of strong reducing agents like hydrazine, of which the trace residue may strongly decrease the performance of rGO-based devices and be harmful to living organisms.13,15,16 Moreover, chemically reduced rGO shows limited solubility as well as formation of irreversible agglomerates during preparation in water and most organic solvents.17 To replace chemical reducing agents, many environmentally friendly and biocompatible reductants have been developed and used for the reduction of GO, including vitamins,18 amino acids,19 reducing sugars,20 tea,21 lysozyme,22 melatonin,23 humanin,24 bacteria,25–27 fungi,6 plant extracts,16,28 as well as enhanced green fluorescent protein (EGFP)29 and a polyphenol compound, resveratrol.30
The nanotoxicity of graphene and GO with different sizes, shapes, composition, and surface functionalization has been studied in various cell lines. Exposure of human neuronal cells (PC12 cells) to different sizes of rGO caused an increase in the activation of caspase-3, release of lactate dehydrogenase, and generation of reactive oxygen species (ROS).31 The cell viability of human lung cells (BEAS-2B) exposed to graphene was significantly decreased in a concentration- and time-dependent manner.32 Exposure of PC12 cells to grapheme resulted in mitochondrial injury that was dose and shape dependent.31 Exposure of blood platelets to rGO caused a strong cumulative response and extensive pulmonary thromboembolism33 and, similarly, exposure of alveolar macrophages and epithelial cells to grapheme generated ROS and led to inflammation, apoptosis, and an increased rate of mitochondrial respiration.34 Our previous studies showed that both GO and rGO induced oxidative stress-mediated cell death in various cell lines, including human breast cancer cell lines MDA-MB-23128 and MCF-7,5 and human ovarian cancer cell line A2780.30
The toxicity of graphene and GO hinders their application in various areas. To minimize toxicity and increase biocompatibility, several approaches have been developed in many laboratories.35 However, the toxicity versus biocompatibility depends on several physicochemical properties, including lateral size, shape, thickness, stiffness, and surface properties,36 the concentration and time of exposure to cells, dispersibility, and the mode of exposure.37 Among these, surface functionalization is essential to determine the toxicity or biocompatibility of the compound. For example, Singh et al38 proved that the biocompatible effect of amine-modified graphene on platelets was likely due to the modification of the surface charges. Aggarwal et al39 demonstrated that PEGylation was another alternative process to modify nanomaterials to reduce nonspecific protein adsorption, preventing flocculation and opsonization as well as the subsequent complement activation. Besides PEGylation, chemical functionalization, or polymer modifications, biological reducing agents have been explored for the development of biocompatible materials using the presence of capping functionalities on the graphene surface.39 Our recent studies suggested that using biological molecules as reducing agents could enhance the biocompatibility of graphene. For instance, rGO synthetized by using trimethylamine and microbial and spinach leaf extracts showed significant biocompatibility with primary mouse embryonic fibroblast cells (MEFs),16 and EGFP-mediated reduction of GO showed excellent biocompatibility with human embryonic kidney 293 cells.29 Studies from intraocular indicated that GO had significant biocompatibility with minimum effects on cell morphology, cell viability, membrane integrity, and apoptosis.40 Glucose-functionalized graphene was biocompatible with higher photothermal therapy.15 GO presented dose-dependent cytotoxicity in human umbilical vein endothelial cells compared to rGO. No toxicity was observed when cells were exposed to doses >100 μg/mL of functionalized rGO.41 The water-soluble oxidized graphene nanoribbons mixed with PEG-DSPE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)]) showed significantly different cytotoxic profile compared to graphene nanoparticles.42 The effects of various two-dimensional graphene nanostructures such as graphene nano-onions, graphene oxide nanoribbons, and graphene oxide nanoplatelets on cell viability and differentiation were evaluated with human mesenchymal stem cells. The results indicated that the cytotoxicity of the two-dimensional graphene nanostructures was dose dependent, not time dependent when the concentration was <50 μg/mL.43 Lalwani et al44 extensively reviewed the toxicological perspectives of graphene both in vitro and in vivo, with a specific focus on the complex interplay of biological responses to graphene.
Because of its excellent properties, graphene has been explored in the area of regenerative medicine. Graphene and its derivatives were used as nanoplatforms for promoting cell proliferation, cell adhesion, and differentiation of various embryonic stem cells (ESCs).45 Although several studies showed the biocompatibility of graphene using different biological models, only few studies reported the biocompatibility of rGO-assisted biomolecules in MEFs to date. In this study, we explored a novel biological molecule called nicotinamide (NAM), a water-soluble amide form of niacin (vitamin B3) involved in energy metabolism. NAM presents antioxidative properties in various types of eukaryotic cells. Therefore, in this study, we utilized this new biomolecule for the synthesis of graphene in potato sprout extracts. The biocompatibility of the resulting rGO was evaluated in MEFs. MEFs could form a link between mesenchymal cells and ESCs because of their ES-like features, in concurrence to developmental hierarchy and the multipotential nature, and are widely used as feeder cells to support the growth of ESCs.46 The objective of this study was to synthesize and characterize the graphene derivative from potato sprout extracts and to investigate the biocompatibility of the resulting rGO with primary MEFs.
Materials and methods
Materials
NAM, penicillin–streptomycin solution, trypsin–EDTA solution, Dulbecco’s Modified Eagle’s Medium (DMEM), RPMI 1,640 medium, and 1% antibiotic–antimycotic solution were obtained from Life Technologies/Gibco (Grand Island, NY, USA). Polyethylene, fetal bovine serum, and the in vitro toxicology assay kit were purchased from Sigma-Aldrich (St Louis, MO, USA). Graphite powder, NaOH, KMnO4, NaNO3, anhydrous ethanol, 98% H2SO4, 36% HCl, 30% H2O2 aqueous solution, and all other chemicals were purchased from Sigma-Aldrich unless otherwise stated. Cell viability assay WST-8, ROS assay kit, 2′,7′-dichlorofluorescin diacetate, and lactate dehydrogenase (LDH) Cytotoxicity Detection Kit were purchased from Dojindo, Sigma, and Takara (Tokyo, Japan), respectively.
GO synthesis, reduction, and characterization
GO was synthesized as described previously with modifications.14,30,47 The reduction of GO was accomplished as described earlier.28,30 Briefly, NAM-rGO was obtained from the reaction of NAM with GO. In a typical reduction experiment, 10 mL NAM (1 mM) was added to 90 mL of 1.0 mg/mL aqueous GO. The mixture was stirred at 40°C for 6 hours. Subsequently, using a magnetostirrer heater, the rGO suspension was stirred at 400 rpm at a temperature of 30°C for 30 minutes. A homogeneous NAM-rGO suspension was obtained without aggregation. The obtained material was washed with distilled water several times to remove the excess NAM residue and redispersed in water by sonication. The suspension was centrifuged at 5,000 rpm for 30 minutes. The final product was collected by vacuum filtration and was vacuum dried. GO and NAM-rGO were characterized according to the methods described previously.28,30,47
Cell viability and cytotoxicity assay
MEF-1 cells were obtained from Sigma-Aldrich. The cell viability and LDH and ROS generation assays were performed as described previously.5,28,30 Briefly, 1×104 cells/well were seeded in a 96-well plate and cultured in DMEM supplemented with 10% fetal bovine serum at 37°C in 5% CO2. The cells were washed twice with 100 μL of serum-free DMEM and incubated with different concentrations of GO and NAM-rGO (0–50 μg/mL). After 24 hours of exposure, the cells were washed twice with serum-free DMEM, and then 100 μL/well of serum-free DMEM was loaded into each well and 15 μL/well of WST-8 solution was added to every well. After 1 hour of incubation at 37°C with 5% CO2, 80 μL/well of the mixture (WST-8 and DMEM) was transferred to a new 96-well plate and the absorbance was measured at 450 nm using a microplate reader.
The membrane integrity of MEFs cells were evaluated according to the manufacturer’s instructions by LDH Cytotoxicity Detection Kit (Takara). Briefly, cells were exposed to various concentrations of GO or NAM-rGO for 24 hours, then the cell-free supernatant was transferred into a 96-well plates (100 μL/well) and 100 μL of the LDH reaction mixture was added to each well. After 3 hours of incubation under standard conditions, the optical density of the solution was determined using a microplate reader at 490 nm wavelength. Three independent experiments were performed in triplicate.
ROS were estimated according to a method described previously.5,28,30 The cells were seeded into a 24-well plate at a density of 5×104 cells/well and cultured for 24 hours. After washing twice with phosphate-buffered saline (PBS), fresh media containing respective concentrations of GO and NAM-rGO were added and the mixtures incubated for 24 hours. The cells were supplemented with 20 μM DCFH-DA and then incubated for 30 minutes at 37°C. The cells were rinsed with PBS once, 2 mL of new PBS solution was added to each well, and the fluorescent intensity was determined using a spectrofluorometer (Gemini EM) with excitation at 485 nm and emission at 530 nm.
Cell proliferation assay
Cell proliferation was evaluated using the trypan blue assay as described earlier.5 PMEF cells were plated in 6-well plates (1×105 cells/well) and incubated for 24 hours. GO or NAM-rGO (10 μg/mL) was added and the cells were incubated for 5 days. Cells cultured in the medium without Go or NAM-rGO were used as controls. Twenty-four hours later, cells were detached with 300 μL trypsin–EDTA solution and both adherent and cells in suspension were collected. The mixture of the supernatant and detached cells was centrifuged at 1,200 rpm for 5 minutes. The pellet was mixed with 700 μL of trypan blue solution. After 5 minutes of staining, cells were counted using a cytometer. The viable cells were unstained, whereas dead cells were stained in blue. Three independent experiments were performed in triplicate. The mean and standard deviation were calculated. Cell proliferation was expressed as the percentage of viable cells.
Mitochondrial membrane potential (MMP)
The MMP was measured according to manufacturer instructions (Molecular Probes, Eugene, OR, USA) as described previously48–50 using a cationic fluorescent indicator JC-1 (Molecular Probes). Ovarian cancer cells and OvCSCs were incubated with 10 μM JC-1 at 37°C for 15 minutes, washed with PBS, resuspended in new PBS solution, and then the fluorescence intensity was measured. MMP was expressed as the ratio of the fluorescence intensity of the JC-1 aggregates to that of monomers.
Real-time qRT-PCR
Total RNA was extracted from cells treated with GO and NAM-rGO using an Arcturus PicoPure RNA isolation kit (EBioscience, San Diego, CA, USA) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using Reverse Transcription Kit (Roche, Basel, Switzerland) in a final volume of 20 μL according to the manufacturer’s instructions. The quantification of gene transcripts (Tjp1, Cldn3, Actr2, Actr3, Tuba1a, and Tuba1b) was carried out in triplicate by real-time qRT-PCR using Lightcycler®FastStart DNA Master SYBR Green I by an ABI Applied Biosystems machine. The primer sequences for each gene are as follows: Tjp1 (5′-CCTGTGAAGCGTCACTGTGT; 3′-CGCGGAGAGAGACAAGATGT), Cldn3 (5′-GCAAGCAGACTGTGTGTCGT; 3′-TACCGTCACCACTACCAGCA), Actr2 (5′-GCTGGGAAGATGTGTTCTGG; 3′-GAACCATGGACAGCCAGG), Actr3 (5′-TGGCAATACATGATGGGATG; 3′-GCCTGTGTGGTGGACTGTG), Tuba1a (5′-CTGTGGAAAACCAAGAAGCC; 3′-CACTACACCATTGGCAAGGA), Tuba1b (5′-AGTAGAGCTCCCAGCAGGC; 3′-TCTCACCCTCGCCTTCTAAC), ALPP (5′ GCTGGCAGTGGTCAGATGTT 3′ CTATCCTGGCTCCGTGCTC), GAPDH (5′ AATCCCATCACCATCTTCCAG, 3′ AAATGAGCCCCAGCCTTC).
The real time gene expression was quantified and analyzed by real-time qRT-PCR method. Target gene expression levels were normalized to GAPDH, which was unaffected by NAM-rGO treatment. Real-time qRT-PCR was performed independently in triplicate for each sample and the data were presented as the mean values of the gene expression levels measured in the NAM-rGO-treated samples versus the controls.
Alkaline phosphatase (ALP) activity
ALP activity was measured according to a method described earlier.6 MEF cells were cultured in 48-well culture dishes at a density of 5×103 cells/well and cultured for 5 days. Then, medium was replaced with treatment medium, which was DMEM containing 5% serum plus GO or NAM-rGO. After 5 days, the ALP activity was measured according to the manufacturer’s instructions (QuantiChrom™ DALP-250 Alkaline Phosphatase Assay Kit; Gentaur, Aachen, Germany).
Statistical analyses
All assays were conducted in triplicate, and each experiment was repeated at least three times. The results are presented as mean ± standard deviation. All experimental data were compared using Student’s t-test. A P-value <0.05 was considered statistically significant.
Results
Synthesis and characterization of NAM-rGO
GO was prepared using graphite powder by a modified Hummers’ method. The prepared GO showed a characteristic UV–vis absorption spectrum at 230 nm, characterized by the π–π* of the C=C plasmon peak about 230 nm and a shoulder about 300 nm, which is often attributed to n–π* transitions of the carbonyl groups,47,51 whereas reduced GO by NAM showed a peak at about 260 nm (Figure 1A). The electronic conjugation was restored after reduction, indicated by the red shift of the peak. It suggests that the GO could be reduced by NAM and the aromatic structure might be gradually restored. Similar findings have been observed using several biological materials as reducing agents.
The X-ray powder diffraction (XRD) patterns of pure GO and NAM-rGO are shown in Figure 1B. After oxidation of graphite, the 002 reflection peak of GO was shifted to the lower angle at 2θ =11.70° (corresponding to a d-spacing of 0.76 nm), compared to the typical peak of pristine graphite exhibited at 2θ =26.6° (d-spacing =0.335 nm), indicating that the d-spacing was increased due to the intercalation of oxygen functionalities in between the basal plane of the graphite. Interestingly, the 002 reflection peak disappeared in the reduced graphene. After reduction by NAM, the broad peak was observed at 2θ about 26.2°, indicating the presence of stacked graphene layers. This is an indication for the formation of a few layers of graphene.
Fourier transform infrared (FTIR) spectroscopy was performed to determine the biologically mediated reduction of GO. As shown in Figure 1C (top panel), the interesting peaks were observed in the spectrum of GO at 3,443 and 1,067 cm−1, due to the presence of O–H stretching vibration. The peak at 1,734 cm−1 is attributed to C=O bonds in carboxylic acid and carbonyl moieties, and 1,179 and 850 cm−1 for asymmetric stretching and bending of –C–O–C– (epoxy) vibrations, respectively. The observed strong peaks at 1,179, 1,067, 850, and 578 cm−1 are due to C–OH stretching, C–O stretching, C–H in plane bending, and possibly C–H out-of-plane wagging vibrations, respectively.52 Interestingly, after NAM reduction, the stretching vibration of C=O disappeared, and other oxygen-containing functional groups in GO were significantly decreased due to the deoxygenation process (Figure 1C, bottom panel).30,53
The size of nanoparticles is an important aspect in nanoparticle toxicity studies. Therefore, we measured the size of GO and NAM-rGO in aqueous dispersions using dynamic light scattering at the scattering angle θ =90°. The size of GO (Figure 1D, left panel) and NAM-rGO (Figure 1D, right panel) was 100 and 120 nm, respectively. NAM-rGO shows a slight increase in size. The surface morphology of GO and NAM-rGO was assessed using scattering electron microscopy (SEM). Figure 1E shows the SEM images of GO and NAM-rGO. GO presented a flaky texture with a relatively large surface area, and their morphology resembled a thin curtain, which suggests the significant exfoliation of graphite during the oxidation process. NAM-rGO showed a relatively wavy, silver-line, leaf-like structure, and a wrinkled and closed surface morphology, indicating that rGO flakes overlapped rather than aggregated.
Raman spectroscopy is a noninvasive technique used for characterizing crystal structure and defects in graphene-based materials.53,54 Figure 1F shows the Raman spectra of GO and rGO. The D peak at 1,363 cm−1 was for GO and at 1,342 cm−1 was for rGO, which was streamed from a defect-induced breathing mode of sp2 rings, and the G peak at about 1,612 cm−1 was for GO and that at 1,583 cm−1 was for NAM-rGO, which resulted from the first-order scattering of the E2g phonon of sp2 C atoms.53,54 The D/G intensity ratio for rGO is larger than that for GO (1.74 for rGO and 1.01 for GO). This suggests that new (or more) graphitic domains are formed and the sp2 cluster number is increased.28 Raman spectrum of the NAM-rGO film showed an increased ratio of D over G band (ID/IG) compared to that of GO (ID/IG) and the appearance of a new two-dimensional band at 2,700 cm−1. This change clearly indicates that the reduction of GO by a biological molecule resulted in a large number of new graphitic domains of smaller dimensions. Moreover, the peak intensities and shift of the D and G bands of NAM-rGO were attributed to the surface-enhanced Raman scattering of rGO.
Effect of NAM-rGO on cell viability, proliferation, and morphology
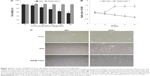
The biocompatibility effect of NAM-rGO was evaluated by assessing the viability and proliferation potential of MEFs using WST-8 and trypan blue exclusion assays, respectively. The results suggest that there was a significant reduction in mitochondrial function when the cells were treated with GO, whereas NAM-rGO significantly enhanced the effect on cell viability up to 20 μg/mL (Figure 2A). The cells treated with GO showed a dose-dependent loss of cell viability, whereas NAM-rGO was more biocompatible in nature. Next, we examined whether a lower concentration of NAM-rGO had a significant effect on proliferation or not. Interestingly, when the cells were treated with 10 μg/mL of NAM-rGO, there was no significant loss in cell number; instead, at lower concentrations, NAM-rGO treatment resulted in an increase in the cell number (Figure 2B). To confirm the cell viability and proliferation data, we determined the effect of GO and NAM-rGO treatments on cell morphology. NAM-rGO did not affect the cell morphology when compared with GO (Figure 2C).
Cytotoxicity of NAM-rGO in MEFs
Cytotoxicity assays such as LDH leakage and ROS generation are frequently used to detect early cytotoxic events. Cytotoxicity has been observed when cells were incubated with different concentrations of GO (Figure 3A). No cytotoxicity was observed when cells were treated with different concentrations of NAM-rGO (Figure 3B). LDH leakage was notable even after exposure of MEFs cells to 20 μg/mL GO. Lower concentrations of GO were enough to induce this effect when the cells were incubated for 24 hours. In contrast, no apparent cytotoxicity was observed when the cells were incubated with NAM-rGO at similar concentrations and for similar periods. Finkel and Holbrook55 reported that ROS was involved in the regulation of various cellular events such as cell growth and cell signaling. GO and rGO induced intracellular ROS production, which implicated it as a mediator of cytotoxicity in cancer cells.56 To investigate the effect of NAM-rGO on intracellular ROS generation in MEFs cells, we measured the intracellular ROS levels after GO and NAM-rGO exposure. GO treatment led to a significant increase in intracellular ROS generation in a dose-dependent manner compared with that in the control groups. Conversely, NAM-rGO had no significant effect on ROS levels (Figure 3B).
Effect of NAM-rGO on mitochondrial dysfunction
Mitochondria are not only the source of energy but also the source of signals that initiate apoptotic cell death; it also is a center of cell metabolism and energy transformation, and so malfunction of mitochondria decreases cell viability.57–59 A decrease in MMP is a cause for apoptosis. A decrease in MMP leads to matrix condensation and exposure of cytochrome c, which plays an important role in cytochrome c-mediated apoptotic cell death.58 Therefore, we examined the effect of NAM-rGO on MMP. Interestingly, NAM-rGO had no significant effect on MMP, whereas GO had a rapid depolarization effect, which was the initial event for apoptosis. These results suggest that NAM could abrogate the effect of GO on the MMP (ΔΨm), which was an early event in GO-induced apoptosis. GO-treated cells showed a JC-1 green/red fluorescence intensity ratio higher than that of control cells. In contrast, NAM-rGO had no effect on the depolarization of the mitochondrial membrane in MEFs (Figure 4A).
Caspase-3 is mainly associated with the mitochondrial pathway. To evaluate the biocompatibility of NAM-rGO, cells were treated with GO and NAM-rGO (10 μg/mL). Interestingly, NAM-rGO had no direct effect on caspase-3 activity, whereas a known inhibitor of caspase-3, Ac-VAD-CMK, a positive control, completely blocked caspase 3 activity (Figure 4B). GO had a significant effect on caspase-3 activity. These results are in-line with those of previous studies performed on ovarian cancer cells using toxic concentrations.30 Similar to other rGO that was produced using microbial biomass, spinach leaf extract, and EGFP, NAM-rGO was considerably biocompatible in the 10–50 μg/mL concentration range.16,26,29
Effect of NAM-rGO on expression of genes encoding proteins involved in tight junctions and the cytoskeleton
To assess the influence of NAM-rGO on tight junctions and cytoskeleton expression, we examined the expression of genes encoding for zonula occludens (ZO) protein-1 (Tjp1) and Cldn3, which is a membrane-associated component of both tight and adherent junctions detected at sites of cell–cell contact. These genes are expressed in epithelia and endothelia and form paracellular barriers and pores that determine tight junction permeability.60,61 Furthermore, we examined genes encoding for cytoskeleton proteins such as ACTB (Actr2, Actr3, Tuba1a, and Tuba1b). Interestingly, NAM-rGO increased the gene levels of the tight junction proteins (TJPs) Tjp1 and Cldn3; however, no significant change was observed for Actr2, Actr3, Tuba1a, and Tuba1b mRNA levels compared to that in the control. Surprisingly, a significant reduction was observed in the expression levels of Actr2, Actr3, Tuba1a, and Tuba1b in GO-treated cells (Figure 5). These results suggest that GO affected the expression of cytoskeleton proteins, resulting in the induction of apoptosis. These results may be associated with the biocompatibility of NAM-rGO rather than with apoptosis.
Effect of NAM-rGO on expression and activity of ALP
In stem cell biology, the measurement of expression of ALP is important. ALP, a marker for differentiation, seems to be a key player of the early-stage differentiation of bone marrow stromal cells, and the expression of ALP is gradually decreased when the cells are subjected to the differentiation process.6 To corroborate the results from cell viability and proliferation, we measured the expression of ALP using RT-PCR. Figure 6A shows the expression of mRNA level of ALP in MEFs treated with GO and NAM-rGO for 5 days. It was clear from these results that low- and high-level expression was associated with GO and NAM-rGO effects, respectively. The expression of ALP appeared to be influenced by NAM-rGO in a positive manner. We observed that the ALP mRNA level was clearly induced by the NAM-rGO compared to that in control. The expression level of ALP was increased in a time-dependent manner. This indicates the ALP mRNA level, which was stimulated by NAM-rGO.
To establish the direct relationship between mRNA level and protein, we measured the specific activity of ALP in cells treated with GO or NAM-rGO for 5 days. Cells treated with GO showed significantly lower ALP activity than that in control as well as in NAM-rGO-treated cells (Figure 6B), which indicated that NAM-rGO was significantly biocompatible. The results were consistent with functional relationship between gene expression and protein. These data suggest the biocompatible nature of NAM-rGO.
Discussion
Graphene and graphene-related materials offer numerous applications in biotechnology and biomedical engineering. Graphene could be considered a wonder material due to its excellent properties compared to those of conventional nanomaterials. However, the mechanistic effects of biologically synthesized graphene in MEFs remain elusive. Therefore, we designed a simple, environment-friendly, fast, cost-effective, and biocompatible method for the synthesis of highly soluble graphene using a novel biomolecule, NAM, and examined the functional properties of graphene in MEFs. To achieve our goal, the synthesized graphene materials were characterized using various analytical techniques such as UV–vis, XRD, FTIR, dynamic light scattering, SEM, and Raman spectroscopy. The synthesized graphene presented characteristic features similar to those of chemically synthesized graphene. For instance, the UV–vis spectrum showed the characteristic red shift at 260 nm. XRD revealed the close d-spacing of NAM-rGO to pristine graphite and disappearance of the peak at 2θ =11.7°, indicating that the oxygen-containing group of GO was efficiently removed during the synthesis of rGO using proteins.24,29 FTIR spectra indicated that a variety of the oxygen-containing functional groups were significantly removed from the GO via biological reduction.26,62,63 The electronic properties of chemically converted materials seemed to be low due to residual defects.62 Furthermore, the major advantage of NAM-rGO was its size, which was about 100 nm. The quantum confinement effect was observed with graphene quantum dots of ≤100 nm.64 The quantum confinement is observed when the particle is too small to be comparable to the wavelength of electron. Therefore, when compared to bulk materials, the reduced size of graphene nanomaterials might have the significant quantum confinement effect, which can be used in all-graphene electronically coherent devices. The data from Raman spectroscopy suggested that the formation of defects supported the biofunctionalization of graphene as reported earlier using Baker’s yeast, humanin, EGFP, and resveratrol.24,29,30,63
The results from cell viability and proliferation assays indicated that the biologically prepared NAM-rGO showed excellent biocompatibility, which was in-line with a previous study reported by Ryoo et al,65 who showed that graphene and carbon nanotubes exhibited a biocompatible effect. As reported earlier, the toxicity or biocompatibility of graphene and graphene derivatives depended on the concentration, dose, solubility, and physical and chemical properties of the compound16,44,66 in both prokaryotes and eukaryotes. Toxicity has been exploited for various biomedical applications such as antibacterial, antiviral, and anticancer effects, drug delivery, and diagnosis. Biocompatibility effects have been exploited for tissue engineering. The behavior of the cells not only depend on physicochemical parameters like wettability and surface-free energy but also on surface roughness.67
Cytotoxicity assays were performed to assess the toxicity of the synthetized compounds and for the general screening of nanomaterials. LDH and ROS generation confirmed the toxic potential of GO and the biocompatibility of NAM-rGO in MEFs. Several studies reported that both GO and rGO could induce the generation of ROS.30,31,68,69 ROS is involved in various signaling pathways including proliferation, metabolic regulation, promoting survival and cell death, the activation of the antioxidant response, the control of iron metabolism, and Ca(2+) signaling.68 Sublethal concentrations of ROS have been involved in the regulation of gene expression, cell growth, and cell differentiation.70,71 NAM-rGO induced the generation of ROS at sublethal levels, which could be involved in promoting cell survival. Liao et al37 reported that a stronger hemolytic activity was observed in GO-treated red blood cells than aggregated graphene sheets, whereas compacted graphene sheets were more damaging to mammalian fibroblasts than less densely packed GO. Another study indicated that lower doses (0.5–31.3 ppm) of graphene had no effect on multiple end points such as metabolic activity, LDH leakage, and ROS production in neuronal cells (PC12).72 Taken together, the end points of all cytotoxicity assays showed that NAM-rGO up to 50 μg/mL had no harmful effect on cells.
The release of cytochrome c from the mitochondrial intermembrane space induces the assembly of the apoptosome, which in turn activates the downstream caspases.73–75 Graphene nanoparticles can induce the dissipation of the MMP, which subsequently increases the generation of intracellular ROS and eventually triggers apoptosis by activating the mitochondrial pathway.76,77 The biocompatible effect of NAM-rGO, which could be associated with its pro-oxidant activity, is responsible for its inhibitory effect on apoptosis by creating a nonpermissive intracellular milieu for caspase activation. Similarly, caspase 3 activation and apoptosis induced by serum deprivation was protected by resveratrol in primary MEFs.78
TJPs serve as a barrier to the diffusion of water and solutes through the paracellular pathway by sealing the intercellular space in epithelial cells.60,61 Our findings indicated that the upregulation of Tjp1 and Cldn3 mRNA by NAM-rGO in MEFs cells may be required for the formation of tight junctions by epithelial cells during normal cell maintenance. It could also play an important role in the differentiation of epithelial cells. Ko et al79 reported that upregulation of ZO-1, occludin, and claudin mRNA expression in human corneal fibroblasts was involved in normal cell maintenance and differentiation. It could also favor the healing of corneal epithelial wounds. Previous studies demonstrated that low concentrations of silver nanoparticles rescued vascular endothelial growth factor and advanced glycation end-products induced vascular permeability through upregulation of ZO-1 and occludin80,81 in porcine retinal endothelial cells. Our data are consistent with previous reports demonstrating that tight junction is important for proper cell function, which can be maintained by the treatment of cells with NAM-rGO.
Cytoskeleton proteins are involved in cell viability, motility, and migration and play a vital role in most cellular processes. Previous studies demonstrated that Actg1−/− MEFs were mildly growth impaired due to reduced cell survival.82 Interestingly, NAM-rGO had no effect on the expression of cytoskeleton genes, whereas GO significantly reduced the expression level of Actr2, Actr3, Tuba1a, and Tuba1b. It indicated that GO resulted in a decrease in cell viability due to the downregulation of cytoskeleton genes. Our results are in-line with a previous study demonstrating that a decrease in β-actin gene expression or the absence of β-actin monomers hinders actin cytoskeleton polymerization, which, in turn, results in a decrease in motility, migration, and proliferation of corneal fibroblasts.83 RT-PCR analysis suggests that a decrease in cytoskeleton gene expression by GO in MEFs could be involved in the disruption of the cytoskeletal structure and functions, including a decrease in cell viability, proliferation, and enhanced apoptosis.
To correlate the enhanced level of expression of TJPs such as TJP1 and Cldn3 with ALP activity in MEFs, we determined both gene expression and protein expression of ALP in GO- and NAM-rGO-treated cells. We found that the presence of NAM-rGO resulted in significant increases in the expression of ALP and genes encoding for the junctional proteins, TJP1 and Cldn3. These results suggest that NAM-rGO plays an important role in the regulation of junctional protein expression and ALP activity. Consistent with our results, recently, Liu et al84 demonstrated that the absence of IAP results in lower levels of the junctional proteins ZO-1, ZO-2, and Occludin in human colon cancer Caco-2 and T84 cells. However, higher IAP levels in human cells are associated with an increased expression of ZO-1 and ZO-2. These findings suggest that the ALP and TJPs might be working together. Downregulation of TJP is associated with many diseases.85 Therefore, maintaining the structure and integrity of TJP is an important factor for paracellular permeability. Therefore, NAM-rGO can be used as scaffolding material for tissue engineering as well as a regulator for TJP levels to maintain the structure and integrity of the membrane.
Several studies reported that GO prepared from graphite by the oxidation method using chemicals containing many oxygen atoms in the forms of carboxyl groups, epoxy groups, and hydroxyl groups86 induced toxicity in various types of cancer cells5,30 and fibroblasts.16 In contrast, the biocompatibility effect of NAM-rGO was enhanced due to the lack of oxides or other functional groups. Our studies are consistent with previous reports demonstrating that biopolymer-functionalized rGO exhibits an ultralow hemolysis ratio and significant cytocompatibility in human umbilical vein endothelial cells, even at a high concentration of 100 μg/mL.29,41 Altogether, these data suggest that graphene can be intrinsically nontoxic, with its toxicity potential only appearing after chemical treatment or increased concentration, incubation time, or size. Besides these factors, surface functionalization is an alternative and suitable approach to improve the biocompatibility of nanomaterials for safer biomedical applications.87 In addition, surface chemistry, surface energy, and hydrophobicity are the major factors controlling the biocompatibility.88 A recent study suggested that smaller particle size and higher oxidation improved the biocompatibility of graphene-based materials.89
Conclusion
We described a simple, easy, green, and biocompatible method for the preparation of smaller size of graphene using a novel biomolecule called NAM. The resulting product (NAM-rGO) shows significant deoxygenation by NAM. NAM-rGO shows significant biocompatibility behavior with MEFs. The cells treated with NAM-rGO have a positive effect on cell viability and proliferation, whereas there is no significant effect on cytotoxicity and mitochondrial dysfunction. Interestingly, NAM-rGO enhanced the expression of genes responsible for TJPs and a differentiation marker called ALP. We conclude that the smaller size of graphene is not only an alternative biocompatibility agent to maintain cell viability and proliferation but that it can also be used to maintain the structure and integrity of TJPs, which play an important role in paracellular permeability. These data provide a strong foundation to develop NAM-rGO as a novel therapy to maintain barrier function. Therefore, NAM-functionalized graphene may represent a safer and excellent biocompatible nanomaterial for future biomedical applications; particularly it can be used as the next scaffolding agent for tissue engineering. However, further studies are required to explain the molecular mechanisms of biocompatibility of NAM-rGO, which would allow not only establishing the biocompatible effect but would also facilitate their proper use in biomedical applications.
Acknowledgments
This study was supported by the KU-Research Professor Program of Konkuk University and by the Science and Technology Research Program of the Department of Education of Hubei Province, People’s Republic of China (D20151701).
Disclosure
The authors report no conflicts of interest in this work.
References
Balandin AA, Ghosh S, Bao WZ, et al. Superior thermal conductivity of single-layer graphene. Nano Lett. 2008;8(3):902–907. | ||
Gardin C, Piattelli A, Zavan B. Graphene in regenerative medicine: focus on stem cells and neuronal differentiation. Trends Biotechnol. 2016;34(6):435–437. | ||
Geim AK, Novoselov KS. The rise of graphene. Nat Mater. 2007;6(3):183–191. | ||
Gomez-Navarro C, Weitz RT, Bittner AM, et al. Electronic transport properties of individual chemically reduced graphene oxide sheets. Nano Lett. 2007;7(11):3499–3503. | ||
Gurunathan S, Han JW, Eppakayala V, Kim JH. Green synthesis of graphene and its cytotoxic effects in human breast cancer cells. Int J Nanomed. 2013;8:1015–1027. | ||
Gurunathan S, Han JW, Park JH, Eppakayala V, Kim JH. Ginkgo biloba: a natural reducing agent for the synthesis of cytocompatible graphene. Int J Nanomedicine. 2014;9:363–377. | ||
Reina G, Tamburri E, Orlanducci S, et al. Nanocarbon surfaces for biomedicine. Biomatter. 2014;4:e28537. | ||
Stankovich S, Dikin DA, Piner RD, et al. Synthesis of graphene-based nanosheets via chemical reduction of exfoliated graphite oxide. Carbon. 2007;45(7):1558–1565. | ||
Chung C, Kim YK, Shin D, Ryoo SR, Hong BH, Min DH. Biomedical applications of graphene and graphene oxide. Acc Chem Res. 2013;46(10):2211–2224. | ||
Dong H, Li Y, Yu J, et al. A versatile multicomponent assembly via beta-cyclodextrin host-guest chemistry on graphene for biomedical applications. Small. 2013;9(3):446–456. | ||
Kim TH, Lee T, El-Said WA, Choi JW. Graphene-based materials for stem cell applications. Materials. 2015;8(12):8674–8690. | ||
Patel SC, Lee S, Lalwani G, Suhrland C, Chowdhury SM, Sitharaman B. Graphene-based platforms for cancer therapeutics. Ther Deliv. 2016;7(2):101–116. | ||
Turcheniuk K, Boukherroub R, Szunerits S. Gold-graphene nanocomposites for sensing and biomedical applications. J Mater Chem B. 2015;3(21):4301–4324. | ||
Hummers WS, Offeman RE. Preparation of graphitic oxide. J Am Chem Soc. 1958;80:1339. | ||
Akhavan O, Ghaderi E, Aghayee S, Fereydooni Y, Talebi A. The use of a glucose-reduced graphene oxide suspension for photothermal cancer therapy. J Mater Chem. 2012;22(27):13773–13781. | ||
Gurunathan S, Han JW, Eppakayala V, Dayem AA, Kwon DN, Kim JH. Biocompatibility effects of biologically synthesized graphene in primary mouse embryonic fibroblast cells. Nanoscale Res Lett. 2013;8(1):393. | ||
Gurunathan S, Han JW, Park JH, et al. Reduced graphene oxide-silver nanoparticle nanocomposite: a potential anticancer nanotherapy. Int J Nanomed. 2015;10:6257–6276. | ||
Fernandez-Merino MJ, Guardia L, Paredes JI, et al. Vitamin C is an ideal substitute for hydrazine in the reduction of graphene oxide suspensions. J Phys Chem C. 2010;114(14):6426–6432. | ||
Gao J, Liu F, Liu YL, Ma N, Wang ZQ, Zhang X. Environment-friendly method to produce graphene that employs vitamin C and amino acid. Chem Mater. 2010;22(7):2213–2218. | ||
Zhu CZ, Guo SJ, Fang YX, Dong SJ. Reducing sugar: new functional molecules for the green synthesis of graphene nanosheets. ACS Nano. 2010;4(4):2429–2437. | ||
Wang Y, Shi ZX, Yin J. Facile synthesis of soluble graphene via a green reduction of graphene oxide in tea solution and its biocomposites. ACS Appl Mater Interfaces. 2011;3(4):1127–1133. | ||
Yang F, Liu YQ, Gao LA, Sun J. pH-Sensitive highly dispersed reduced graphene oxide solution using lysozyme via an in situ reduction method. J Phys Chem C. 2010;114(50):22085–22091. | ||
Esfandiar A, Akhavan O, Irajizad A. Melatonin as a powerful bio-antioxidant for reduction of graphene oxide. J Mater Chem. 2011;21(29):10907–10914. | ||
Gurunathan S, Han J, Kim JH. Humanin: a novel functional molecule for the green synthesis of graphene. Colloid Surf B. 2013;111:376–383. | ||
Akhavan O, Ghaderi E. Escherichia coli bacteria reduce graphene oxide to bactericidal graphene in a self-limiting manner. Carbon. 2012;50(5):1853–1860. | ||
Gurunathan S, Han JW, Eppakayala V, Kim JH. Microbial reduction of graphene oxide by Escherichia coli: a green chemistry approach. Colloid Surf B. 2013;102:772–777. | ||
Salas EC, Sun ZZ, Luttge A, Tour JM. Reduction of graphene oxide via bacterial respiration. ACS Nano. 2010;4(8):4852–4856. | ||
Gurunathan S, Han J, Park JH, Kim JH. An in vitro evaluation of graphene oxide reduced by Ganoderma spp. in human breast cancer cells (MDA-MB-231). Int J Nanomed. 2014;9:1783–1797. | ||
Gurunathan S, Han JW, Kim E, Kwon DN, Park JK, Kim JH. Enhanced green fluorescent protein-mediated synthesis of biocompatible graphene. J Nanobiotechnol. 2014;12:41. | ||
Gurunathan S, Han JW, Kim ES, Park JH, Kim JH. Reduction of graphene oxide by resveratrol: a novel and simple biological method for the synthesis of an effective anticancer nanotherapeutic molecule. Int J Nanomed. 2015;10:2951–2969. | ||
Zhang Y, Ali SF, Dervishi E, et al. Cytotoxicity effects of graphene and single-wall carbon nanotubes in neural phaeochromocytoma-derived PC12 cells. ACS Nano. 2010;4(6):3181–3186. | ||
Vallabani NVS, Mittal S, Shukla RK, et al. Toxicity of graphene in normal human lung cells (BEAS-2B). J Biomed Nanotechnol. 2011;7(1):106–107. | ||
Singh SK, Singh MK, Nayak MK, et al. Thrombus inducing property of atomically thin graphene oxide sheets. ACS Nano. 2011;5(6):4987–4996. | ||
Duch MC, Budinger GR, Liang YT, et al. Minimizing oxidation and stable nanoscale dispersion improves the biocompatibility of graphene in the lung. Nano Lett. 2011;11(12):5201–5207. | ||
Xu M, Zhu JQ, Wang FF, et al. Improved in vitro and in vivo biocompatibility of graphene oxide through surface modification: poly(acrylic acid)-functionalization is superior to PEGylation. ACS Nano. 2016;10(3):3267–3281. | ||
Yang K, Feng LZ, Shi XZ, Liu Z. Nano-graphene in biomedicine: theranostic applications. Chem Soc Rev. 2013;42(2):530–547. | ||
Liao KH, Lin YS, Macosko CW, Haynes CL. Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts. ACS Appl Mater Interfaces. 2011;3(7):2607–2615. | ||
Singh SK, Singh MK, Kulkarni PP, Sonkar VK, Gracio JJA, Dash D. Amine-modified graphene. Thrombo-protective safer alternative to graphene oxide for biomedical applications. ACS Nano. 2012;6(3):2731–2740. | ||
Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Deliv Rev. 2009;61(6):428–437. | ||
Yan L, Wang Y, Xu X, et al. Can graphene oxide cause damage to eyesight? Chem Res Toxicol. 2012;25(6):1265–1270. | ||
Cheng C, Nie SQ, Li S, et al. Biopolymer functionalized reduced graphene oxide with enhanced biocompatibility via mussel inspired coatings/anchors. J Mater Chem B. 2013;1(3):265–275. | ||
Mullick Chowdhury S, Lalwani G, Zhang K, Yang JY, Neville K, Sitharaman B. Cell specific cytotoxicity and uptake of graphene nanoribbons. Biomaterials. 2013;34(1):283–293. | ||
Talukdar Y, Rashkow JT, Lalwani G, Kanakia S, Sitharaman B. The effects of graphene nanostructures on mesenchymal stem cells. Biomaterials. 2014;35(18):4863–4877. | ||
Lalwani G, D’Agati M, Khan AM, Sitharaman B. Toxicology of graphene-based nanomaterials. Adv Drug Deliv Rev. 2016;105(Pt B):109–144. | ||
Bressan E, Ferroni L, Gardin C, et al. Graphene based scaffolds effects on stem cells commitment. J Transl Med. 2014;12. | ||
Yusuf B, Gopurappilly R, Dadheech N, Gupta S, Bhonde R, Pal R. Embryonic fibroblasts represent a connecting link between mesenchymal and embryonic stem cells. Dev Growth Differ. 2013;55(3):330–340. | ||
Gurunathan S, Han JW, Dayem AA, Eppakayala V, Kim JH. Oxidative stress-mediated antibacterial activity of graphene oxide and reduced graphene oxide in Pseudomonas aeruginosa. Int J Nanomed. 2012;7:5901–5914. | ||
Gurunathan S, Jeong JK, Han JW, Zhang XF, Park JH, Kim JH. Multidimensional effects of biologically synthesized silver nanoparticles in Helicobacter pylori, Helicobacter felis, and human lung (L132) and lung carcinoma A549 cells. Nanoscale Res Lett. 2015;10:1–17. | ||
Sareen D, van Ginkel PR, Takach JC, et al. Mitochondria as the primary target of resveratrol-induced apoptosis in human retinoblastoma cells. Invest Ophthalmol Vis Sci. 2006;47(9):3708–3716. | ||
White RJ, Reynolds IJ. Mitochondrial depolarization in glutamate-stimulated neurons: an early signal specific to excitotoxin exposure. J Neurosci. 1996;16(18):5688–5697. | ||
Luo ZT, Lu Y, Somers LA, Johnson ATC. High yield preparation of macroscopic graphene oxide membranes. J Am Chem Soc. 2009;131(3):898. | ||
Acik M, Lee G, Mattevi C, Chhowalla M, Cho K, Chabal YJ. Unusual infrared-absorption mechanism in thermally reduced graphene oxide. Nat Mater. 2010;9(10):840–845. | ||
Choi YJ, Kim E, Han J, Kim JH, Gurunathan S. A novel biomolecule-mediated reduction of graphene oxide: a multifunctional anti-cancer agent. Molecules. 2016;21(3):375. | ||
Ferrari AC, Meyer JC, Scardaci V, et al. Raman spectrum of graphene and graphene layers. Phys Rev Lett. 2006;97(18):187401. | ||
Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. | ||
Gurunathan S, Han JW, Kim JH. Green chemistry approach for the synthesis of biocompatible graphene. Int J Nanomed. 2013;8:2719–2732. | ||
Gottlieb RA. Role of mitochondria in apoptosis. Crit Rev Eukar Gene. 2000;10(3–4):231–239. | ||
Gottlieb RA. Mitochondrial signaling in apoptosis: mitochondrial daggers to the breaking heart. Basic Res Cardiol. 2003;98(4):242–249. | ||
Gogvadze V, Orrenius S. Mitochondrial regulation of apoptotic cell death. Chem-Biol Interact. 2006;163(1–2):4–14. | ||
Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141(7):1539–1550. | ||
Smalley KSM, Brafford P, Haass NK, Brandner JM, Brown E, Herlyn M. Up-regulated expression of zonula occludens protein-1 in human melanoma associates with N-cadherin and contributes to invasion and adhesion. Am J Pathol. 2005;166(5):1541–1554. | ||
Guo HL, Wang XF, Qian QY, Wang FB, Xia XH. A green approach to the synthesis of graphene nanosheets. ACS Nano. 2009;3(9):2653–2659. | ||
Khanra P, Kuila T, Kim NH, Bae SH, Yu DS, Lee JH. Simultaneous bio-functionalization and reduction of graphene oxide by baker’s yeast. Chem Eng J. 2012;183:526–533. | ||
Shen H, Zhang LM, Liu M, Zhang ZJ. Biomedical applications of graphene. Theranostics. 2012;2(3):283–294. | ||
Ryoo SR, Kim YK, Kim MH, Min DH. Behaviors of NIH-3T3 fibroblasts on graphene/carbon nanotubes: proliferation, focal adhesion, and gene transfection studies. ACS Nano. 2010;4(11):6587–6598. | ||
Akhavan O, Ghaderi E. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano. 2010;4(10):5731–5736. | ||
Ponsonnet L, Reybier K, Jaffrezic N, et al. Relationship between surface properties (roughness, wettability) of titanium and titanium alloys and cell behaviour. Mat Sci Eng C Bio S. 2003;23(4):551–560. | ||
Lammel T, Boisseaux P, Fernandez-Cruz ML, Navas JM. Internalization and cytotoxicity of graphene oxide and carboxyl graphene nanoplatelets in the human hepatocellular carcinoma cell line Hep G2. Part Fibre Toxicol. 2013;10:27. | ||
Martinez-Reyes I, Cuezva JM. The H(+)-ATP synthase: a gate to ROS-mediated cell death or cell survival. Biochimica et Biophysica Acta. 2014;1837(7):1099–1112. | ||
Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82(1):47–95. | ||
Hancock JT, Desikan R, Neill SJ. Role of reactive oxygen species in cell signalling pathways. Biochem Soc T. 2001;29:345–350. | ||
Hong SW, Lee JH, Kang SH, et al. Enhanced neural cell adhesion and neurite outgrowth on graphene-based biomimetic substrates. Biomed Res Int. 2014;2014:212149. | ||
Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15(22):2922–2933. | ||
Adrain C, Creagh EM, Martin SJ. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 2001;20(23):6627–6636. | ||
Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407(6805):770–776. | ||
Li Y, Liu Y, Fu Y, et al. The triggering of apoptosis in macrophages by pristine graphene through the MAPK and TGF-beta signaling pathways. Biomaterials. 2012;33(2):402–411. | ||
Sasidharan A, Panchakarla LS, Sadanandan AR, et al. Hemocompatibility and macrophage response of pristine and functionalized graphene. Small. 2012;8(8):1251–1263. | ||
Ahmad KA, Clement MV, Pervaiz S. Pro-oxidant activity of low doses of resveratrol inhibits hydrogen peroxide-induced apoptosis. Ann N Y Acad Sci. 2003;1010:365–373. | ||
Ko JA, Liu Y, Yanai R, Chikama TI, Takezawa T, Nishida T. Upregulation of tight-junctional proteins in corneal epithelial cells by corneal fibroblasts in collagen vitrigel cultures. Invest Ophth Vis Sci. 2008;49(1):113–119. | ||
Sheikpranbabu S, Kalishwaralal K, Lee KJ, Vaidyanathan R, Eom SH, Gurunathan S. The inhibition of advanced glycation end-products-inuced retinal vascular permeability by silver nanoparticles. Biomaterials. 2010;31(8):2260–2271. | ||
Sheikpranbabu S, Kalishwaralal K, Venkataraman D, Eom SH, Park J, Gurunathan S. Silver nanoparticles inhibit VEGF-and IL-1beta-induced vascular permeability via Src dependent pathway in porcine retinal endothelial cells. J Nanobiotechnol. 2009;7:8. | ||
Bunnell TM, Ervasti JM. Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton. 2010;67(9):564–572. | ||
Joseph R, Srivastava OP, Pfister RR. Downregulation of beta-actin and its regulatory gene HuR affect cell migration of human corneal fibroblasts. Mol Vis. 2014;20:593–605. | ||
Liu W, Hu D, Huo H, et al. Intestinal alkaline phosphatase regulates tight junction protein levels. J Am Coll Surg. 2016;222(6):1009–1017. | ||
Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. | ||
Dreyer DR, Park S, Bielawski CW, Ruoff RS. The chemistry of graphene oxide. Chem Soc Rev. 2010;39(1):228–240. | ||
Nel AE, Madler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. | ||
Sanchez VC, Jachak A, Hurt RH, Kane AB. Biological interactions of graphene-family nanomaterials: an interdisciplinary review. Chem Res Toxicol. 2012;25(1):15–34. | ||
Pinto AM, Goncalves C, Sousa DM, et al. Smaller particle size and higher oxidation improves biocompatibility of graphene-based materials. Carbon. 2016;99:318–329. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.