Back to Archived Journals » Research and Reports in Biology » Volume 7
Binding of MCM-interacting proteins to ATP-binding site in MCM6
Authors Hosoi A, Sakairi T, Yukio I
Received 11 September 2015
Accepted for publication 15 December 2015
Published 9 March 2016 Volume 2016:7 Pages 31—40
DOI https://doi.org/10.2147/RRB.S96215
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Zvi Kelman
Atsutoshi Hosoi, Taku Sakairi, Yukio Ishimi
Graduate School of Science and Engineering, Ibaraki University, Mito, Ibaraki, Japan
Abstract: The function of MCM2–7 complex that is a DNA helicase in DNA replication may be regulated by various MCM-interacting proteins, including CDC45, RPA, TIM, TIPIN, Claspin, MCM10, and MCM-BP. It has been shown by immunoprecipitation that human MCM6 interacts with all these proteins in coexpressed insect cells. To determine the region in MCM6 to interact with these proteins, we prepared various truncated forms of MCM6 and examined the interaction of these MCM6 fragments with the MCM-interacting proteins. All these proteins bound to C-terminal half of MCM6, and CDC45, RPA2, TIM, TIPIN, MCM-BP, and MCM10 bound to the fragments containing ATP-binding motifs. CDC45 and RPA2 bound to the smallest fragment containing Walker motif A. Only MCM-BP is bound to the N-terminal half of MCM6. Site-directed mutagenesis study suggests that hydrophobic interaction is involved in the interaction of MCM6 with CDC45 and TIM. These results suggest a possibility that MCM-interacting proteins regulate MCM2–7 function by modulating the ATP-binding ability of the MCM2–7.
Keywords: DNA helicase, DNA replication, checkpoint, MCM2–7 proteins
Introduction
It is probable that MCM2–7 complex functions as a DNA helicase that unwinds duplex DNA at the DNA replication forks.1–4 The function of MCM2–7 is regulated by various proteins. CDC45 binds to MCM2–7 with GINS complex to form CMG complex, and the cumulative data suggest that the CMG complex functions as a DNA helicase at the forks.5–8 MCM10, which may function to coordinate DNA unwinding and DNA synthesis by interacting with the CMG complex and DNA polymerase α,9,10 is required for the activation of CMG complex at replication origins.11–13 RPA, a single-stranded DNA-binding protein, is required for the progression of DNA replication fork by stabilizing single-stranded region. TIM, TIPIN, and Claspin, which are involved in checkpoint signaling, are required for stabilizing stalled DNA replication forks probably by inhibiting MCM2–7 function.14–20 MCM-BP, which is required for sister-chromatid cohesion,21,22 may be involved in detaching MCM2–7 complex from the forks at the termination of DNA replication.23,24 Thus, it is evident that MCM2–7 function is regulated with various MCM-interacting proteins, but the molecular details of the regulation are not well understood.
All the MCM2–7 members contain conserved ATP-binding motifs, including Walker A and B and sensors I and II in the central region,25 and each member-specific region is present near N- and C-terminus. The N-terminal half-region containing a Zn-finger motif of MCM from Archaea is involved in the assembly of MCM into hexamer.26 MCM2–7 heterohexamer from Saccharomyces cerevisiae exhibits DNA helicase activity under the selected conditions,27 and human CMG complex exhibits robust DNA helicase in vitro.6 MCM4/6/7 trimer, which consists of a core structure of MCM2–7 complex, forms a dimer, and the resultant MCM4/6/7 hexamer exhibits DNA helicase activity.28,29 The results indicate that ATP-binding sites in MCM4, 6, and 7 proteins play different roles in exhibiting the DNA helicase activity and high-affinity binding of MCM6 with ATP is required for the activity. It is possible that MCM4, 6, and 7 proteins play a central role in the exhibition of DNA helicase activity of MCM2–7 complex. We systematically examined the interaction of MCM2–7 proteins with MCM-interacting proteins by immunoprecipitation using extracts prepared from the insect cells where one human MCM protein is overexpressed with one MCM-interacting protein.30–32 The MCM-interacting proteins specifically interacted with the MCM2–7 proteins. However, they all interacted with MCM6 in these systems.
In this article, we fragmented human MCM6 protein and used these MCM6 fragments in immunoprecipitation experiments to determine the region required for interaction with MCM-interacting proteins. The results indicate that all the MCM-interacting proteins bind to the C-terminal half-fragment of MCM6. CDC45, RPA2, TIM, TIPIN, MCM-BP, and MCM10 bound to a smaller fragment containing ATP-binding motifs. CDC45 and RPA2 bound to the smallest fragment containing Walker motif A. These results suggest that the MCM-interacting proteins regulate MCM2–7 function by modulating the ATP-binding ability of MCM2–7.
Materials and methods
Preparation of MCM6 fragments
Various human MCM6 cDNA fragments were amplified by polymerase chain reaction (Toyobo, Japan), where nucleotides generating a myc-tag were attached to forward primer and they were cloned into pVL1392 between EcoRI and BamHI sites. The use of the human material in this study was approved by the Ibaraki University Bioethics Committee. They were N1+C2 (amino acids 1–572), N1+490 (1–490), N1 (1–325), C1 (326–821), C2 (326–572), C3 (573–821), and C4 (326–413). MCM2 fragments (469–618 and 469–672) were also cloned with nucleotides for a flag-tag in forward primer and a myc-tag in reverse primer. The DNAs purified by endotoxin-free system (Qiagen) were cotransfected with linealized baculovirus DNA into Sf9 cells, according to the procedures described by Pharmingen (BaculoGold) or AB vector (ProEasy). Site-directed mutagenesis in the C2 fragment was performed by using QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies). The primer for converting leucine to alanine at amino acid 370 was CGGGGTGTCCTGGCGATGCTCTTTGGTG, that for converting leucine to alanine at 429 was CAGTGCTGCTGGCGCAACAGCAGCTGTTG and that for converting leucine to alanine at 449 was GAGGCTGGAGCTGCGATGTTGGCTGATAATGG.
Interaction of MCM-interacting proteins with MCM2 and MCM6 fragments
We examined interactions of MCM-interacting proteins with MCM2 and MCM6 fragments by immunoprecipitation using anti-MCM-interacting protein antibodies.32 Typically, 100 µL of recombinant baculovirus stock solution for expression of MCM-interacting proteins and 200 µL of stock solution for expression of the MCM2 or MCM6 protein fragments were added to High5 cells (4×106). The cells were cultured for 2 days at 27°C. They were suspended in 200 µL of lysis buffer containing 10 mM Tris-HCl (pH 7.5), 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM sodium phosphate buffer, 10 mM Na4P2O7, and protease inhibitor cocktails (8 µg/mL benzamidine-HCl, 5 µg/mL phenanthroline, 5 µg/mL aprotinin, 5 µg/mL leupeptin, 5 µg/mL pepstatin A, and 0.5 mM phenylmethylsulfonyl fluoride). The mixture was incubated for 30–45 minutes on ice, and insoluble components were separated by centrifugation at 14,000 rpm for 20 minutes at 4°C. The supernatant containing Triton-soluble proteins (S) and Triton-insoluble proteins (P) was recovered. The recovered supernatant was mixed for 2 hours ~overnight at 4°C with protein G-Sepharose-antibody beads that had been prepared by incubation with the culture supernatant (1.2 mL) of hybridoma cells producing anti-MCM-interacting protein antibodies with protein G-Sepharose (30 µL) (Amersham Biosciences Corp.) overnight at 4°C. The supernatant after centrifugation was recovered (U). The beads were washed seven times with 200 µL of phosphate-buffered saline containing 0.05% Triton X-100, and the supernatant was recovered after a final spin was recovered (F). Proteins bound to the beads were eluted three times with 30 µL of elution buffer (0.1 M glycine, pH 2.5, and 0.15 M NaCl) (E1, 2, and 3). The eluate was neutralized by addition of 1/10 volume of 2 M Tris-HCl, pH 8.0. Proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. After proteins in the gel were transferred to an Immobilon-P transfer membrane (EMD Millipore, Billerica, MA, USA), the membrane was incubated for 1 hour at room temperature with blocking buffer (Block Ace; Yukijirusi Co. Ltd., Sapporo, Japan) diluted threefold with Tris-buffered saline (50 mM Tris–HCl, pH 7.5, 150 mM NaCl); the membrane was then incubated overnight at 4°C with 0.1–0.2 µg/mL of primary antibody or 3,000-fold diluted antiserum in blocking buffer (EzBlock; ATTO, Tokyo, Japan) diluted sixfold with Tris-buffered saline containing 0.1% Triton X-100. To detect MCM-interacting proteins, a tenfold diluted culture supernatant of hybridoma cells was used. After the membrane was washed with Tris-buffered saline containing Triton X-100, it was incubated for 2 hours at 27°C with secondary antibody conjugated to horseradish peroxidase (Bio-Rad Laboratories Inc., Hercules, CA, USA). The membrane was then washed and incubated with EzWestLumi plus (ATTO), and chemiluminescent signals were detected using Light-Capture (ATTO). The signals were detected using a reagent of EzWest Blue (ATTO). Antibodies against Claspin (4D9),30 TIM (7D2),32 TIPIN (13B4),31 CDC45 (21F4 and 35C9),30 MCM-BP (18A6),30 and MCM10 (1E5 and 3E8)30 were used. Anti-MCM3, MCM4, and MCM6 antiserum were prepared as reported.24 Anti-myc (sc-40; 9E10; Santa Cruz Biotechnology Inc., Dallax, TX, USA) and anti-flag (M2; Sigma-Aldrich Co., St Louis, MO, USA) antibodies were purchased. Anti-RPA2 mouse antibody (9H9-H4) was kindly provided by J Hurwitz in the Memorial Sloan Kettering Cancer Center.
Purification of proteins and their binding
Human His-MCM2/MCM3/His-MCM4/His-MCM5/MCM6/His-MCM7 complex was purified from coexpressed High5 cells as reported.24 Flag-CDC4530 and Flag-MCM-BP24 were purified as reported from the lysate of infected High5 cells. In 100 µL of phosphate-bufferred saline, ~1 µg of CDC45 or MCM-BP was mixed with 1 µ g of MCM2–7 and incubated for 1–2 hours at 4°C. Protein G beads premixed with anti-CDC45 or anti-MCM-BP antibodies were added to the reaction, and the incubation was continued for 1–2 hours. After washing the beads, proteins bound to the beads were eluted, and they were examined for Western blotting as described earlier.
Results
Binding of MCM6 fragments with MCM-interacting proteins
We reported the possible interaction of human MCM2–7 proteins with MCM-interacting proteins,30–32 and the results are summarized in Table 1. The interaction of MCM2–7 proteins with MCM10 is shown in Figure S1. In these experiments, one of the MCM2–7 proteins was expressed with an MCM-interacting protein in the insect cells. The MCM-interacting protein in the cell lysate was immunoprecipitated, and coimmunoprecipitation of MCM2–7 proteins was examined by Western blotting analysis. Among MCM2–7 proteins, MCM6 was the protein that most frequently interacted with the MCM-interacting proteins. ATP-binding sites in MCM6 play important roles in exhibiting DNA helicase activity of MCM4/6/7 complex.28,29 Thus, we chose MCM6 to determine the region of interaction with the MCM-interacting proteins. MCM6 was first divided into two fragments: N-terminal half containing Zn-finger motif (N1) and C-terminal half containing ATP-binding motifs (C1).25 Smaller C2 fragment contains Walker motifs A and B and sensor I region, and C3 fragment contains C-terminal region containing sensor II region. Structural partition between sensor I and sensor II was suggested for the analysis of human MCM2 fragments digested with trypsin.33 The smallest fragment of C4 only contains Walker A region. N1+C2 fragment contains N-terminal half and Walker motifs A and B and sensor I in the C-terminal region. N1+490 fragment contains N-terminal half and regions of Walker motifs A and B.
  | Table 1 Summary of interaction of replication proteins with MCM2–7 |
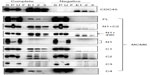
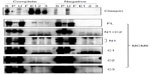
These MCM6 fragments were expressed with CDC45 in the insect cells for immunoprecipitation (Figure 1, complete). As a control, the same immunoprecipitation was performed in the absence of expression of CDC45 (negative). In all combinations, CDC45 expression was confirmed by detecting the bands in Triton-soluble (S) and Triton-insoluble (P) fractions, and precipitated CDC45 was detected in elution fractions (E1, 2, and 3). The CDC45 bands were not detected in these fractions in the control experiment. Full-length MCM6 was coprecipitated with CDC45 in the complete experiment but not in the control experiment. N-terminal N1 fragment, which was detected as two bands by unknown reason, was not coprecipitated, but C-terminal half C1 fragment was coprecipitated. Addition of ATP-binding motifs to N1 fragment gave CDC45-binding ability (N1+C2 and N1+490), suggesting the region containing ATP-binding motifs is required for binding with CDC45. C2 fragment containing Walker motifs A and B and sensor I itself was able to bind to CDC45, but C3 containing the C-terminal end region was not. The smallest C4 fragment containing Walker motif A was coprecipitated with CDC45. Consistent with these findings, direct binding of CDC45 and MCM2–7 was observed by incubation of purifying these two proteins (Figure S2). Also, interaction of CDC45 and myc-MCM6 fragments (C2 and C4) was confirmed by immunoprecipitation with anti-myc antibody, instead of anti-CDC45 antibody (Figure S3).
Interaction of RPA2, a subunit of RPA complex, with MCM6 fragments was examined (Figure 2). RPA2 was precipitated with the antibody. Coprecipitation of MCM6 was detected for C1 fragment but not for N1 fragment. Addition of the region containing ATP-binding motifs to N1 fragment gave RPA2-binding ability (N1+C1, N1+490). C2 itself was able to bind to RPA2, but C3 did not bind to RPA2. Coprecipitated C4 fragment was faintly detected. Although the levels of sensitivity to detect coprecipitated MCM6 fragments vary among the fragments, all these data are essentially comparable to those on the CDC45 binding; that is, ATP-binding motifs of MCM6 containing Walker motif A are involved in the interaction with CDC45 and RPA2.
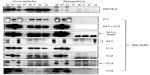
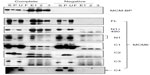
Binding of TIM with MCM6 fragments was examined (Figure 3). C1 fragment but not N1 fragment bound to TIM. Addition of ATP-binding motifs to N1 fragment gave TIM-binding ability (N1+C2, N1+490). Coprecipitation of C2 fragment with TIM was detected but that of C3 and C4 was not. Direct binding of TIM and MCM2–7 was observed by incubation of purifying these two proteins (Figure S2). Essentially, the same results to TIM were obtained for TIPIN (Figure 4). Thus, the region containing Walker motifs A and B is involved in binding with TIM and TIPIN. As to Claspin, faint coprecipitated bands were detected for full length and C1 fragment of MCM6 (Figure 5). However, coprecipitation was not detected for N1+C2, N1, C2, and C3 fragments. These results suggest that the entire region of C1 is required for binding to Claspin.
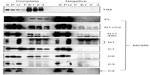
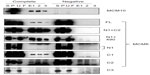
Interaction of MCM-BP with MCM6 fragments was examined (Figure 6). Both N1 and C1 fragments were coprecipitated with MCM-BP, as reported earlier.24 Coprecipitation of C2 fragment but not of C3 and C4 fragments was detected. These data suggest that the two separated regions of N-terminal half and ATP-binding motifs in the C-terminal half of MCM6 are involved in binding with MCM-BP. Binding to N-terminal half-fragment was only detected for MCM-BP among the MCM-interacting proteins examined in this study. Binding of MCM10 to the MCM6 fragments was examined (Figure 7). Coprecipitation of C1 fragment with MCM10 but not of N1 fragment was detected. Addition of C2 fragment or the region of 326–490 to the N1 fragment gave MCM10-binding ability. Binding of C2 fragment was weakly detected but that of C3 fragment was not. Thus, the data suggest that MCM10 has affinity to C2 region, more stringently to the region containing Walker motifs A and B. However, van Deursen et al13 reported that Mcm10 binds to the MCM domain located at the C-terminus in S. cerevisiae using two-hybrid system. These data are not consistent with the present results, and further experiments are required to clarify this apparent discrepancy.
MCM4 is a subunit that is localized next to MCM6 in MCM2–7 hexamer.34 Binding of MCM4 to the MCM6 fragments was examined (Figure 8). In this experiment, pull down of His-MCM4 using Ni-beads was carried out instead of immunoprecipitation. Binding of MCM6 N1 and C1 fragments was weakly detected, although faint bands in eluates were detected in the control experiment in these experiments. The presence of C2 fragment or the region of 326–490 appears to increase the binding ability of the N1 fragment. Corecovery of C2 fragment with MCM4 was weakly detected but that of C3 fragment was not.
  | Figure 8 Binding of MCM4 with MCM6 fragments. |
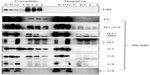
To understand the mode of the interaction, a mutated C2 fragment where three conserved leucines at 370, 429, and 449 in the C2 fragment were converted to alanines was prepared. These amino acids are highly conserved, since these are conserved in MCM4 in S. cerevisiae and in Sulfolobus solfataricus MCM. The leucine at 370 is located near the external hairpin region and those at 429 and 449 are near the helix 2 region.26 Binding of the mutated C2 fragment to CDC45 and TIM proteins was examined (Figure 9). The decreased levels (approximately one-fourth of wild type) of the bands in the eluates were detected for CDC45 and TIM. These results suggest that hydrophobic binding is involved in the interaction of these two proteins with C2 fragment of MCM6.
Discussion
All data are summarized in Figure 10A. Distinct binding to the C2 fragment containing Walker motifs A and B and sensor I was observed for CDC45, RPA2, TIM, TIPIN, MCM-BP, and MCM10. Schematic view of binding of each MCM-interacting protein to MCM6 is shown in Figure 10B. Almost all proteins except for Claspin bound to central region of MCM6 where ATP-binding motifs are located. The central region of MCM is conserved among MCM2–7 proteins, suggesting a possibility that these MCM-interacting proteins interact to all the MCM2–7 proteins. These results suggest that the MCM-interacting proteins examined here regulate MCM2–7 complex by modulating ATP-binding ability of the complex.
MCM4/6/7 hexamer exhibits DNA helicase activity in vitro.28 Specific binding of ATP with MCM6 was observed when mouse MCM4/6/7 complex was incubated with radiolabeled ATP. Both the high-affinity binding of ATP to MCM6 and the DNA helicase activity of the complex were inhibited by introducing mutation at Walker motif A or B of MCM6. Other activities of single-stranded DNA binding and ATPase activity of the complex were not inhibited by the MCM6 mutation. These results indicate that the binding of ATP to MCM6 is required for DNA helicase activity of the MCM4/6/7 complex. We examined whether the interaction of the MCM-interacting proteins, including CDC45 with MCM complexes, modulates ATP-binding ability of MCM proteins, including MCM6 (Figure S4). The results indicate that CDC45 did not interfere with the binding of ATP to MCM2–7 proteins in the MCM2–7 complex, but it inhibited the binding of ATP to MCM6 in the MCM4/6/7 complex under these conditions. To understand functional significance of the findings, we examined the effect of these proteins on the DNA helicase activity of the MCM4/6/7 complex (Figure S5). The results indicate that MCM-BP and TIM/TIPIN can inhibit the activity and CDC45 is slightly inhibitory.
CDC45 bound to the shortest fragment of C4 containing Walker motif A. The amino-acid sequence in C4 fragment is conserved among MCM2–7 proteins. This finding can explain that CDC45 can bind all the MCM2–7 proteins in the system similar to that described in this study.32 Electron microscopic observation of the isolated CMG complex shows that CDC45 interacts with Mcm2 and Mcm5 in the Mcm2–7 hexamer.35,36 This interaction may bridge the gap between Mcm2 and Mcm5 in the hexamer. Thus, it is possible that CDC45 mainly interacts with the region containing Walker motif A in Mcm2 and Mcm5 in the isolated Mcm2–7 complex. Thus, we examined the interaction of MCM2 fragments and MCM-interacting proteins. First, we examined the interaction of an MCM2 fragment (469–672), which is an equivalent of C2 fragment of MCM6, with MCM-interacting proteins, including CDC45 (Figure S6). The results indicate that the MCM2-C2 fragment is able to bind to CDC45 and TIM but not to MCM-BP. When binding of a smaller fragment of MCM2 (469–618) containing Walker motifs A and B was examined, the MCM2 fragment bound to CDC45 but not to MCM-BP (Figure S7).
We reported that TIM, TIPIN, and RPA2 bind to MCM3-7 proteins but hardly bind to MCM2 in the similar systems;30,31 this is in contrast to the binding mode of CDC45 to MCM2–7.32 TIM and TIPIN required wider region of MCM6 than CDC45 that binds to the C4 fragment containing Walker motif A. It appears that CDC45 binds to C4 fragment more strongly than RPA2. These different binding modes between CDC45 and other three proteins may explain the binding of CDC45 to MCM2.32 TIM/TIPIN/Claspin complex is involved in inhibiting MCM helicase at the fork when DNA synthesis is prevented.14 Consistent with this notion, it has been reported that TIM/TIPIN complex inhibits DNA helicase activity of MCM2–7 in vitro.20 It is possible that TIM/TIPIN disturbs ATP-binding ability of MCM2–7 complex. It has been reported that Mrc1, a homologue of Claspin, binds to the C-terminal region of MCM6, and the region is sufficient for binding to Mrc1 in S. cerevisiae.17 The result appears not to be consistent with those of this study, since the entire region in the C-terminal half is required for binding to Claspin. Also, we did not observe the specific binding of MCM10 to the C-terminal region of MCM6.13 Further analysis is required to clarify these apparent discrepancies.
It is suggested that MCM-BP is involved in dislodging MCM complex from the replication forks at termination of DNA replication.23 Related with this notion, we reported that MCM-BP mainly interacts with MCM7 in the S and G2 phases of HeLa cells.24 It has also been shown that MCM-BP mainly binds to MCM3, MCM5, and MCM7 when it was coexpressed with MCM2–7 proteins in the insect cells. It has been shown that MCM-BP binds to the amino-terminal region of MCM6 in addition to the carboxyl-terminal region in this study. This binding mode of MCM-BP may be consistent with the notion that MCM-BP disassembles MCM2–7 complex at the forks. It is also possible that MCM-BP regulates MCM function by disrupting the interaction of MCM6 with other replication proteins.
Recently, structural analysis of MCM2–7 prepared from yeast at the G1 phase was reported.37 The results suggest that hydrophobic interaction between contacting ATPase domains plays an important role in the intersubunit interaction. Conformational differences lies at the six ATPase centers of the ATPase domain. Two active centers, MCM2:MCM6 and MCM5:MCM2, are outliers, and ATP-binding pockets are less compact in MCM2 and MCM6. The nucleotide occupancies at the centers of MCM3:MCM5 and MCM6:MCM4 dimers are comparatively low, and only dimers of MCM7:MCM3 and MCM4:MCM7 are active. This structural information would be important to consider the possibility that MCM2–7 helicase activity is regulated by the interaction of MCM-interacting proteins with MCM6.
Acknowledgments
We thank N Nozaki from the MAB Institute for preparing monoclonal antibodies. This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Disclosure
The authors report no conflicts of interest in this work.
References
Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. | |
Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–131. | |
Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda H. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. | |
Araki H. Initiation of chromosomal DNA replication in eukaryotic cells; contribution of yeast genetics to the elucidation. Genes Genet Syst. 2011;86:141–149. | |
Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. | |
Kang Y-H, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase ε in rolling circle DNA synthesis. Proc Natl Acad Sci U S A. 2012;109:6042–6047. | |
Gambus A, Jones RC, Sanchez-Diaz A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. | |
Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucl Acids Res. 2009;37:2087–2095. | |
Ricke RM, Bielinsky A-K. Mcm10 regulates the stability and chromatin association of DNA polymerase-a. Mol Cell. 2004;16:173–185. | |
Zhu W, Ukomadu C, Jha S, et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. | |
Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. | |
Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31:2182–2194. | |
van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. | |
Katou Y, Kanoh Y, Bando M, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2007;424:1078–1083. | |
Unsal-Kaçmaz K, Chastain PD, Qu PP, et al. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. | |
Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2007;282:2729–2740. | |
Komata M, Bando M, Araki H, Shirahige K. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Mol Cell Biol. 2009;29:5008–5019. | |
Bando M, Katou Y, Komata M, et al. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 2009;284:34355–34365. | |
Lema AR, Noguchi C, Lee CY, Noguchi E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci. 2010;123:660–670. | |
Cho WH, Kang YH, An YY, Tappin I, Hurwitz J, Lee JK. Human Tim-Tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc Natl Acad Sci U S A. 2013;110:2523–2527. | |
Sakwe AM, Nguyen T, Athanaspoulos V, Shire K, Flappier L. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol Cell Biol. 2007;27:3044–3055. | |
Takahashi N, Quimbaya M, Schubert V, et al. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 2010;6:e1000817. | |
Nishiyama A, Flappier L, Mechali M. MCM-BP regulates unloading of the MCM2-7 helicase in late S phase. Genes Dev. 2011;25:165–175. | |
Kusunoki S, Ishimi Y. Interaction of human minichromosome maintenance protein-binding protein with minichromosome maintenance 2-7. FEBS J. 2013;281:1057–1067. | |
Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperon-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. | |
Brewster AS, Wang G, Yu X, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105:20191–20196. | |
Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. | |
You Z, Ishimi Y, Masai H, Hanaoka F. Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J Biol Chem. 2002;277:42471–42479. | |
Lee J-K, Hurwitz J. Isolation and characterization of various complexes of the minichromosome maintenance protein of Scizosaccharomyces pombe. J Biol Chem. 2000;275:18871–18878. | |
Nakaya R, Takaya J, Onuki T, Moritani M, Nozaki N, Ishimi Y. Identification of proteins that may directly interact with human RPA. J Biochem. 2010;148:539–547. | |
Numata Y, Ishihara S, Hasegawa N, Nozaki N, Ishimi Y. Interaction of human MCM2-7 proteins with TIM, TIPIN and Rb. J Biochem. 2010;147:917–927. | |
Takaya J, Kusunoki S, Ishimi Y. Protein interaction and cellular localization of human CDC45. J Biochem. 2013;153:381–388. | |
Komamura-Kohno Y, Tanaka R, Omori A, Kohno T, Ishimi Y. Biochemical characterization of fragmented human MCM2. FEBS J. 2008;275:727–738. | |
Yu Z, Feng D, Liang C. Pairwise interactions of the six human MCM protein subunits. J Mol Biol. 2004;340:1197–1206. | |
Costa A, Ilves I, Tamberg N, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struc Mol Biol. 2011;18:471–477. | |
Costa A, Renault L, Swuec P, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014;3:e03273. | |
Li N, Zhai Y, Zhang Y, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524:186–191. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.